Abstract
Soil microbial communities contain the highest level of prokaryotic diversity of any environment, and metagenomic approaches involving the extraction of DNA from soil can improve our access to these communities. Most analyses of soil biodiversity and function assume that the DNA extracted represents the microbial community in the soil, but subsequent interpretations are limited by the DNA recovered from the soil. Unfortunately, extraction methods do not provide a uniform and unbiased subsample of metagenomic DNA, and as a consequence, accurate species distributions cannot be determined. Moreover, any bias will propagate errors in estimations of overall microbial diversity and may exclude some microbial classes from study and exploitation. To improve metagenomic approaches, investigate DNA extraction biases, and provide tools for assessing the relative abundances of different groups, we explored the biodiversity of the accessible community DNA by fractioning the metagenomic DNA as a function of (i) vertical soil sampling, (ii) density gradients (cell separation), (iii) cell lysis stringency, and (iv) DNA fragment size distribution. Each fraction had a unique genetic diversity, with different predominant and rare species (based on ribosomal intergenic spacer analysis [RISA] fingerprinting and phylochips). All fractions contributed to the number of bacterial groups uncovered in the metagenome, thus increasing the DNA pool for further applications. Indeed, we were able to access a more genetically diverse proportion of the metagenome (a gain of more than 80% compared to the best single extraction method), limit the predominance of a few genomes, and increase the species richness per sequencing effort. This work stresses the difference between extracted DNA pools and the currently inaccessible complete soil metagenome.
The soil microbial community is relatively diverse (9, 31), with arguably the highest level of prokaryotic diversity of any environment (32, 41). One gram of soil has been reported to contain up to 10 billion microorganisms and thousands of different species (20). This soil species pool represents a goldmine for genes involved in pharmaceutical and industrial applications (42) and in the biodegradation of human-made pollutants (4, 13). Currently, less than 1% of this diversity is considered to be cultivable by traditional techniques (34), a problem that can be circumvented by metagenomic approaches. Metagenomic approaches have been applied to study a range of soil environments (8, 10, 15, 17, 28), and comparisons with cultivation techniques should include biases in the methods used to extract DNA from soil. Different DNA extraction methods are widely used, although they each have biases that restrict the diversity of the so-called metagenomic DNA (6, 12, 18, 22, 24, 25). Therefore, the total microbial diversity of soil might still be underestimated, independent of the method used to calculate the species (or operational taxonomic unit [OTU]) diversity in a soil. Indeed, the relative dominance of certain groups in DNA extracted from soil will mask less abundant species, thus confounding estimates of soil microbial community structure.
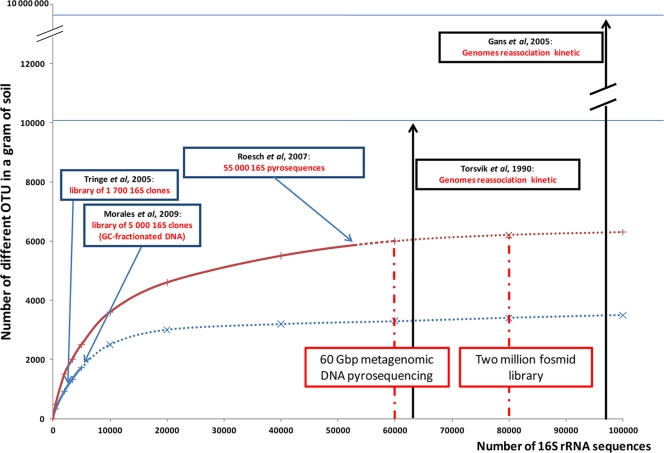
Recently developed technologies provide relatively quick and deep sequencing of metagenomic DNA samples at a moderate cost (19, 35), although metagenomic DNA sequencing, however completely sequenced, depends on the DNA extracted. Deciphering soil function based on soil metagenome sequencing (such as that proposed previously by the Terragenome International Consortium [43]) requires extraction of the DNA from all members of the soil microbial community. The difficulty is that every protocol facilitates the extraction of part of the microbially diverse population to the detriment of the rest. Biodiversity estimates from a variety of methods (Fig. 1) already range from 104 species (32, 38) to 107 species (14) per gram of soil. Therefore, a measure of the dependence of biodiversity estimates on metagenomic access would aid in an understanding of whether sequencing depth or DNA extraction diversity is driving diversity estimations.
FIG. 1.
Theoretical contribution of the Terragenome Initiative to soil diversity exploration, which starts with 60 “454” titanium plates and the construction of a 2-million-fosmid (40-kb inserts) clone library in the context of soil microbial diversity estimation studies (Metasoil Project). (Based on data from references 14, 26, 32, 37, and 39.)
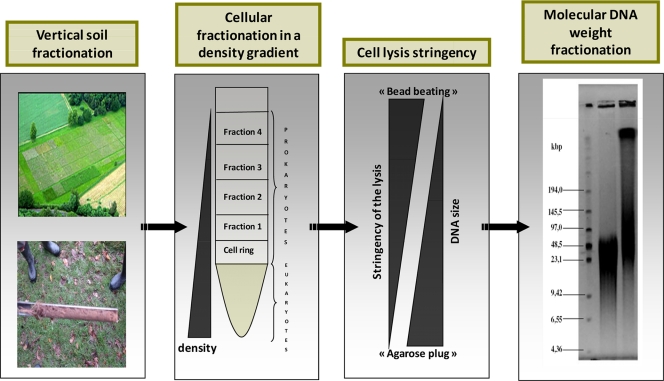
Our approach was to combine different methods to recover different spectra of community diversity in order to increase access to the biodiverse soil community. We applied four classes of DNA (or microbial) separation techniques that significantly resolve DNA diversity. These techniques are based on (i) vertical soil sampling, (ii) cell separation in a density gradient, (iii) cell lysis stringency, and (iv) DNA fragment size distribution (Fig. 2). Although the respective methods used are not without some overlap, we have shown that they can be adjusted to increase the relative diversity of the final DNA pool. In other words, by varying the conditions of the four methods and applying a phylogenetic technique to track relative diversity and the less represented species, the final DNA pool can be optimized for increased nucleic acid diversity. This strategy was compared to other more common approaches (including the individual application of one of the methods used here) in order to illustrate the advantages of this approach. Although applying these four variables might improve the already distorted view of the relative abundance of species, the aim here is to enhance species and gene discovery by maximizing the identification of the genetic diversity of a DNA pool before high-throughput sequencing efforts or the construction of libraries is performed.
FIG. 2.
Schematic of the different classes of DNA separation methods, starting with physical distance in the field and then density differences in Nycodenz gels, resistance to cell lysis, and finally DNA size separation by pulsed-field gel electrophoresis.
MATERIALS AND METHODS
Soil samples.
Samples were collected from two sites: Ecully, France, and an untreated control plot (plot “3d”) of Park Grass (lat 51.481481°N, long 0.222231°E), Rothamsted, England (see http://www.rothamsted.ac.uk/for further information), in October 2008 and March 2009, respectively. The Park Grass soil is an internationally recognized resource and is targeted as a reference soil for soil metagenomic studies (43). It is classified as chromic luvisol according to FAO guidelines (11) and is a silty clay loam overlying clay with flints with a pH of 5.2 (measured in H2O). Park Grass covers 249 m2 (13.28 by 18.75 m), and the sampling strategy consisted of taking randomized soil samples in four areas of the plot (horizontal sampling) and at seven depths (vertical sampling, each 3 cm between 0 and 21 cm). The Ecully soil (silty topsoil) was sampled in a grassland area (lat 45.470759°N, long 4.460152°E) at the same seven depths. Samples were placed into plastic bags and transported on ice. Soil was homogenized manually by thorough physical mixing. All tools and materials used were washed and sterilized.
DNA extraction methods.
DNA extraction from soil is a key step in the metagenomic approach (3, 12, 21). Two different methods are routinely used. In the first method, direct extraction, cells are lysed within the soil sample (27, 40, 44). We used two direct DNA extraction protocols that involve bead beating: a method described previously Griffiths et al. (16) that uses the FastPrep lysing matrix (MP bead beating; Bio101 Biomedical) and the MoBio UltraClean soil DNA isolation kit. For both protocols, DNA was extracted from 0.5 g of soil. For the alternative method, cells were first removed from the soil (60 g) and then lysed (2). This method is commonly called indirect extraction and has been reported to separate prokaryotic from eukaryotic cells via a Nycodenz density gradient (1, 7, 23). During the centrifugation, the Nycodenz gradient is stabilized at a density of 1.3 g/ml and should isolate prokaryotes to form a cellular fraction called the cell ring (Fig. 2). We fractionated the gradient into six parts, each 5 ml (four fractions above the cell ring, the cell ring, and one below the cell ring [total of 30 ml]), by varying the centrifugation speed (1,000 × g, 2,000 × g, 5,000 × g, and 9,000 × g for 40 min). After centrifugation at each speed, the Nycodenz gradient was subsampled from the top down by pipetting out 5-ml samples. The cell ring was within the fifth subsample.
After cell separation in the gradient, we used different cell lysis protocols, which have various degrees of stringency: the MP bead-beating protocol, the Epicentre Gram-positive kit, the Nucleospin tissue kit, and five agarose plug protocols called protocols A, B, C, D, and E.
Agarose plugs.
The extraction of soil bacteria was performed on fresh soil samples as previously described by Bertrand et al. (3), using the Nycodenz gradient separation method. The collected bacterial cell fraction was washed with ultrapure water and then centrifuged for 10 min at 12,000 × g. The cell pellet was then resuspended in a 50 mM Tris (pH 8)-100 mM EDTA buffer, mixed with an equal volume of molten 1.6% Incert agarose, and then transferred into disposable plug molds (Bio-Rad). The lysis of the soil bacteria was then performed with agarose. After the different lysis methods were used, agarose plugs were equilibrated in a 10 mM Tris (pH 8.0)-1 mM EDTA storage buffer.
(i) Protocol A.
For protocol A, agarose plugs were first transferred into 3 ml of G− lysis buffer (1% lauroyl sarcosine, 500 mM EDTA-Na2 [pH 9.5]) with 0.5 mg/ml of lysozyme and incubated at 37°C for 12 h. The agarose plugs were then incubated in 3 ml of G− lysis buffer with 500 μg/ml of proteinase K at 56°C for 12 h.
(ii) Protocol B.
For protocol B, agarose plugs were first transferred into 45 ml of LA lysis buffer (50 mM Tris [pH 8.0], 100 mM EDTA, 5 mg of lysozyme/ml, 0.5 mg of achromopeptidase/ml) and incubated at 37°C for 6 h. The agarose plugs were then incubated in 45 ml of SP lysis buffer (50 mM Tris [pH 8.0], 100 mM EDTA, 1% lauryl sarcosyl, 2 mg of proteinase K/ml) at 55°C for 24 h. An additional incubation for 24 h was performed with fresh SP buffer.
(iii) Protocol C.
For protocol C, agarose plugs were first transferred into 3 ml of G+ lysis buffer (6 mM Tris-HCl, 100 mM EDTA-Na2, 1 M NaCl, 0.5% Brij 58, 0.2% sodium deoxycholate, 0.5% lauroyl sarcosine [pH 7.5]) with 0.5 mg/ml of lysozyme and incubated at 37°C for 12 h. The agarose plugs were then incubated in 3 ml of G− lysis buffer with 500 μg/ml of proteinase K at 56°C for 12 h.
(iv) Protocol D.
For protocol D, agarose plugs were incubated in 45 ml of SP lysis buffer (50 mM Tris [pH 8.0], 100 mM EDTA, 1% lauryl sarcosyl, 2 mg of proteinase K/ml) at 55°C for 24 h. An additional incubation for 24 h was performed with fresh SP buffer.
(v) Protocol E.
For protocol E, agarose plugs were transferred into 45 ml of LA lysis buffer (50 mM Tris [pH 8.0], 100 mM EDTA, 5 mg of lysozyme/ml, 0.5 mg of achromopeptidase/ml) and incubated at 37°C for 6 h.
These plug protocols for differential DNA recovery have also been used for fosmid library construction that requires high-molecular-weight DNA to create clone libraries with different sequence diversities.
DNA size separation.
Pulsed-field gel electrophoresis (PFGE) was used to separate the metagenomic DNA as a function of the fragment size distribution (1% low-melting-point agarose and 0.5× Tris-borate-EDTA [TBE], with a program of 2 s, 20 s, and 15 h). The DNA was then extracted from the gel by using agarase I (New England BioLabs Inc.). For the Rothamsted soil samples, a portion of DNA was physically sheared to generate a range of fragments that were smaller than those in the undisrupted portion, as demonstrated by the differential migration of the smears (Fig. 2).
Ribosomal intergenic spacer analysis (RISA).
The intergenic spacer (IGS) region between the small (16S) and the large (23S) subunits of ribosomal sequences were amplified by PCR using primers 5′-TGCGGCTGGATCCCCTCCTT-3′ (forward) and 5′-CCGGGTTTCCCCATTCGG-3′ (reverse) (29). For the PCR mix, 2 μl of DNA (10 μM) was mixed with 1.25 μl of reverse and forward primers (10 μM) and 20.5 μl of distilled water (DH2O). PCR cycles consisted of 95°C for 10 min and then 30 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 1 min, followed by 72°C for 15 min, with a Biometra thermocycler. One microliter of the PCR mix was then loaded into an Agilent DNA 7500 Lab on a Chip, and electropherograms were analyzed and data were normalized by using an Agilent 2100 Bioanalyzer. An example of different replicates is shown in Fig. S1 in the supplemental material in order to demonstrate the reproducibility of this fingerprint approach.
Phylochip analyses.
The microarray format used in these experiments was that from Agilent Sureprint Technologies. The format used consisted of 8 blocks of 15,000 spots each on a standard glass slide format, 1 in. by 3 in. (25 mm by 75 mm). Each spot was formed by the in situ synthesis of 20-mer oligonucleotide probes. Each oligonucleotide probe occurred at least in triplicate within each block. All blocks were identical. This format provides for the hybridization of eight samples at the same time and on the same slide. The use of multiple slides was necessary for the hybridization of over eight samples. Probes were designed to target the rrs gene and to cover a wide part of the Bacteria and Archaea phylogenic tree. Probes were designed with the ARB software package and PhylArray (24a). We have chosen to design 20-mer probes with a melting temperature range of 65°C ± 5°C and with a weighted mismatch of less than 1.5. Our design includes oligonucleotide probes at different taxonomic levels. This microarray covers over 400 genera and 400 OTUs (“species” or “hits”).
The rrs genes were amplified by PCR from total DNA by using universal primer pA (TAATACGACTCACTATAGAGAGTTTGATCCTGGCTCAG) and pH-T7 (AAGGAGGTGATCCAGCCGCA) (5) (universal for most members of the Bacteria and some of the Archaea) under standard conditions. The amplification of DNA was performed with a 48-μl PCR mixture using 5 U of Ex Taq titanium polymerase. PCR was conducted at 94°C for 4 min and then with 35 cycles of 94°C for 45 s, 55°C for 45 s, and 68°C for 90 s, followed by 68°C for 5 min. Amplified PCR products were electrophoresed on a 1% agarose gel, and the desired 1.5-kb bands were removed and purified by using GFX PCR DNA and a gel band purification kit (Amersham Biosciences). Purified PCR products were then transcribed onto RNA using T7 RNA polymerase (Invitrogen) with the incorporation of labeled Cy3-UTP. Cy3 is a fluorescent dye, emitting light at 532 nm. RNA purification was performed by using the Qiagen RNeasy minikit according to the manufacturer's instructions. RNA fragmentation was achieved by the addition of 1.14 μl of Tris-Cl (1 mM) and 4.57 μl of ZnSO4 (100 mM) to 40 μl of labeled RNA sample and incubation for 30 min at 60°C. Chemically fragmented labeled RNA was then hybridized to the phylochips.
Microarray scanning and data processing.
An Innoscan (Carbonne, France) 700 scanner was used for scanning microarray slides according to the manufacturer's instructions. Raw hybridization fluorescence signals for each spot were determined based on the signal-to-noise ratio (SNR), which was calculated by using the following formula: SNR = (signal intensity − background)/standard deviation of the background. Hybridization fluorescence signals for all probes, including negative controls, were transformed by calculating the log2 of the signal. Since at least three replicates exist for all oligonucleotide probes, outliers were eliminated when any individual spot was greater than 2 standard deviations from the average of all replicates. Analysis of variance (ANOVA) was used to evaluate positive probes from the results for all microarray data from one experiment. Since the probes have different phylogenetic depths, the genera described here were those for which all relevant probes were positive. While all of the thousands of probes could not be independently verified, many of the probes were validated by the application of DNA from a single bacterium (33).
RESULTS
Two different soils were employed for the elaboration of the DNA-recovering strategy: Ecully, France, and Park Grass (plot 3d), Rothamsted, England. Different approaches were tested to separate and increase the metagenomic DNA extracted at one time and one place, and the diversity of the different samples was estimated with ribosomal intergenic spacer analysis (RISA) fingerprinting (electropherogram profiles are shown in Fig. 3). After preliminary tests, four methods appeared to separate metagenomic DNA into the most diverse fractions for the two soils: vertical soil sampling, density gradient (cell separation), cell lysis stringency, and DNA fragment size distribution (Fig. 2). The different methods could be applied sequentially to maximize differential DNA extraction (Fig. 2). Clearly, all the extracted DNA pools have distinct species diversities and distributions. However, the cell lysis stringency appeared to have the most influence on the diversity of the extracted DNA pool, as discussed below.
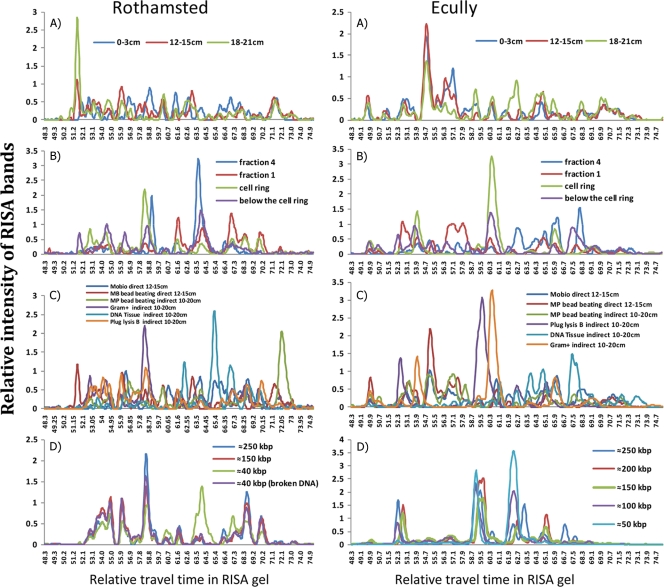
FIG. 3.
Multiple examples of ribosomal intergenic spacer analysis (RISA) electropherograms of DNA from the Ecully and Park Grass, Rothamsted, soils illustrating the differences in the diversities of microbial community DNA as a function of the applied separation technique. Graphs represent relative RISA band intensities as a function of travel time in the gel. (A) Physical separation with MP bead-beating direct DNA extraction of soil samples from different depths (0 to 3, 12 to 15, and 18 to 21 cm deep). (B) Cellular fractionation in a density gradient (Gram-positive lysis after centrifugation at 5,000 × g). (C) Cell lysis with MP bead-beating and MoBio kit direct DNA extractions (depth, 12 to 15 cm) and Epicentre Gram-positive, bead-beating, and DNA tissue indirect DNA extractions (depth, 0 to 10 cm). (D) Metagenomic DNA fractionation by PFGE after extraction by plug protocol B (depth, 10 to 20 cm).
RISA profiles.
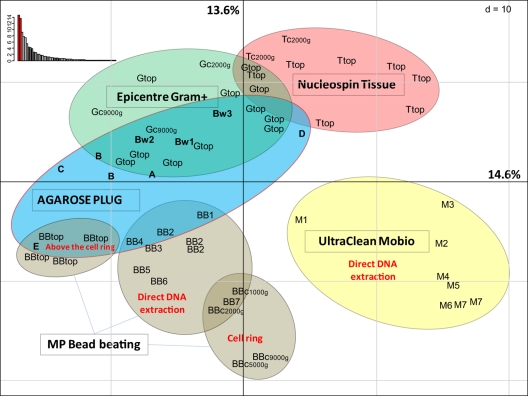
RISA fingerprints representing electropherograms demonstrate the presence and absence of different populations within the DNA extracted from the microbial community (Fig. 3). The different DNA extraction methods applied to the two soils are compared in four categories (vertical samples [Fig. 3A and A′], Nycodenz separation [Fig. 3B and B′], the cell lysis procedure [Fig. 3C and C′], and DNA size differences [Fig. 3D and D′]). The fingerprints of the microbial community extracted by the different methods are all different (in contrast to the similar profiles seen with replicate samples) (see Fig. S1 in the supplemental material), although the differences are more pronounced for those methods that include different lysis procedures (Fig. 3C and C′). Soil sample depth (Fig. 3A and A′) and DNA size (Fig. 3D and D′) showed the fewest differences, although extreme size classes were noticeably different (i.e., 40 kb in C and 250 kb in C′). The use of different lysis procedures had the greatest impact on RISA diversity, together covering the entire spectrum of possible RISA peaks (Fig. 3C and C′), in contrast to DNA fractionated according to size, which has several areas without peaks (Fig. 3D and D′). In order to evaluate the differences between the different RISA profiles, the Rothamsted profiles were quantified and the different samples were compared with a principal component analysis (PCA). The PCA separated the groups principally as a function of the cell lysis procedure (Fig. 4). Within these large groups, other parameters are regrouped, such as fractions from different depths in the soil core, the Nycodenz gradient fractions (where “top” refers to different samples from above the cell ring), and the PFGE smear (where Bw1 is about 40 kb, Bw2 is about 100 kb, and Bw3 is about 250 kb). In addition, the MP bead-beating extraction method was applied to both the soil (direct cell extraction) and the Nycodenz cell ring. While the bead beating produced somewhat similar RISA profiles, the direct and indirect (“cell ring”) samples were differentiated by the PCA (Fig. 4). Some replicates are provided in order to evaluate the relative importance of the RISA profiles. For example, the MoBio kit method was performed twice on the deepest soil sample (18 to 21 cm deep), and the bead beating was performed three times on the second depth fraction (4 to 6 cm). All of the replicates grouped relatively closely together (Fig. 4).
FIG. 4.
Principal component analysis (showing the first and second components) of the matrix data for the RISA analysis from each DNA separation method. The percentages of variance of all axes are shown in the upper left corner. BB, bead beating; A, B, C, D, and E, agarose plug protocols; Bw1, Bw2, and Bw3, low-, medium-, and high-molecular-weight DNA extracted with plug protocol B; M, MoBio Ultraclean kit; G, Epicentre G+ kit; c, cell ring from the Nycodenz density gradient separation; top, DNA recovered from the different fractions above the cell ring. The numbers 1 to 7 refer to the depth intervals (3 cm deep each) of the soil samples from the soil core, with 1 being 0 to 3 cm and 2 being 4 to 6 cm, etc.; 1,000g refers to DNA recovered from the cell ring in the Nycodenz gradient when the centrifuge was operated at 1,000 × g rather than the usual 9,000 × g.
Taxonomic comparisons.
For the Rothamsted soil, the difference between the metagenomic DNAs extracted by these different methods was further explored with the phylogenetic microarray in order to determine which genera were selectively extracted by one approach or the other. Comparisons of the microarray responses were therefore made between different extraction protocols.
In addition, the same DNA extraction protocol (MP bead beating) was used to evaluate the microbial diversity differences as a function of depth (vertical soil sampling). Phylochip analysis using 16S rRNA gene (rrs) hybridization showed significant diversity variations, with the frequency of Bacillus spp. increasing and that of Mesorhizobium species decreasing with depth. Some genera were detected in only one fraction. For example, Sandarakinotalea was detected only at the 3- to 6-cm depth; Alkalibacillus and Ammoniphilus were detected at the lowest depth (see Fig. S2 in the supplemental material). After centrifugation at a relatively low speed (2,000 × g), the density gradient was subsampled in six fractions (four fractions above the cell ring, one at the cell ring, and one below the cell ring of 5 ml each). One DNA extraction protocol (Epicentre Gram-positive kit) was used for phylochip comparisons. The frequency of detection of the genera Glycomyces and Legionella increased with depth in the Nycodenz gradient. Moreover, the populations of some genera were relatively isolated in one fraction and undetected or at very low levels in all others (e.g., Marinobacter, Pseudoxanthomonas, Fervidobacterium, and Treponema), emphasizing the value of varying the centrifugation speed to access different metagenomic DNAs (see Fig. S3 in the supplemental material).
After the soil and cell separation, different cellular lysis protocols were used to separate the metagenomic DNA as a function of the cell wall resistance to lysis. Seven different protocols were applied. In addition, two direct extraction protocols (DNA extracted directly from the soil), the MP bead-beating protocol and the MoBio Ultraclean soil DNA kit, were applied to the soils. The seven other protocols were indirect extraction protocols (cells extracted before lysis), the same MP bead-beating protocol, the Epicentre Gram-positive kit, the Nucleospin tissue kit, and five different agarose plug protocols, by varying the lysis stringency. Each lysis method facilitated the DNA extraction of a part of the microbially diverse population to the detriment of the rest. For example, MP bead-beating direct DNA extraction (fraction of 0 to 3 cm) facilitated the extraction of the genera Brevundimonas and Mesorhizobium but not the genera Sphingobium (detected only with plug lysis protocol E) or Pseudomonas. On the other hand, indirect bead-beating DNA extraction accessed more members of the Pseudomonas genus but not Mesorhizobium or Gloeobacter (see Fig. S4 in the supplemental material).
Finally, after an in-plug lysis (protocol B), DNA was separated as a function of its size distribution by pulsed-field gel electrophoresis. This separated DNA based on its molecular weight. The low-molecular-weight (30- to 50-kb) fraction was extracted and analyzed directly, and the DNA was fragmented so that the 250-kb fraction was fragmented down to the same size (30 to 50 kb) and then analyzed by phylochip analysis. Some genera were clearly unevenly represented in these two DNA samples (see Fig. S5 in the supplemental material). Notably, the genera Sulfurimonas, Xylella, and Leuconostoc were undetected in the low-molecular-weight (30- to 50-kb) fraction but were easily detected in the high-molecular-weight fraction. On the other hand, the genera Marinobacter and Rhodopirellula were detected only in the low-molecular-weight fraction (30 to 50 kb). These results demonstrate the variation in genetic diversity in the soil metagenomic DNA smear and might explain some of the bias found in the fosmid clone libraries, as the DNA selected is generally between 25 and 40 kb.
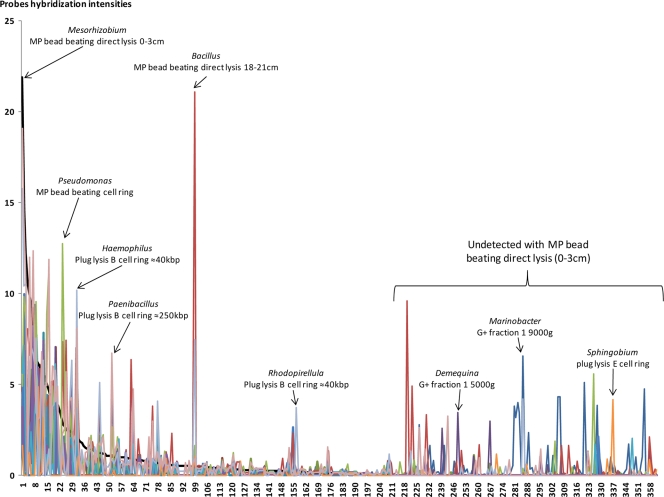
The relative phylogenetic distributions (based on probe hybridization intensities) of soil DNA pools extracted as a function of all four parameters (soil depth, Nycodenz gradient depth, cell lysis stringency, and DNA size) were also compared. The presence or absence of different genera and their relative fluorescence intensities from the different DNA pools were plotted against those for the MP bead-beating direct lysis of the top soil fraction (Fig. 5, black line). Thus, this pool of DNA defines the order (descending) of the genera (not listed here) along the x axis from most abundant to least abundant (Fig. 5). This DNA pool had 218 identified genera, which is why the genera after the 218th genus were not detected in the MP bead-beating top soil fraction of “0 to 3 cm” but were detected in other DNA extracts. All other DNA pools were thus compared to this pool, and where there are peaks above the black line, the pool in question has more of a given genus, and where there are valleys, the given pool has less of a given genus than those determined by MP bead beating. For example, Mesorhizobium (Fig. 5, far left) is the most predominant genus in the reference DNA pool (MP bead-beating direct lysis of the top soil fraction of 0 to 3 cm), more so than in any other extraction method's DNA pool. Other examples include Pseudomonas in the DNA pool from the MP bead beating applied to the Nycodenz cell ring after centrifugation at 9,000 × g and Bacillus in the DNA pool from the MP bead-beating direct lysis on the bottom soil sample (Fig. 5). Note that when the reference pool (MP bead-beating direct lysis of the top soil sample) does not detect certain genera at all (Fig. 5, right), several different extraction DNA pools have relatively high levels of these genera (e.g., Marinobacter with the Gram-positive extraction of the Nycodenz cell ring at 1,900 × g and Sphingobium with cell lysis procedure E). Many genera were not detected by using a single DNA extraction protocol but were revealed by applying other protocols. While some protocols, like direct and indirect MP bead beating, access more genera than some of the more specific extraction protocols, the relative proportions are not the same. In any case, no single protocol accesses the entire microbial community metagenome. When the phylogenetic probes on the microarray are quantified by extraction techniques, the numbers of phyla, classes, genera, and potential species (“hits”) vary considerably (e.g., from 50 to 214 genera) between protocols (Table 1). However, while some protocols detected relatively low numbers of genera (e.g., lysis procedure E), these protocols add to the overall recovery of diversity. For example, if all 23 different protocols were used, then 385 different genera would be detected (Table 1).
FIG. 5.
Phylogenetic distribution (genus level) of 14 DNA pools for 360 different genera. The genus order is based on the decreasing percentage of those detected in the DNA pool extracted with MP bead-beating direct lysis of the surface (0- to 3-cm) soil sample (black line) from genera 1 to 218. The order from genera 218 to 360 (where the genera were not detected in the reference DNA pool) is alphabetical.
TABLE 1.
Potential microbial biodiversity detected from the Rothamsted soil as a function of the extraction techniquea
| Method | No. of phyla | No. of classes | No. of orders | No. of families | No. of genera | Total no. of hits |
|---|---|---|---|---|---|---|
| MP bead-beating direct DNA extraction, 0-3 cm | 19 | 29 | 60 | 107 | 210 | 1,892 |
| MP bead-beating direct DNA extraction, 18-21 cm | 18 | 28 | 58 | 107 | 214 | 2,024 |
| G+ fraction 4, 2,000 × g, 0-10 cm | 12 | 18 | 29 | 39 | 51 | 333 |
| G+ cell ring, 9,000 × g, 0-10 cm | 17 | 24 | 49 | 75 | 121 | 1,201 |
| G+ cell ring, 2,000 × g, 0-10 cm | 15 | 22 | 47 | 72 | 121 | 974 |
| Plug lysis protocol B cell ring, 0-10 cm, ∼40 kbp | 18 | 26 | 58 | 102 | 203 | 2,130 |
| Plug lysis protocol B cell ring, 0-10 cm, ∼250 kbp | 17 | 26 | 56 | 95 | 182 | 1,887 |
| Plug lysis protocol D cell ring, 10-23 cm | 16 | 23 | 47 | 63 | 109 | 869 |
| Plug lysis protocol E cell ring, 10-23 cm | 12 | 16 | 27 | 32 | 50 | 270 |
| 23 different DNA extraction approaches | 23 | 36 | 71 | 148 | 385 | 3,940 |
The value of 2,000 × g refers to DNA recovered from the cell ring in the Nycodenz gradient when the centrifuge was operated at 2,000 × g rather than at the usual 5,000 × g.
DISCUSSION
The exploration of the biodiversity in soils requires metagenomic approaches that extract DNA from all the Bacteria and Archaea present as comprehensively as is possible. The scale of the spatial variation of the microbial diversity in a soil must influence any attempts to recover the genomes of all members of the microbial community. RISA profiles showed that Park Grass diversity varies both horizontally and vertically; however, the vertical variation appeared to be greater. To increase the level of biodiversity recovered from soil, we applied a range of approaches to access the metagenomic DNA pool. These approaches were dependent on soil depth, cell separation in density gradients, cell lysis stringency, and DNA molecular weight. The often-applied strategy of sampling different locations at the site was not the most significant factor in increasing the level of diversity of DNA extracted from the soils tested here (Fig. 3 and 4). Rather, the most critical strategies were those applied to the soil samples in the laboratory to extract and fractionate cells and DNA. This implies that the sample size (roughly 100 g) was sufficient to capture the majority of the microbial community metagenome. Nevertheless, all of the different approaches, including vertical soil sampling, altered the accessible biodiversity. The relevant issue was the relative improvement achieved with every additional DNA extraction protocol.
While all cell lysis protocols have numerous biases that limit the diversity of the metagenomic DNA extracted (12), we used these biases to our advantage in order to access different soil microbial communities with different proportions of species represented. This approach separated the metagenomic DNA as a function of cell wall resistance to lysis. RISA analyses showed important differences between lysis methods. The PCA corresponding to RISA profiles of some Rothamsted soil DNA samples emphasized the importance of this step (Fig. 4). The lysis protocol was the major driving force in grouping microbially diverse communities and thus was a crucial step for DNA extraction differences. These different lysis methods had significant effects on the metagenomic DNA extracted from a soil, with different microbial populations being represented in each sample (Fig. 5). Furthermore, we made an effort to access different diverse populations with the agarose plug protocols (five different lysis protocols) so that this strategy could be coupled with fosmid clone library production.
No one protocol can provide an accurate determination of species distribution, and therefore, different DNA extraction protocols, more or less stringent, could be employed, and the DNA pools could then be mixed together to maximize the number of different species represented and to decrease the proportion of the dominant species with a consequent increase in the final level of metagenomic diversity. The true relative abundance of different species is not currently determinable, and both microarray approaches and attempts to validate “16S” clone libraries by quantitative PCR are unfortunately dealing with the same DNA extraction pool (e.g., see reference 26) and, thus, the same extraction bias. Nevertheless, improved knowledge of the species present in the soil will aid in our understanding of soil function independent of their relative abundances. Since the majority of microorganisms are probably underrepresented in soil (30, 36), they are not easily accessible for study. Our approach was to maximize the representation of different species in DNA extracted from the same soil using four different techniques in order to improve our understanding of soil biodiversity.
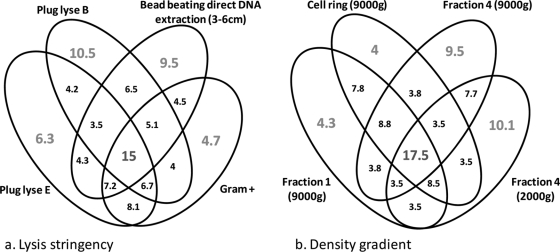
To visualize the impact of our strategy on accessing different levels of biodiversity in soil, sample DNA was analyzed with a phylochip containing the 20-mer complementary strands of the 16S rRNA gene (rrs). The different strategies clearly extracted different relative numbers of genera (Fig. 5 and Table 1), with some not detecting the presence of certain genera (Fig. 5). These rather large differences confirm the requirement for multiple approaches when high levels of microbial diversity are sought. Clearly, there is some overlap between different DNA extraction strategies (Fig. 3 and 4). In the case of different lysis stringencies or cellular fractions in a density gradient, at least 15% of the biodiversity (as measured by positive phylogenetic microarray probes) was detected in all DNA extraction method variations. On the other hand, over 20% of the biodiversity was detected only in individual pools of extracted DNA (Fig. 6). The different approaches tested appear to access variable quantities of phyla, classes, families, genera, and species (corresponding to different “hits” in the NCBI database) (Table 1). Some of the methods accessed a maximum amount of diversity (e.g., MP direct DNA extraction and plug protocol B indirect DNA extraction), while others provide in-depth information on diversity (e.g., fraction 4 of the density gradient with the Gram-positive Epicentre kit or plug protocol E indirect DNA extraction), which can help metagenomic DNA assemblages and provide access to generally unrepresented genetic resources. Combining the outputs from the different methods provides a greater level of biodiversity than any individual approach, increasing the number of hypothetical species by 83.5% in comparison to the best individual DNA extraction method tested.
FIG. 6.
Venn diagram showing percentages of probe hybridization coverage (out of over 3,000 total) between DNA extraction protocols as a function of the lysis stringency (a) and location in a Nycodenz density gradient at different centrifugation speeds (b).
None of the different extraction protocols described here are suitable for high-throughput sequencing, although PCR approaches can be easily applied to prokaryote community studies. The yield is particularly low when DNA is extracted from the cell density gradient fractions above the cell ring and when the DNA is extracted from agarose gels. In theory, it is possible to use whole-genome amplification to increase yields, but the inherent bias in this method would considerably limit the utility of sequencing these fractionated parts of a soil metagenome. There is some anecdotal evidence that the lower the DNA yield, the more the DNA sample represents unique phyla. The challenge is to accumulate sufficient DNA with low-yield approaches to enable high-throughput sequencing. Sequencing may not be appropriate for comparisons across many samples but is likely to be crucial when species richness and diversity within a small number of soil samples need to be defined in detail.
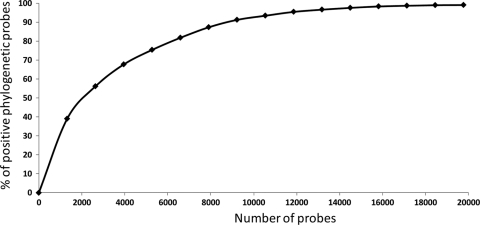
We have defined a strategy for increasing the level of detection of metagenomic DNA diversity in two soils by employing multiple DNA extraction methods. By comparing these multiple methods, we showed that the spatial distance between soil samples did not have a major impact on the genetic diversity that was determined, in contrast to both depth and the different DNA extraction and purification methods. The mixed metagenomic DNA containing products from different soil depths and with different extraction factors (density gradient, cell lysis stringency, and DNA molecular weight) will maximize the representation of different species, although it may distort their relative abundance at the nucleic acid level. However, the “true” distribution is unknown, and no existing method provides this information. To the contrary, most methods provide limited views of the true soil biodiversity, and it is only by adopting a range of extraction and lysis methods that rare species are captured, thus increasing the number of species detected (Table 1). The increase in the phylochip probe diversity from these different DNA fractions follows standard rarefaction curves (Fig. 7). These results imply that the level of soil diversity is greater than estimations based on one DNA extraction method (e.g., see references 14, 32, and 38). Therefore, considerable efforts and technologies are needed to access not only DNA pools but also an entire metagenome for unbiased microbial ecology studies.
FIG. 7.
Rarefaction curve based on phylogenetic microarray analyses of 15 different (based on extraction methods) DNA pools from the Rothamsted soil samples. The percentage of positive probes is plotted against the number of probes tested over multiple microarrays used to test different DNA pools.
Supplementary Material
Acknowledgments
We thank the French National Research Agency (ANR GMGE Metasoil). We also thank Libragen for its help and collaboration.
T.O.D. was funded by the Rhône-Alpes region. Rothamsted Research receives grant-aided support from the Biotechnology and Biological Sciences Research Council of the United Kingdom.
Footnotes
Published ahead of print on 23 December 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Bakken, L. R. 1985. Separation and purification of bacteria from soil. Appl. Environ. Microbiol. 49:1482-1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berry, A. E., C. Chiocchini, T. Selby, M. Sosio, and E. M. Wellington. 2003. Isolation of high molecular weight DNA from soil for cloning into BAC vectors. FEMS Microbiol. Lett. 223:15-20. [DOI] [PubMed] [Google Scholar]
- 3.Bertrand, H., et al. 2005. High molecular weight DNA recovery from soils prerequisite for biotechnological metagenomic library construction. J. Microbiol. Methods 62:1-11. [DOI] [PubMed] [Google Scholar]
- 4.Boubakri, H., M. Beuf, P. Simonet, and T. M. Vogel. 2006. Development of metagenomic DNA shuffling for the construction of a xenobiotic gene. Gene 375:87-94. [DOI] [PubMed] [Google Scholar]
- 5.Bruce, K. D., et al. 1992. Amplification of DNA from native populations of soil bacteria by using the polymerase chain reaction. Appl. Environ. Microbiol. 58:3413-3416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carrig, C., O. Rice, S. Kavanagh, G. Collins, and V. O'Flaherty. 2007. DNA extraction method affects microbial community profiles from soils and sediment. Appl. Microbiol. Biotechnol. 77:955-964. [DOI] [PubMed] [Google Scholar]
- 7.Courtois, S., et al. 2001. Quantification of bacterial subgroups in soil: comparison of DNA extracted directly from soil or from cells previously released by density gradient centrifugation. Environ. Microbiol. 3:431-439. [DOI] [PubMed] [Google Scholar]
- 8.Courtois, S., et al. 2003. Recombinant environmental libraries provide access to microbial diversity for drug discovery from natural products. Appl. Environ. Microbiol. 69:49-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Curtis, T. P., W. T. Sloan, and J. W. Scannell. 2002. Estimating prokaryotic diversity and its limits. Proc. Natl. Acad. Sci. U. S. A. 99:10494-10499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Demaneche, S., et al. 2008. Antibiotic-resistant soil bacteria in transgenic plant fields. Proc. Natl. Acad. Sci. U. S. A. 105:3957-3962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.FAO. 2006. Guidelines for soil description. FAO, Rome, Italy. ftp://ftp.fao.org/agl/agll/docs/guidel_soil_descr.pdf.
- 12.Frostegård, A., et al. 1999. Quantification of bias related to the extraction of DNA directly from soil. Appl. Environ. Microbiol. 65:5409-5420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Galvao, T. C., W. W. Mohn, and V. de Lorenzo. 2005. Exploring the microbial biodegradation and biotransformation gene pool. Trends Biotechnol. 23:497-506. [DOI] [PubMed] [Google Scholar]
- 14.Gans, J., M. Wolinsky, and J. Dunbar. 2005. Computational improvements reveal great bacterial diversity and high metal toxicity in soil. Science 309:1387-1390. [DOI] [PubMed] [Google Scholar]
- 15.Ginolhac, A., et al. 2004. Phylogenetic analysis of polyketide synthase I domains from soil metagenomic libraries allows selection of promising clones. Appl. Environ. Microbiol. 70:5522-5527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Griffiths, R. I., A. S. Whitely, A. G. O'Donnell, and M. J. Bailey. 2000. Rapid method for coextraction of DNA and RNA from natural environments for analysis of ribosomal DNA- and rRNA-based microbial community composition. Appl. Environ. Microbiol. 66:5488-5491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Handelsman, J., M. R. Rondon, S. F. Brady, J. Clardy, and R. M. Goodman. 1998. Molecular biological access to the chemistry of unknown soil microbes: a new frontier for natural products. Chem. Biol. 5:R245-R249. [DOI] [PubMed] [Google Scholar]
- 18.Head, I. M., J. R. Saunders, and R. W. Pickup. 1998. Microbial evolution, diversity, and ecology: a decade of ribosomal RNA analysis of uncultivated microorganisms. Microb. Ecol. 35:1-21. [DOI] [PubMed] [Google Scholar]
- 19.Kahvejian, A., J. Quackenbush, and J. F. Thompson. 2008. What would you do if you could sequence everything? Nat. Biotechnol. 26:1125-1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Knietch, A., T. Waschkowitz, S. Bowien, A. Henne, and R. Daniel. 2003. Metagenomes of complex microbial consortia derived from different soils as sources for novel genes conferring formation of carbonyls from short-chain polyols on Escherichia coli. J. Microbiol. Biotechnol. 5:46-56. [DOI] [PubMed] [Google Scholar]
- 21.Lakay, F. M., A. Botha, and B. A. Prior. 2007. Comparative analysis of environmental DNA extraction and purification methods from different humic acid-rich soils. J. Appl. Microbiol. 102:265-273. [DOI] [PubMed] [Google Scholar]
- 22.LaMontagne, M. G., F. C. Michel, P. A. Holden, and C. A. Reddy. 2002. Evaluation of extraction and purification methods for obtaining PCR- amplifiable DNA from compost for microbial community analysis. J. Microbiol. Methods 49:255-264. [DOI] [PubMed] [Google Scholar]
- 23.Lefevre, F., et al. 2008. Drugs from hidden bugs: their discovery via untapped resources. Res. Microbiol. 159:153-161. [DOI] [PubMed] [Google Scholar]
- 24.Martin-Laurent, F., et al. 2001. DNA extraction from soils: old bias for new microbial diversity analysis methods. Appl. Environ. Microbiol. 67:2354-2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24a.Milton, C., et al. 2007. PhylArray: phylogenetic probe design algorithm for microarray. Bioinformatics 23:2550-2557. [DOI] [PubMed] [Google Scholar]
- 25.Morales, S. E., T. F. Cosart, J. V. Johnson, and W. E. Holben. 2008. Extensive phylogenetic analysis of a soil bacterial community illustrates extreme taxon evenness and the effects of amplicon length, degree of coverage, and DNA fractionation on classification and ecological parameters. Appl. Environ. Microbiol. 75:668-675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morales, S. E., and W. E. Holben. 2009. Empirical testing of 16S rRNA gene PCR primer pairs reveals variance in target specificity and efficacy not suggested by in silico analysis. Appl. Environ. Microbiol. 75:2677-2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ogram, A., G. S. Sayler, and T. Barbay. 1987. The extraction and purification of microbial DNA from sediments. J. Microbiol. Methods 7:57-66. [Google Scholar]
- 28.Rajendhran, J., and P. Gunasekaran. 2008. Strategies for accessing soil metagenome for desired applications. Biotechnol. Adv. 26:576-590. [DOI] [PubMed] [Google Scholar]
- 29.Ranjard, L., E. Brothier, and S. Nazaret. 2000. Sequencing bands of ribosomal intergenic spacer analysis fingerprints for characterization and microscale distribution of soil bacterium populations responding to mercury spiking. Appl. Environ. Microbiol. 66:5334-5339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rappé, M. S., and S. J. Giovannoni. 2003. The uncultured microbial majority. Annu. Rev. Microbiol. 57:369-394. [DOI] [PubMed] [Google Scholar]
- 31.Robe, P., R. Nalin, C. Capellano, T. M. Vogel, and P. Simonet. 2003. Extraction of DNA from soil. Eur. J. Soil Biol. 39:183-190. [Google Scholar]
- 32.Roesch, L. L., et al. 2007. Pyrosequencing enumerates and contrasts soil microbial diversity. ISME J. 1:283-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sanguin, H., et al. 2006. Potential of a 16S rRNA-based taxonomic microarray for analyzing the rhizosphere effects of maize on Agrobacterium spp. and bacterial communities. Appl. Environ. Microbiol. 72:4302-4312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schloss, P. D., and J. Handelsman. 2003. Biotechnological prospects from metagenomics. Curr. Opin. Biotechnol. 14:303-310. [DOI] [PubMed] [Google Scholar]
- 35.Shendure, J., and J. Hanlee. 2008. Next-generation DNA sequencing. Nat. Biotechnol. 26:1135-1145. [DOI] [PubMed] [Google Scholar]
- 36.Sogin, M. L., et al. 2006. Microbial diversity in the deep sea and the underexplored ‘′rare biosphere.'’ Proc. Natl. Acad. Sci. U. S. A. 103:12115-12120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Torsvik, V., J. Goksoyr, and F. L. Daae. 1990. High diversity in DNA of soil bacteria. Appl. Environ. Microbiol. 56:782-787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Torsvik, V., L. Ovreas, and T. F. Thingstad. 2002. Prokaryotic diversity—magnitude, dynamics, and controlling factors. Science 296:1064-1066. [DOI] [PubMed] [Google Scholar]
- 39.Tringe, S. G., et al. 2005. Comparative metagenomics of microbial communities. Science 308:554-557. [DOI] [PubMed] [Google Scholar]
- 40.Van Elsas, J. D., V. Mantynen, and A. C. Wolters. 1997. Soil DNA extraction and assessment of the fate of Mycobacterium cholorophenolicum strain PC-1 in different soils by 16S ribosomal gene sequence based most probable number PCR and immunofluorescence. Biol. Fertil. Soils 24:188-195. [Google Scholar]
- 41.Van Elsas, J. D., J. K. Jansson, and J. T. Trevors. 2006. Modern soil microbiology II. CRC Press, Boca Raton, FL.
- 42.Van Elsas, J. D., et al. 2008. The metagenomics of disease-suppressive soils—experiences from the Métacontrol project. Trends Biotechnol. 26:591-601. [DOI] [PubMed] [Google Scholar]
- 43.Vogel, T. M., et al. 2009. TerraGenome: a consortium for the sequencing of a soil metagenome. Nat. Rev. Microbiol. 7:252. [Google Scholar]
- 44.Zhou, J., M. A. Bruns, and J. M. Tiedje. 1996. DNA recovery from soils of diverse composition. Appl. Environ. Microbiol. 62:316-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.