Abstract
Mannosylphosphorylation of N- and O-glycans, which confers negative charges on the surfaces of cells, requires the functions of both MNN4 and MNN6 in Saccharomyces cerevisiae. To identify genes relevant to mannosylphosphorylation in the dimorphic yeast Yarrowia lipolytica, the molecular functions of five Y. lipolytica genes showing significant sequence homology with S. cerevisiae MNN4 and MNN6 were investigated. A set of mutant strains in which Y. lipolytica MNN4 and MNN6 homologues were deleted underwent glycan structure analysis. In contrast to S. cerevisiae MNN4 (ScMNN4), the Y. lipolytica MNN4 homologue, MPO1 (YlMPO1), encodes a protein that lacks the long KKKKEEEE repeat domain at its C terminus. Moreover, just a single disruption of YlMPO1 resulted in complete disappearance of the acidic sugar moiety in both the N- and O-linked glycan profiles. In contrast, even quadruple disruption of all ScMNN6 homologues, designated YlKTR1, YlKTR2, YlKTR3, and YlKTR4, resulted in no apparent reduction in acidic sugar moieties. These findings strongly indicate that YlMpo1p performs a significant role in mannosylphosphorylation in Y. lipolytica with no involvement of the Mnn6p homologues. Mutant strains harboring the YlMPO1 gene disruption may serve as useful platforms for engineering Y. lipolytica glycosylation pathways for humanized glycans without any yeast-specific acidic modifications.
Yeast species are important hosts for the production of therapeutic secretory proteins owing to their ability to secrete and glycosylate proteins, their rapid growth to high cell density, and the ease with which they can be genetically manipulated. However, the glycosylation pathway of yeast cells is known to be different from that of mammalian cells, and yeast glycans could induce immunological responses in humans (5). In the traditional yeast Saccharomyces cerevisiae, the typical characteristics of the Asn (N)-linked glycan include hypermannosylation (Man50-150GlcNAc2), mannosylphosphorylation in the core and outer regions, and termination with the α1,3-linked mannose residue. These yeast-specific sugar moieties could prove problematic in the production of therapeutic glycoproteins; hypermannosylation can impair protein activity, and mannosylphosphate residues and terminal α1,3-linked mannoses may elicit an antigenic response (14).
Several nonconventional yeast species, including the methylotrophic species Pichia pastoris and Hansenula polymorpha, have emerged as alternative systems for the production of recombinant therapeutic proteins. Advantages over S. cerevisiae include reduced hypermannosylation (Man8-14GlcNAc2), no terminal α1,3-linked mannose of N-linked oligosaccharides, and efficient heterologous protein secretion (18, 20, 21, 22). As another alternative expression system for the production of human-derived therapeutic glycoproteins, the dimorphic yeast Yarrowia lipolytica has recently drawn attention since this yeast naturally secretes several enzymes, including proteases, lipases, esterases, and RNases, at elevated levels and its posttranslational modification ability is similar to that of mammalian systems (3, 26, 29).
Recently, we reported that the N-linked glycans of Y. lipolytica are composed of neutral and acidic sugars lacking a terminal α1,3-linked mannose (38). The neutral sugars of N-linked glycans are high-mannose oligosaccharides, principally Man7-12GlcNAc2, and the acidic sugars of N-linked glycans are composed of monomannosylphosphorylated Man7-9GlcNAc2 sugars. In the case of S. cerevisiae, at least four mannosylphosphorylation loci have been detected in the core and outer chain regions of N-linked glycans, and the mannosylphosphorylation of N-linked glycans in the Golgi apparatus requires both S. cerevisiae Mnn4p (ScMnn4p) and ScMnn6p (19). Deletion of ScMNN4 or ScMNN6 induced a significant reduction in alcian blue intensity, which reflects the presence of negatively charged molecules such as mannosylphosphate. However, the level of acidic oligosaccharides in N-linked core oligosaccharides is not reduced in the Scmnn6Δ strain, which implies the existence of additional genes required for core oligosaccharide phosphorylation. Moreover, the Scmnn4Δ strain also retains approximately 30% of its acidic oligosaccharide content, thereby indicating the presence of an Mnn4p-independent mannosylphosphorylation pathway (19).
In this study, we have identified and deleted Y. lipolytica homologues to ScMNN4 and ScMNN6 in various combinations, and analyzed the N- and O-linked glycan profiles in the mutants in order to identify the gene(s) responsible for the mannosylphosphorylation of Y. lipolytica glycans. We demonstrate that mannosylphosphorylation of Yarrowia N- and O-linked glycans is mediated primarily by a single gene, Y. lipolytica MPO1 (mannosyl phosphorylation of oligosaccharides 1) (YlMPO1) without the involvement of other MNN6 homologues. Thus, the results of this study lead us to suggest that a Yloch1Δ Ylmpo1Δ double mutant strain might be a useful host for the production of glycoproteins lacking yeast-specific hypermannosylation and mannosylphosphorylation.
MATERIALS AND METHODS
Strains, media, and culture conditions.
The Y. lipolytica strains employed in this study are described in Table 1. The yeast strains were grown routinely in YPD medium (1% yeast extract [BD, Franklin Lakes, NJ], 2% peptone [BD], 2% glucose [Junsei Chemical Co., Tokyo, Japan]) and synthetic complete (SC) medium (0.67% yeast nitrogen base without amino acids [BD], 2% glucose, dropout amino acid mixture, including all of the required amino acids [Sigma, St. Louis, MO]) at 28°C. When necessary, 0.625 mg/ml 5′-fluoroorotic acid (5′-FOA; MP Bio, Solon, OH) was added to the YPD agar medium for the selection of strains auxotrophic for Ura (Ura−). Phenotypic analysis was conducted on YPD solid medium containing 10 μg/ml calcofluor white (CFW; Sigma), 50 μg/ml Congo red (CR; Sigma), 7.5 mM sodium orthovanadate (Van; Sigma), or 40 μg/ml hygromycin B (Hyg B; Sigma). YPDm medium (38) was used for the expression of Trichoderma reesei endoglucanase I (EGI).
TABLE 1.
Strains used in this study
| Strain | Genotype | Reference |
|---|---|---|
| Y. lipolytica | ||
| SMS397A | MATA ade1 ura3 xpr2 | 38 |
| Ylmpo1Δ | MATA ade1 ura3 xpr2 mpo1::tc | This study |
| Ylmpo1Δ::YlMPO1 | MATA ade1 ura3 xpr2 mpo1::tc MPO1::URA3 | This study |
| Ylktr1Δ | MATA ade1 ura3 xpr2 ktr1::tc | This study |
| Ylktr2Δ | MATA ade1 ura3 xpr2 ktr2::tc | This study |
| Ylktr3Δ | MATA ade1 ura3 xpr2 ktr3::tc | This study |
| Ylktr4Δ | MATA ade1 ura3 xpr2 ktr4::tc | This study |
| Ylktr1,2,3,4Δ | MATA ade1 ura3 xpr2 ktr1::tc ktr2::tc ktr3::tc ktr4::tc | This study |
| Yloch1Δ | MATA ade1 ura3 xpr2 och1::tc | 38 |
| Yloch1Δ Ylmpo1Δ | MATA ade1 ura3 xpr2 och1::tc mpo1::tc | This study |
| Yloch1Δ Ylktr1,2,3,4Δ | MATA ade1 ura3 xpr2 och1::tc ktr1::tc ktr2::tc ktr3::tc ktr4::tc | This study |
| Yloch1Δ Ylmpo1Δ Ylktr1,2,3,4Δ | MATA ade1 ura3 xpr2 och1::tc mpo1::tc ktr1::tc ktr2::tc ktr3::tc ktr4::tc | This study |
| Yloch1Δ Ylmpo1Δ::YlMPO1 | MATA ade1 ura3 xpr2 och1::tc mpo1::tc MPO1::URA3 | This study |
Recombinant DNA techniques and gene disruption.
Recombinant DNA techniques and one-step transformation of Y. lipolytica were conducted as previously described by Sambrook and Russell (37) and Chen et al. (6), respectively. The oligonucleotides used for this study are listed in Table S1 in the supplemental material. The Y. lipolytica target genes were disrupted via the PCR-based gene disruption method described by Song et al. (38). To disrupt the YlMPO1 gene, primer set YlMPO1-NF and YlMPO1-NR and primer set YlMPO1-CF and YlMPO1-CR were designed to amplify the 5′ and 3′ flanking regions (571 bp for the 5′ region and 477 bp for the 3′ region) of the YlMPO1 gene, respectively. The PCR fragments amplified by Taq polymerase (Bioneer, Daejeon, South Korea) were fused via a linker sequence (AGATCTACGGATCCATGG) using the YlMPO1-NF and YlMPO1-CR primers, and the product (1,066 bp) was subcloned into the pDrive vector (Qiagen, Hilden, Germany). The BamHI/BglII-treated TcR-YlURA3-TcR cassette from pYIUB (38) was inserted into the BglII site of the linker sequence of the fused PCR product. This disruption cassette was linearized via digestion with BamHI/HindIII and used to transform the Y. lipolytica SMS397A strain. Correct disruption was confirmed by PCR using the primer set YlMPO1-NCF and YlMPO1-CCR. The YlURA3 gene of the integrated TcR-YlURA3-TcR cassette was deleted from the Ylmpo1Δ (URA3) strain by growth on YPD medium containing 5′-FOA at 28°C for 3 days. Additionally, mutants with the YlKTR (KRE two-related) gene deletion were constructed using a strategy that was the same as that used for the detection of the YlMPO1 gene.
In order to reintegrate the YlMPO1 gene into the Ylmpo1Δ mutant, the YlMPO1 gene, including its own promoter (1,000 bp) and terminator (426 bp) regions, was amplified via PCR using TaKaRa Ex Taq polymerase (Takara Bio, Shiga, Japan) with the appropriate primers (primers YlMPO1_prom and YlMPO1_term) and inserted into the pDrive cloning vector. The YlURA3 gene was used as an auxotrophic marker and was amplified from the pIMR53-AUX plasmid as a template (38) and then inserted into the EcoRI/BamHI sites in pUC18 (Invitrogen, Carlsbad, CA), generating pUC18-YlURA3. The cloned YlMPO1 DNA fragment was then cut by XbaI and ligated with XbaI-treated pUC18-YlURA3, yielding pUC18-YlURA3-YlMPO1. The recombinant plasmid linearized by DraIII was introduced into the Ylmpo1Δ and Yloch1Δ Ylmpo1Δ strains. Correct transformants were selected on Ura− minimal medium and confirmed by PCR using gene-specific primers.
Analysis of N-linked oligosaccharides.
The secretory recombinant EGI protein was purified as described previously by Song et al. (38), and the crude cell wall mannoproteins (CWPs) were obtained and purified using hot citrate buffer and concanavalin A (ConA)-Sepharose beads (GE Healthcare, Uppsala, Sweden) as previously described (7). Two hundred micrograms of the purified EGI and 100 μg of the CWPs were treated with 3 μl of peptide:N-glycosidase F (500 U/μl; NEB, Ipswich, MA) for 16 h at 37°C, and the N-linked glycans were purified using a Carbograph Extract-Clean column (150 mg; Alltech, Deerfield, IL). The isolated sugars were then labeled with 2-aminopyridine (PA) or 2- aminobenzoic acid (2-AA). Labeled glycans were analyzed by normal-phase high-pressure liquid chromatography (NP-HPLC). Detailed procedures of sample preparation and analysis are described in the Materials and Methods section in the supplemental material. Matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF) analysis of the N-linked oligosaccharides from the CWPs of the Yloch1Δ and Yloch1Δ Ylmpo1Δ mutant strains was conducted as previously described (38).
Analysis of O-linked glycans.
O-linked glycans were obtained by a modified hydrazinolysis reaction as previously described (30). In brief, dried CWPs (100 to 200 μg) were completely dissolved in 10 to 20 μl of hydrazine monohydrate (Tokyo Chemical Industry, Tokyo, Japan), and the mixture was incubated in a heat block (60°C) for 6 h. After the reactants were dried without heating, they were dissolved in 10 μl of saturated NaHCO3, mixed with 4 μl of (CH3CO)2O, and incubated on ice for 30 min. Next, other contaminants, such as peptides and salts, were removed using Sepabeads SP-20SS (Supelco, Bellefonte, PA) and Dowex 50WX8-400 (H+ form; Sigma) resins, and the purified O-glycans were dried in a vacuum concentrator. The 2-AA labeling, purification, and HPLC analysis of the O-glycans were conducted as described in the Materials and Methods section in the supplemental material. To identify the acidic O-glycans, the glycans underwent mild acid hydrolysis with 0.02 M HCl and dephosphorylation with alkaline phosphatase (1 U/μl; Promega, Madison, WI). The processed glycan was analyzed by HPLC, as described in the Materials and Methods section in the supplemental material.
Capillary electrophoresis.
The O-glycans isolated from the yeast strains were labeled with 1 μl of 8-aminopyrene-1,3,6-trisulfonic acid (APTS; Invitrogen, Carlsbad, CA) labeling solution (1 μl of 100 mM APTS, 4 μl of 1.2 M citric acid, and 5 μl of 1 M sodium cyanoborohydride dissolved in dimethyl sulfoxide) and incubated overnight at 37°C. The reaction was halted by adding 4 μl of water. The APTS-labeled glycans were purified using Sephadex G-10 resin (GE Healthcare), dried, and dissolved in 5 μl of water. After the above purification step was repeated, the glycans were diluted with water (1:10, vol/vol) and analyzed using an ABI GA3130 DNA sequencer and GeneMapper software (Applied Biosystems, Foster City, CA) as previously described (24).
Alcian blue staining.
Yeast colonies were seeded and cultivated in YPD medium for 16 h at 28°C in a shaking incubator. The cells were inoculated at an optical density at 600 nm (OD600) of 0.1 and incubated for 24 h at 28°C. Following the incubation, the cells were harvested, adjusted to an OD600 of 10, washed in 1 ml of washing solution A (0.9% NaCl), and mixed with 100 μl of an alcian blue staining solution (0.1% alcian blue 8GX [Sigma] dissolved in 0.02 N HCl [pH 3.0]). After the yeast cells were allowed to stand for 10 min at room temperature, they were harvested and washed twice with 100 μl of washing solution B (0.02 N HCl, pH 3.0). The cells were transferred into test tubes and analyzed for color as previously described (10).
RESULTS
Sequence analysis of Y. lipolytica homologues to ScMNN4 and ScMNN6.
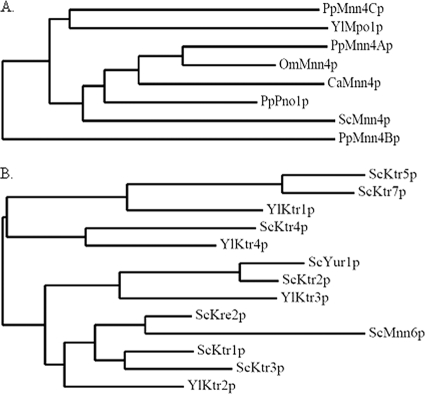
We initially searched for Y. lipolytica homologues of S. cerevisiae MNN4 in the Yarrowia genome database (http://cbi.labri.fr/Genolevures/elt/YALI/). A single Y. lipolytica homologue (YALI0D24101g) was predicted to encode a type II membrane protein showing an overall 40% identity with ScMnn4p that would localize to the Golgi apparatus (http://www.cbs.dtu.dk/services/TMHMM/, http://bioapps.rit.albany.edu/pTARGET/; see Fig. S1 and Table S2 in the supplemental material). The predicted product of YALI0D24101g has a region with significant homology to the LicD domain found in the LICD protein family (PF04991), a member of recently identified groups of nucleotidyltransferase fold proteins. The LICD protein family includes ScMnn4p and bacterial LicD phosphotransferase which use GDP-mannose and CDP-choline as substrates, respectively (23). However, the Y. lipolytica homologue does not have at its C terminus the long KKKKEEEE repeat region, which has been reported to be an important domain for the function of ScMnn4p (32). Moreover, phylogenetic analysis with other yeast ScMnn4p homologues revealed that the Y. lipolytica homologue was distantly related to ScMnn4p and other yeast homologues (Fig. 1A). Therefore, we designated the Y. lipolytica MNN4 homologue MPO1.
FIG. 1.
Phylogenetic analysis of yeast Mnn4p and Mnn6p homologues. (A) Phylogenetic tree analysis of YlMpo1p and other yeast Mnn4p homologues was carried out as previously described (13) using the PhyML (version 3.0) and TreeDyn 198 program tools (http://www.phylogeny.fr/). (B) Phylogenetic tree of the Y. lipolytica Mnn6p homologues and ScKTR family. OmMnn4p, Ogataea minuta Mnn4p.
It has been proposed that ScMnn6p may be a main enzyme involved in mannosylphosphorylation and ScMnn4p may be a regulator of ScMnn6p (31, 32, 40). ScMnn6p, which is also referred to as ScKtr6p, belongs to the ScKTR family (characterized by mannosyltransferase activity), playing roles in the outer chain elaboration of N-glycans and/or O-glycans (25). Our BLAST search for homologues to S. cerevisiae MNN6 identified four putative Y. lipolytica open reading frames (YALI0F25817g, YALI0A06589g, YALI0B01672g, and YALI0E01892g) (see Table S3 in the supplemental material). As shown in Fig. S2 in the supplemental material, the Yarrowia Mnn6p homologues, with the exception of YALI0A06589g, encoded proteins with a type II transmembrane topology. However, the four Yarrowia homologues showed higher sequence identity with the other ScKTR family of genes than with the S. cerevisiae MNN6 gene. The predicted proteins of Yarrowia homologues shared 41% to 54% identities with the ScKTR proteins, including ScKre2p, ScKtr1p, ScKtr2p, ScKtr3p, ScKtr4p, ScKtr5p, ScKtr7p, and ScYur1p. On the other hand, the four Yarrowia proteins shared 28% to 33% identities with ScMnn6p (see Table S3 in the supplemental material). On the basis of the phylogenetic tree analysis (Fig. 1B) as well as the higher similarity of the four Y. lipolytica homologues to the ScKTR proteins than to ScMnn6p, we designated the four Y. lipolytica homologues, YALI0F25817g, YALI0A06589g, YALI0B01672g, and YALI0E01892g, as YlKTR1, YlKTR2, YlKTR3, and YlKTR4, respectively.
YlMPO1 is responsible for mannosylphosphorylation of N-linked oligosaccharides in Y. lipolytica.
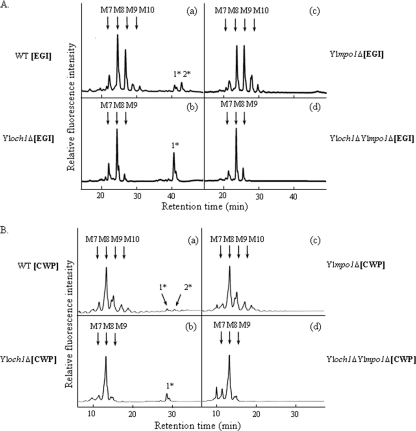
In an effort to evaluate the functions of the Y. lipolytica Mnn4p homologue YlMpo1p in the mannosylphosphorylation of N-linked glycans, we deleted the YlMPO1 gene from the wild-type and Yloch1Δ strains, generating mutants Ylmpo1Δ and Yloch1Δ Ylmpo1Δ, respectively. The N-linked oligosaccharides from a model secretory glycoprotein, the recombinant Trichoderma reesei EGI, and from CWPs were obtained from the wild-type and mutant strains for structural analysis with NP-HPLC. Similar to a previous report (38), the N-linked oligosaccharides of the recombinant EGI secreted from the Yloch1Δ strain displayed an elevated acidic glycan peak (monomannosylphosphorylated Man8GlcNAc2) compared to that generated from the wild-type strain (Fig. 2A, panels a and b). The same pattern of glycan profiles with an increased acidic glycan peak was also observed in the N-linked oligosaccharides of CWPs from the Yloch1Δ strain, reflecting the general presence of phosphorylated N-glycans in endogenous Y. lipolytica glycoproteins (Fig. 2B, panels a and b). Notably, the deletion of YlMPO1 in the wild-type or Yloch1Δ strain completely eliminated the acidic glycan peaks of the N-glycan profiles in the secreted EGI (Fig. 2A, panels c and d) and in the cell wall proteins (Fig. 2B, panels c and d).
FIG. 2.
HPLC analysis of the N-linked oligosaccharides of EGI and cell wall mannoproteins from wild-type (WT) and Ylmpo1Δ mutant strains. (A) N-linked oligosaccharides from the recombinant EGI secreted from wild-type (a), Yloch1Δ (b), Ylmpo1Δ (c), and Yloch1Δ Ylmpo1Δ (d) strains were labeled with PA and analyzed by NP-HPLC. (B) N-linked oligosaccharides from the CWPs of wild-type (a), Yloch1Δ (b), Ylmpo1Δ (c), and Yloch1Δ Ylmpo1Δ (d) strains were labeled with 2-AA and analyzed by NP-HPLC. The elution times of the peaks were compared with those of authentic PA- or AA-sugar chains (Man7GlcNAc2-AA and Man8GlcNAc2-AA) of known structure (indicated by arrows): M7, Man7GlcNAc2-PA; M8, Man8GlcNAc2-PA; M9, Man9GlcNAc2-PA; M10, Man10GlcNAc2-PA; 1*, monomannosylphosphorylated Man8GlcNAc2-PA; 2*, monomannosylphosphorylated Man9GlcNAc2-PA.
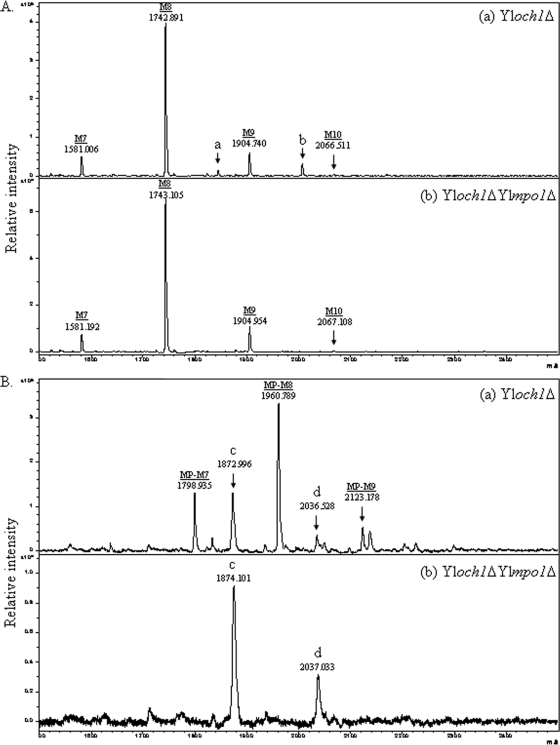
The dramatic disappearance of acidic glycan moieties resulting from the deletion of YlMPO1 was confirmed via MALDI-TOF mass spectrometry analysis. The N-linked oligosaccharides derived from the CWPs of the Yloch1Δ and Yloch1Δ Ylmpo1Δ mutant strains were analyzed using MALDI-TOF, both in the positive reflector mode for the detection of neutral sugars (Fig. 3A) and in the negative linear mode for the detection of acidic sugars (Fig. 3B). The major peaks of the Yloch1Δ mutant detected in the negative mode corresponded to mannosylphosphorylated Man7GlcNAc2 and Man8GlcNAc2 (Man8PGlcNAc2 and Man9PGlcNAc2, respectively) species, with their molecular masses exceeding those of the neutral sugars. These peaks were clearly eliminated via the disruption of the YlMPO1 gene. The results clearly demonstrate the crucial function of YlMpo1p in the mannosylphosphorylation of N-linked glycans in Y. lipolytica.
FIG. 3.
MALDI-TOF mass spectrometry of the N-linked oligosaccharides from CWPs of Yloch1Δ and Yloch1Δ Ylmpo1Δ mutant strains. (A) Mass spectra analyzed in the positive reflector mode for the detection of neutral sugars of Yloch1Δ (a) and Yloch1Δ Ylmpo1Δ (b) strains. The intermediate peaks, designated a and b, are assumed to represent the monomannosylphosphorylated forms of Man7GlcNAc2 and Man8GlcNAc2 on the basis of their m/z values of 1844.583 and 2006.425, respectively. M7, Man7GlcNAc2; M8, Man8GlcNAc2; M9, Man9GlcNAc2; M10, Man10GlcNAc2. (B) Mass spectra analyzed in the linear negative mode for the detection of acidic sugars of the Yloch1Δ (a) and Yloch1Δ Ylmpo1Δ (b) strains. The intermediate peaks, designated c and d, are assumed to represent Man9GlcNAc2 and Man10GlcNAc2, respectively. MP-M7, monomannosylphosphorylated Man7GlcNAc2; MP-M8, monomannosylphosphorylated Man8GlcNAc2; MP-M9, monomannosylphosphorylated Man9GlcNAc2.
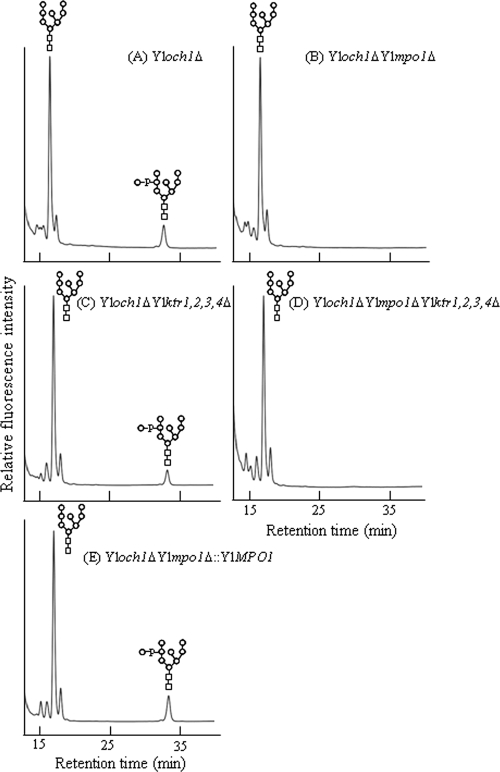
We also attempted to determine whether the Y. lipolytica MNN6 homologues, YlKTR1, YlKTR2, YlKTR3, and YlKTR4, are involved with the mannosylphosphorylation of N-glycans in Y. lipolytica. Interestingly, all mutant strains from which each YlKTR gene was deleted showed mannosylphosphorylation in N-glycans similar to that for the wild-type strain (see Fig. S3 in the supplemental material). Comparable results were observed in the Yloch1Δ background strain, which showed a more definite acidic sugar profile (Fig. 4). Additionally, even the quadruple deletion of all YlKTR genes (YlKTR1, YlKTR2, YlKTR3, and YlKTR4; strain Yloch1 ΔYlktr1,2,3,4Δ) did not influence mannosylphosphorylation of N-glycans (Fig. 4C). Moreover, no changes in the overall glycan profiles of the Ylktr mutant strains were observed, thereby indicating that these genes do not participate in the elongation of N-glycans. The reintroduction of YlMPO1 resulted in the reappearance of the acidic glycan peak (Fig. 4E), demonstrating that YlMPO1 may perform an essential function in the mannosylphosphorylation of N-linked oligosaccharides without the involvement of YlKTR1, YlKTR2, YlKTR3, and YlKTR4.
FIG. 4.
HPLC analysis of the N-linked oligosaccharide of CWPs from the YlktrΔ mutant strains. The N-linked oligosaccharide profiles of CWPs from Yloch1Δ (A), Yloch1Δ Ylmpo1Δ (B), Yloch1Δ Ylktr1,2,3,4Δ (C), Yloch1Δ Ylmpo1Δ Ylktr1,2,3,4Δ (D), and Yloch1Δ Ylmpo1Δ::YlMPO1 (E) mutant strains were compared. Man8GlcNAc2-AA was used as a standard AA-sugar chain.
YlMPO1 is also involved in the mannosylphosphorylation of O-linked glycans in Y. lipolytica.
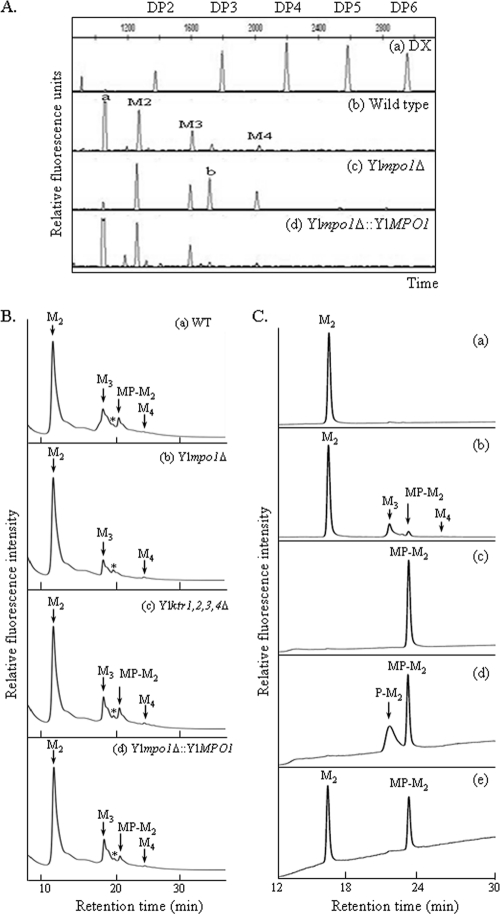
ScMnn4p and ScMnn6p are required for the mannosylphosphorylation of both N- and O-oligosaccharides in S. cerevisiae (19). Thus, we evaluated the possible functions of the YlMpo1p or YlKtr protein in the mannosylphosphorylation of O-glycans in Y. lipolytica. O-glycans of CWPs from the wild-type and Ylmpo1Δ strains were labeled with APTS fluorescent dye and analyzed with a DNA sequencer. As seen in Fig. 5A, the most rapidly moving peak in the wild-type strain (Fig. 5A, trace b) was not detected in the Ylmpo1Δ strain (Fig. 5A, trace c) but reappeared following the reintroduction of the YlMPO1 gene into the Ylmpo1Δ strain (Fig. 5A, trace d). Considering the negative charge properties of APTS, acidic oligosaccharides are expected to move more rapidly than neutral oligosaccharides in the DNA sequencer. Thus, the fast-moving peak detected in the wild-type strain was speculated to represent an acidic O-glycan with mannosylphosphate residues.
FIG. 5.
Analysis of the O-linked oligosaccharides from Ylmpo1Δ and YlktrΔ mutant strains. (A) O-linked oligosaccharides of CWPs from wild-type (b), Ylmpo1Δ (c), and Ylmpo1Δ::YlMPO1 (d) strains were analyzed with a DNA sequencer. (a) Profile of the APTS-maltooligosaccharide ladder, which served as a size reference. The number of glucose units (degrees of polymerization [DP]) is indicated along the x axis. (B) O-linked oligosaccharides of CWPs from the wild-type (WT) (a), Ylmpo1Δ (b), Ylktr1,2,3,4Δ (c), and Ylmpo1Δ::YlMPO1 (d) strains were labeled with 2-AA and analyzed by NP-HPLC. O-linked oligosaccharides were acquired via hydrazinolysis, labeled with 2-AA, and subsequently analyzed via NP-HPLC with a TSKgel Amide-80 column. (C) Analysis of MP-M2 in acidic O-linked glycans of CWPs. α1,2-Mannobiose-AA was used as a standard (a). O-glycans of CWPs from the wild-type strain were analyzed (b). The peak corresponding to the putative MP-M2 was collected (c), hydrolyzed with mild acid (d), and treated with alkaline phosphatase (e). M3, α-linked mannotriose; M4, α-linked mannotetraose.
To evaluate this possibility, O-glycans from several mutant strains were analyzed via NP-HPLC with an amide column. As shown in Fig. 5B, O-glycans from the wild-type Y. lipolytica strain were composed of Man2-4, as suggested by the DNA sequencer analysis (Fig. 5A, trace b). The peak at 20.5 min in the wild-type strain (Fig. 5B, trace a), which was also detected in the Ylktr1,2,3,4Δ strain (Fig. 5B, trace c), disappeared in the Ylmpo1Δ strain (Fig. 5B, trace b). This peak reappeared in the Ylmpo1Δ::YlMPO1 strain into which the functional YlMPO1 was reintegrated (Fig. 5B, trace d), thereby suggesting that it may represent a monomannosylphosphorylated mannobiose (MP-M2). To confirm the identity, the peak in Fig. 5B was collected, hydrolyzed with mild acid, treated with alkaline phosphatase (31), and analyzed via HPLC (Fig. 5C). Mild acid hydrolysis of the collected peak (Fig. 5C, trace c) generated an additional peak, monophosphorylated mannobiose (P-M2) (Fig. 5C, trace d). If P-M2 resulted from hydrolysis of MP-M2, then treatment with alkaline phosphatase would be anticipated to yield a peak corresponding to α-linked mannobiose (M2). Indeed, alkaline phosphatase treatment generated a novel peak at a retention time of 16.4 min (Fig. 5C, trace e), corresponding to the retention time of α1,2-Man2 (Fig. 5C, trace a). These results clearly indicate that the peak missing from the Ylmpo1Δ strain is an acidic O-glycan, a monomannosylphosphorylated mannobiose form, and that the YlMPO1 gene participates in the mannosylphosphorylation of O-glycans as well as N-glycans in Y. lipolytica. However, the Ylktr1,2,3,4Δ strain showed no changes in the O-glycan profiles. This suggests that none of the Y. lipolytica Mnn6p homologues are involved in the addition of mannosylphosphate to O-glycans, in contrast to the case in S. cerevisiae Mnn6p.
Phenotypic characteristics of Ylmpo1Δ and YlktrΔ mutant strains.
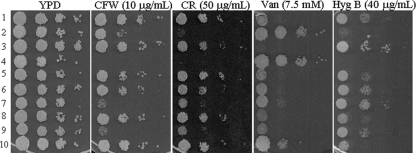
It has been demonstrated that glycosylation pathway-defective yeast strains change their sensitivities to cell wall synthesis inhibitors, including CFW and CR (34, 35). CFW functions by binding to the chitin polymer, one of the components of the fungal cell wall, and inhibiting the assembly enzymes that link chitin to β1,3- and β1,6-glucans (34, 35). CR binds preferentially to β1,3-glucan and to chitin to a lesser degree. To evaluate the roles of the YlMPO1 and YlKTR genes in cell wall integrity, each mutant strain was spotted onto YPD plates containing CFW or CR. The Yloch1Δ strain, which is sensitive to CFW and CR, was used as a positive control in the assay. As shown in Fig. 6, the Ylmpo1Δ strain was slightly more sensitive to CFW and CR than the wild-type strain, and the deletion of YlMPO1 in the Yloch1Δ mutant increased the sensitivity to CFW and CR. These results indicate that YlMpo1p is required, but not to a great extent, for cell wall integrity in Y. lipolytica. Another interesting finding from this analysis was that the Ylktr3Δ strain was as sensitive to CFW and CR as the Yloch1Δ mutant. The data imply that although YlKTR3 does not appear to be involved in N- or O-linked glycosylation, the functions of YlKTR3 might be relevant to cell wall integrity.
FIG. 6.
Phenotypic analyses of the Ylmpo1Δ and YlktrΔ mutant strains. Wild-type (row 1), Yloch1Δ (row 2), Ylmpo1Δ (row 3), Yloch1Δ Ylmpo1Δ (row 4), Ylktr1Δ (row 5), Ylktr2Δ (row 6), Ylktr3Δ (row 7), Ylktr4Δ (row 8), Ylktr1,2,3,4Δ (row 9), and Ylmpo1Δ::YlMPO1 (row 10) cells were spotted onto YPD medium plates containing 10 μg/ml of CFW, 50 μg/ml of CR, 7.5 mM Van, or 40 μg/ml of Hyg B and incubated for 3 to 4 days at 28°C.
Glycosylation-defective mutants are generally resistant to Van but sensitive to Hyg B (2, 12). As anticipated, the Yloch1Δ strain showed markedly increased resistance to vanadate but reduced sensitivity to an aminoglycoside antibiotic, hygromycin B, compared to the susceptibilities of the wild type. It is worthwhile to note that the Ylmpo1Δ strain displayed phenotypes opposite those of the Yloch1Δ strain; it became rather sensitive to vanadate, but the level of hygromycin B resistance increased. Reintroduction of YlMPO1 into the Ylmpo1Δ strain completely restored the expected phenotype but with a slightly altered sensitivity and resistance relative to the susceptibilities of the wild-type strain. It has been proposed that the vanadium contained in sodium orthovanadate has a conformation similar to that of phosphorus and thus competes with phosphorus in phosphorylated compounds (39). Therefore, the increased sensitivity of Ylmpo1Δ to vanadate further supports the notion that YlMpo1p plays a role in phosphorylation-related processes.
The Y. lipolytica mutant strains were stained with alcian blue dye, which binds via electrostatic forces to negatively charged molecules such as mannosylphosphate (9). The Ylmpo1Δ strain exhibited the most dramatic reduction in staining intensity (Table 2), consistent with lowered levels of mannosylphosphate on the cell surfaces. Interestingly, the deletion of YlKTR4 (with a yet-undetermined function) also resulted in reduced staining. The stain intensity increased significantly in the Yloch1Δ strain compared to that in the wild type, suggesting that the absence of the outer chain might enhance the efficiency of mannosylphosphorylation by YlMpo1p or the binding of core N-glycans with the dye.
TABLE 2.
Alcian blue staining of various Y. lipolytica strains
| Strain | Relevant characteristic(s) | Alcian blue staininga |
|---|---|---|
| SMS397A | xpr2 | +++ |
| Yloch1Δ | xpr2 och1 | ++++ |
| Ylmpo1Δ | xpr2 mpo1 | − |
| Yloch1Δ Ylmpo1Δ | xpr2 och1 mpo1 | + |
| Ylmpo1Δ::YlMPO1 | xpr2 mpo1 MPO1 | +++ |
| Ylktr1Δ | xpr2 ktr1 | +++ |
| Ylktr2Δ | xpr2 ktr2 | +++ |
| Ylktr3Δ | xpr2 ktr3 | +++ |
| Ylktr4Δ | xpr2 ktr4 | +++ |
The colors that developed on the cell pellets in the alcian blue assay were classified as follows: −, white; +, light blue; +++, blue; and ++++, dark blue.
DISCUSSION
In this study, we evaluated the molecular functions of the Y. lipolytica MPO1 gene, which encodes a protein that exhibits an overall identity of 40% to ScMnn4p but which lacks a long C-terminal KE repeat domain crucial for the function of ScMnn4p (32). One of the most noticeable results was that, in contrast to S. cerevisiae and P. pastoris, a single deletion of the YlMPO1 gene proved to be sufficient for completely eliminating mannosylphosphorylation of both N- and O-linked glycans in Y. lipolytica. Thus, it can be surmised that YlMpo1p controls the majority of the mannosylphosphorylation of N- and O-linked glycans in Y. lipolytica.
Many yeast species harbor multiple Mnn4p homologues (or hypothetical proteins) to ScMnn4p. Two Mnn4-like genes exist in S. cerevisiae (19), four exist in P. pastoris (4, 27) and Ogataea minuta (1), and eight exist in Candida albicans (17). However, in Y. lipolytica, only a single gene (YALI0D24101g) was predicted to encode a type II membrane protein showing homology with ScMnn4p. In S. cerevisiae, the inability of the Scmnn4Δ mutant to remove mannosylphosphorylation entirely from glycans (32) implies that an additional gene(s) may participate in mannosylphosphorylation. In P. pastoris, both the P. pastoris PNO1 (phosphomannosylation of N-linked oligosaccharides) (PpPNO1) gene and the PpMNN4B gene perform roles in the mannosylphosphorylation of N-linked oligosaccharides (4). Among the eight MNN4-like sequences in the C. albicans genome, the C. albicans MNN4 (CaMNN4) gene is the closest homologue to the ScMNN4 gene and harbors a KE repeat region. CaMNN4 appears to be primarily responsible for mannosylphosphorylation, on the basis of the observation that the Camnn4Δ null mutant was unable to bind to alcian blue dye and was devoid of acid-labile β1,2-linked oligomannosaccharides in the outer cell wall (17). However, detailed structural analysis data demonstrating the lack of mannosylphosphorylated residues in the N- and O-glycans derived from the Camnn4Δ mutant have not been performed. It is worth noting that the NCBI BLASTP searches using YlMpo1p as a query identified several hypothetical and putative fungal proteins, besides C. albicans Mnn4p (AAL86704), as YlMpo1p homologues with greater sequence similarities than the sequence similarity of ScMnn4p. Interestingly, most of these homologues possess a predicted LicD region, strongly indicating their possible roles in nucleotidyl transfer.
ScMnn4p is regarded as a positive regulator of ScMnn6p, a mannosylphosphate transferase (19). However, the relationship between the MNN4 and MNN6 genes has yet to be clearly elucidated. If YlMpo1p also participates in mannosylphosphorylation together with a putative mannosylphosphate transferase, we anticipate that a gene encoding mannosylphosphate transferase should exist in Y. lipolytica. However, the YlKTR1, YlKTR2, YlKTR3, and YlKTR4 genes, initially detected to be the closest ScMNN6 homologues, are not involved in the mannosylphosphorylation of Y. lipolytica glycans (Fig. 4; see also Fig. S3 in the supplemental material). In C. albicans, no ScMnn6p homologues were detected in the genome database. However, a very recent study demonstrated that the double deletion of CaMNT3 and CaMNT5, which are involved in N-glycosylation, resulted in a reduction in the ability to bind to alcian blue for staining. This suggests the presence of multifunctional mannosyltransferases with redundant activities in both the N-glycosylation and phosphomannosylation of C. albicans (28). Aside from phosphomannosyltransferase activity per se, the extent of mannosylphosphorylation of glycans might be affected indirectly by several factors. This could include the activities of other mannosyltransferases, which can generate more optimal substrates or compete for a common substrate, as well as vacuolar targeting and actin/cytoskeleton organization in the secretory pathway, which may influence the localization of enzymes (11). Therefore, further studies will attempt to determine which protein(s) besides the YlKtr1, YlKtr2, YlKtr3, and YlKtr4 proteins has mannosylphosphate transferase activity as a ScMnn6p functional homologue. Future work will also examine whether YlMpo1p alone can function in mannosylphosphate transfer in association with other multifunctional glycosyltransferases, as has been suggested in C. albicans.
Despite the fact that the YlKTR1, YlKTR2, YlKTR3, and YlKTR4 genes appeared to play no role in the mannosylphosphorylation or protein glycosylation of Y. lipolytica (Fig. 4; see also Fig. S3 in the supplemental material), the Ylktr3Δ mutant exhibited a profound sensitivity to CFW and CR (Fig. 6). Ruiz-Herrera et al. (36) reported that an increase in either the sensitivity or resistance of Y. lipolytica to certain levels of CFW is indicative of alterations in cell wall structure independent of the level of chitin. Considering that the Ylktr3Δ mutant strain displayed defective hyphal development (data not shown), we speculate that YlKTR3 might be associated with the glycosylation process of glycolipids which is required for morphogenesis in Y. lipolytica (3).
Several yeast and fungal species have been used in the development of secretion hosts producing therapeutic glycoproteins with human complex-type N-glycans (15, 16, 33). Recently, pioneering work on the engineering of the O-glycosylation pathway expressing human proteins in S. cerevisiae was reported (8). As one of the nonconventional yeasts, Y. lipolytica exhibits some advantages as a potential host for the secretory production of human-derived recombinant glycoproteins, including greatly reduced hypermannosylation (Man7-12GlcNAc2), the absence of a terminal α1,3-mannose epitope in both N- and O-linked oligosaccharides, and posttranslational modification processes similar to those in mammalian systems (26). Therefore, we anticipate that the combined deletion of YlMPO1 and YlOCH1, the factor responsible for yeast-specific outer chain initiation in N-linked glycans (38), should serve as a foundation for engineering a humanized glycosylation pathway in Y. lipolytica with no yeast-specific glycan modification.
Supplementary Material
Acknowledgments
We thank Y. Shimma and Y. Chiba at AIST, Japan, for their valuable comments on designing experiments in this study.
This work was supported by grants from the Korean Ministry of Education and Science (Microbial Genomics and Applications R&D Program) and from the Korean Ministry of Knowledge and Economy (Next Generation New Technology Development Program).
Footnotes
Published ahead of print on 23 December 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Akeboshi, H., et al. 2009. Production of human beta-hexosaminidase A with highly phosphorylated N-glycans by the overexpression of the Ogataea minuta MNN4 gene. Glycobiology 19:1002-1009. [DOI] [PubMed] [Google Scholar]
- 2.Ballou, L., R. A. Hitzeman, M. S. Lewis, and C. E. Ballou. 1991. Vanadate-resistant yeast mutants are defective in protein glycosylation. Proc. Natl. Acad. Sci. U. S. A. 88:3209-3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barth, G., and C. Gaillardin. 1997. Physiology and genetics of the dimorphic fungus Yarrowia lipolytica. FEMS Microbiol. Rev. 19:219-237. [DOI] [PubMed] [Google Scholar]
- 4.Bobrowicz, P., T. A. Stadheim, and S. Wildt. August 2007. Methods for eliminating mannosylphosphorylation of glycans in the production of glycoproteins. U.S. patent 7,259,007.
- 5.Böer, E., G. Steinborn, G. Kunze, and G. Gellissen. 2007. Yeast expression platforms. Appl. Microbiol. Biotechnol. 77:513-523. [DOI] [PubMed] [Google Scholar]
- 6.Chen, D.-C., J.-M. Beckerich, and C. Gaillardin. 1997. One-step transformation of the dimorphic yeast Yarrowia lipolytica. Appl. Microbiol. Biotechnol. 48:232-235. [DOI] [PubMed] [Google Scholar]
- 7.Chiba, Y., et al. 1998. Production of human compatible high mannose-type (Man5GlcNAc2) sugar chains in Saccharomyces cerevisiae. J. Biol. Chem. 273:26298-26304. [DOI] [PubMed] [Google Scholar]
- 8.Chigira, Y., T. Oka, T. Okajima, and Y. Jigami. 2008. Engineering of a mammalian O-glycosylation pathway in the yeast Saccharomyces cerevisiae: production of O-fucosylated epidermal growth factor domains. Glycobiology 18:303-314. [DOI] [PubMed] [Google Scholar]
- 9.Conde, R., G. Pablo, R. Cueva, and G. Larriba. 2003. Screening for new yeast mutants affected in mannosylphosphorylation of cell wall mannoproteins. Yeast 20:1189-1211. [DOI] [PubMed] [Google Scholar]
- 10.Conde, R., R. Cueva, and G. Larriba. 2007. Rsc14-controlled expression of MNN6, MNN4 and MNN1 regulates mannosylphosphorylation of Saccharomyces cerevisiae cell wall mannoproteins. FEMS Yeast Res. 7:1248-1255. [DOI] [PubMed] [Google Scholar]
- 11.Corbacho, I., I. Olivero, and L. M. Hernández. 2005. A genome-wide screen for Saccharomyces cerevisiae nonessential genes involved in mannosyl phosphate transfer to mannoprotein-linked oligosaccharides. Fungal Genet. Biol. 42:773-790. [DOI] [PubMed] [Google Scholar]
- 12.Dean, N. 1995. Yeast glycosylation mutants are sensitive to aminoglycosides. Proc. Natl. Acad. Sci. U. S. A. 92:1287-1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dereeper, A., et al. 2008. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 36(Web Server issue):W465-W469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gemmill, T. R., and R. B. Trimble. 1999. Overview of N- and O-linked oligosaccharide structures found in various yeast species. Biochim. Biophys. Acta 1426:227-237. [DOI] [PubMed] [Google Scholar]
- 15.Gerngross, T. U. 2004. Advances in the production of human therapeutic proteins in yeasts and filamentous fungi. Nat. Biotechnol. 22:1409-1414. [DOI] [PubMed] [Google Scholar]
- 16.Hamilton, S. R., et al. 2006. Humanization of yeast to produce complex terminally sialylated glycoproteins. Science 313:1441-1443. [DOI] [PubMed] [Google Scholar]
- 17.Hobson, R. P., et al. 2004. Loss of cell wall mannosylphosphate in Candida albicans does not influence macrophage recognition. J. Biol. Chem. 279:39628-39635. [DOI] [PubMed] [Google Scholar]
- 18.Ilgen, C., J. Lin-Cereghino, and J. M. Cregg. 2005. Production of recombinant proteins, p. 143-162. In G. Gellissen (ed.), Pichia pastoris. Wiley-VCH, Weinheim, Germany.
- 19.Jigami, Y., and T. Odani. 1999. Mannosylphosphate transfer to yeast mannan. Biochim. Biophys. Acta 1426:335-345. [DOI] [PubMed] [Google Scholar]
- 20.Kang, H. A., and G. Gellissen. 2005. Production of recombinant proteins, p. 111-142. In G. Gellissen (ed.), Hansenula polymorpha. Wiley-VCH, Weinheim, Germany.
- 21.Kim, M. W., et al. 2006. Functional characterization of the Hansenula polymorpha HOC1, OCH1, and OCR1 genes, as members of the yeast OCH1 mannosyltransferase family involved in protein glycosylation. J. Biol. Chem. 281:6261-6272. [DOI] [PubMed] [Google Scholar]
- 22.Kim, M. W., et al. 2004. Characterization of N-linked oligosaccharides assembled on secretory recombinant glucose oxidase and cell wall mannoproteins from the methylotrophic yeast Hansenula polymorpha. Glycobiology 14:243-251. [DOI] [PubMed] [Google Scholar]
- 23.Kuchta, K., L. Knizewski, L. S. Wyrwicz, L. Rychlewski, and K. Ginalski. 2009. Comprehensive classification of nucleotidyltransferase fold proteins: identification of novel families and their representatives in human. Nucleic Acids Res. 37:7701-7714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee, K. J., et al. 2009. High-throughput quantitative analysis of plant N-glycan using a DNA sequencer. Biochem. Biophys. Res. Commun. 380:223-229. [DOI] [PubMed] [Google Scholar]
- 25.Lussier, M., A.-M. Sdicu, and H. Bussey. 1999. The KTR and MNN1 mannosyltransferase families of Saccharomyces cerevisiae. Biochim. Biophys. Acta 1426:323-334. [DOI] [PubMed] [Google Scholar]
- 26.Madzak, C., C. Gaillardin, and J.-M. Beckerich. 2004. Heterologous protein expression and secretion in the non-conventional yeast Yarrowia lipolytica: a review. J. Biotechnol. 109:63-81. [DOI] [PubMed] [Google Scholar]
- 27.Miura, M., M. Hirose, T. Miwa, S. Kuwae, and H. Ohi. 2004. Cloning and characterization in Pichia pastoris of PNO1 gene required for phosphomannosylation of N-linked oligosaccharides. Gene 324:129-137. [DOI] [PubMed] [Google Scholar]
- 28.Mora-Montes, H. M., et al. 2010. A multifunctional mannosyltransferase family in Candida albicans determines cell wall mannan structure and host-fungus interactions. J. Biol. Chem. 285:12087-12095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Müller, S., T. Sandai, P. Kamp-Hansen, and H. Dalbøge. 1998. Comparison of expression systems in the yeasts Saccharomyces cerevisiae, Hansenula polymorpha, Kluyveromyces lactis, Schizosaccharomyces pombe and Yarrowia lipolytica. Cloning of two novel promoters from Yarrowia lipolytica. Yeast 14:1267-1283. [DOI] [PubMed] [Google Scholar]
- 30.Nakakita, S., W. Sumiyoshi, N. Miyanishi, and J. Hirabayashi. 2007. A practical approach to N-glycan production by hydrazinolysis using hydrazine monohydrate. Biochem. Biophys. Res. Commun. 362:639-645. [DOI] [PubMed] [Google Scholar]
- 31.Nakayama, K., Y. Feng, A. Tanaka, and Y. Jigami. 1998. The involvement of mnn4 and mnn6 mutations in mannosylphosphorylation of O-linked oligosaccharides in yeast Saccharomyces cerevisiae. Biochim. Biophys. Acta 1425:255-262. [DOI] [PubMed] [Google Scholar]
- 32.Odani, T., Y. Shimma, A. Tanaka, and Y. Jigami. 1996. Cloning and analysis of the MNN4 gene required for phosphorylation of N-linked oligosaccharides in Saccharomyces cerevisiae. Glycobiology 6:805-810. [DOI] [PubMed] [Google Scholar]
- 33.Oh, D.-B., et al. 2008. Glycoengineering of the methylotrophic yeast Hansenula polymorpha for the production of glycoproteins with trimannosyl core N-glycan by blocking core oligosaccharide assembly. Biotechnol. J. 3:659-668. [DOI] [PubMed] [Google Scholar]
- 34.Ram, A. F., A. Wolters, R. Ten Hoopen, and F. M. Klis. 1994. A new approach for isolating cell wall mutants in Saccharomyces cerevisiae by screening for hypersensitivity to calcofluor white. Yeast 10:1019-1030. [DOI] [PubMed] [Google Scholar]
- 35.Ram, A. F., and F. M. Klis. 2006. Identification of fungal cell wall mutants using susceptibility assays based on calcofluor white and Congo red. Nat. Protoc. 1:2253-2256. [DOI] [PubMed] [Google Scholar]
- 36.Ruiz-Herrera, J., P. García-Maceira, L. C. Castillo-Barahona, E. Valentín, and R. Sentandreu. 2003. Cell wall composition and structure of Yarrowia lipolytica transposon mutants affected in calcofluor sensitivity. Antonie Van Leeuwenhoek 84:229-238. [DOI] [PubMed] [Google Scholar]
- 37.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 38.Song, Y., et al. 2007. Engineering of the yeast Yarrowia lipolytica for the production of glycoproteins lacking the outer-chain mannose residues of N-glycans. Appl. Environ. Microbiol. 73:4446-4454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thiebault, F., F. Kiss, R. Bonaly, and J. Coulon. 2002. Effect of sodium orthovanadate on glycosylation of the phosphopeptidomannans involved in the cell-cell aggregation of the yeast Kluyveromyces bulgaricus. Carbohydr. Res. 337:1873-1877. [DOI] [PubMed] [Google Scholar]
- 40.Wang, X.-H., K. Nakayama, Y. Shimma, A. Tanaka, and Y. Jigami. 1997. MNN6, a member of the KRE2/MNT1 family, is the gene for mannosylphosphate transfer in Saccharomyces cerevisiae. J. Biol. Chem. 272:18117-18124. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.