Abstract
To inspect the norovirus contamination of groundwater in South Korea, a nationwide study was performed in the summer (June to August) and winter (October to December) of 2008. Three-hundred sites designated by the government ministry were inspected. Water samples were collected for analysis of water quality, microorganism content, and viral content. Water quality was assessed by temperature, pH, turbidity, residual chlorine, and nitrite nitrogen content. Microorganism contents were analyzed bacteria, total coliforms, Escherichia coli, and bacteriophage. Virus analyses included panenterovirus and norovirus. Two primer sets were used for the detection of norovirus genotypes GI and GII, respectively. Of 300 samples, 65 (21.7%) were norovirus positive in the summer and in 52 (17.3%) were norovirus positive in the winter. The genogroup GI noroviruses that were identified were GI-1, GI-2, GI-3, GI-4, GI-5, GI-6, and GI-8 genotypes; those in the GII genogroup were GII-4 and GII-Yuri genotypes. The analytic data showed correlative relationships between the norovirus detection rate and the following parameters: water temperature and turbidity in physical-chemical parameters and somatic phage in microbial parameters. It is necessary to periodically monitor waterborne viruses that frequently cause epidemic food poisoning in South Korea for better public health and sanitary conditions.
As many studies have revealed, gastroenteritis has been, and still is, a major public health problem worldwide. Recently, the development of sensitive diagnostic techniques has provided evidence that noroviruses (NoVs) are the leading cause of acute nonbacterial gastroenteritis outbreaks in humans. In developed countries—including the United States, Europe, and Japan—up to 93% of such outbreaks, along with 60 to 85% of all gastroenteritis outbreaks, were associated with NoVs (15, 24). Mortality due to acute gastroenteritis was estimated at 2.1 million in the year 2000, and mortality due to gastroenteritis in children was higher in developing countries than in developed countries (4).
A member of the Caliciviridae family, NoVs have a positive-sense, single-stranded RNA (7.6 kb). There are five NoV genogroups (genogroup I [GI] to GV) and three human NoV genogroups (GI, GII, and GIV). Each of the human NoV genogroups is further classified into genotypes; there are 8, 17, and 1 genotypes in GI, GII, and GIV, respectively (24).
NoVs exhibit broad genomic diversity, and human NoV genogroups (GI, GII, and GIV) are comprised of at least 25 genotypes and many subgroups (7). The diversity of NoVs is maintained by the accumulation of point mutations from the error-prone nature of RNA replication and by genetic recombination due to sequence exchange between related RNA viruses (22).
The rapid, extensive spread of disease outbreak in closed settings such as hospitals, hotels, cruise ships, and among troops might be explained by transmission through infectious vomitus, which could be passed on from person to person either on environmental surfaces (i.e., through hand/mouth contact) or through aerosolization (2, 3, 10, 13, 19, 23). Currently, an increasing number of outbreaks and sporadic cases are caused by food-borne and waterborne NoVs (1, 10).
The primary transmission route of human NoVs is fecal-to-oral spread, but infectious vomitus also plays a role. In addition, NoV has the following characteristics that facilitate spread (7). (i) The first is an extremely low infectious dose of approximately 18 to 1,000 viral particles. This trait facilitates its spread through inconspicuous routes including droplets, fomites, person-to-person contact, and environmental contamination. The low infectious dose is reflected in the secondary attack rate of 30% or more for those in close contact with an infected individual. (ii) The second characteristic is a prolonged shedding period of NoVs. Thus, the risk of secondary spread increased, thus explaining the epidemiologic NoV spread from food handlers that have already recovered from symptoms and from family members without gastroenteritis. (iii) The third characteristic is strong stability. This characteristic supports the resistance of NoV in a wide range of temperatures, from freezing to 60°C, and in various environmental conditions, including recreational, drinking water, and a variety of food items such as raw oysters, fruits, and vegetables. (iv) The great diversity of NoV strains can bring lifelong, repeated infections due to the lack of complete cross-protection and long-term immunity. (v) Antigenic shift and recombination from frequent NoV genome mutations can endow new strains with the capacity to infect susceptible hosts.
Several cases of broad-range contamination by human enteric viruses have been reported in South Korean water resources (12). It can be implied, therefore, that there is a necessary for close monitoring of human NoV content in the environmental waters of South Korea.
Several small-scale studies have been launched previously to detect and monitor NoVs in river, waste, and sewage water, but limitations stand in that these studies covered only certain local areas (5, 11, 12).
This study is significant because it is the first large-scale research project focusing on the inspection of NoV contamination in groundwaters being utilized as major sources of water. Due to insufficient cell culture techniques in the detection of human NoVs, nested reverse transcription-PCR (RT-PCR) was used because of its higher sensitivity. In the present study, groundwater samples were collected nationwide from 300 sites in South Korea (specifically, in the capital, six metropolises, and nine provinces) in the summer and winter seasons, and the NoV contamination levels were monitored for the purpose of clarifying the relationship between water quality and microbial content.
MATERIALS AND METHODS
Water sample collection and processing.
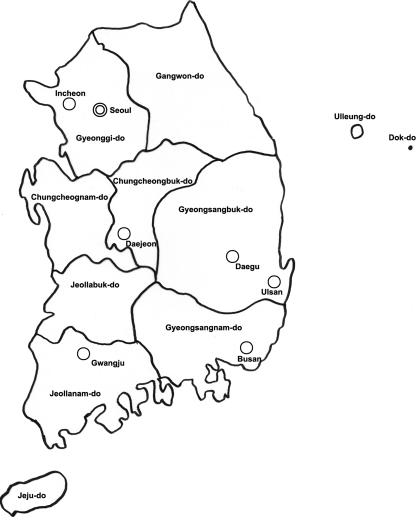
Groundwater samples from 300 sites were collected in the summer (June to August) and winter (October to December) of 2008 in seven metropolitan areas along with the capital, and nine provinces in South Korea (Fig. 1). The 300 sites were selected by the National Institute of Environmental Research from candidates recommended by government agencies (Korea Center for Disease Control and Prevention; Food and Drug Administration; Ministry of Education, Science, and Technology; and Ministry of National Defense) and by municipal governments, as well as sites included in the Groundwater Quality Network.
FIG. 1.
Sample collection sites in South Korea. Symbols: double circle, capital (Seoul); smaller circles, metropolises; divided areas, provinces.
Water sampling and concentration measurements were conducted by the National Institute of Environmental Research in Incheon, Republic of Korea, using standard procedures (6, 14). Samples of 570 to 1,400 liters were collected, depending on the turbidity of the water, which ranged from 2.3 to 5.5 nephelometric turbidity units. All samples were collected with a filter apparatus that contained a NanoCeram filter (Argonide Corp., Sanford, FL). The samples were eluted and further concentrated for NoV assays.
Water quality analysis and physical-chemical parameters.
The temperature and pH were measured with portable electrode-carrying devices (pH 300 series; Cyberscan, Eutech Instruments, Singapore). The nitrite nitrogen was measured with a pocket colorimeter (Hach Company, Loveland, CO). Turbidity was measured with a portable Turbidimeter 2100P (Hach).
Microorganism analysis.
The pour plate method was applied to count heterotrophic bacteria as described for standard methods (5). Briefly, 1-ml of the water sample was mixed with sterile, molten (44 to 46°C) plate count agar in a sterile petri dish (100-mm diameter). It was allowed to solidify at room temperature and then inverted and incubated at 35 ± 0.5°C for 48 ± 2 h. The resulting colonies were counted and expressed in CFU/milliliters. The total coliforms and Escherichia coli in the water were analyzed as the bacterial indicator. Enumeration of total coliforms and E. coli in water samples was performed with the Colilert test kit (Idexx Laboratories, Westbrook, ME) according to the manufacturer's instructions. The total somatic coliphage and male-specific coliphage contents were taken as viral indicators and measured according to the EPA method 1602 (18).
NoV and pan-enterovirus detection.
Water processed with the NanoCeram filter produced a final elute of 20 ml. Viral RNA was extracted from 140 μl of this elute with a QIAamp viral RNA minikit (Qiagen, Hilden, Germany) according to the manufacturer's protocol to obtain the final volume of 60 μl.
RT-PCR was conducted with a One-Step RT-PCR kit (Qiagen). To detect the NoV genotypes, two seminested RT-PCR amplifications were applied with previously described primer sets (NV-GIF1M/NV-GIR1M and SRI-1/SRI-3 for NoV GI; NV-GIIF1M/NV-GIIR1M and SRII-1/SRII-3 for NoV GII) (Table 1). We used 5 μl of viral RNA as the template and 20 μl of the premixed kit solution. The PCR was carried out in a PCR System Px2 thermal cycler (Thermo Hybaid, Middlesex, United Kingdom) according to the following protocol: one initial RT step at 50°C for 30 min, followed by PCR activation at 95°C for 15 min; followed by 35 cycles of amplification at 45 s at 94°C, 50 s at 58°C, and 45 s at 72°C; with a final extension step of 10 min at 72°C. Then, 2 μl of the products from this reaction was used for the templates in the nested PCR with 18 μl of HiPi PCR Premix (Elpis Biotech, South Korea) and the primer sets (NV-GIF2/NV-GIR1M and SRI-2/SRI-3 for NoV GI; NV-GIIF3M/NV-GIIR1M and SRII-2/SRII-3 for NoV GII). The nested PCR protocol was as follows: 94°C for 5 min; followed by 25 cycles of amplification at 30 s at 94°C, 30 s at 55°C, and 90 s at 72°C; with a final extension step of 10 min at 72°C. The PCR products were run on 1.5% agarose gels, stained with ethidium bromide, and visualized under UV light. The products were extracted from the agarose gel with a QiaQuick PCR purification kit (Qiagen), and the sequences were analyzed by Cosmogenetech (Seoul, South Korea).
TABLE 1.
Primers used for nested RT-PCR assays
| Genogroup, region, and size (bp) | Primer/polarity | Sequence (5′-3′) | Positiona |
|---|---|---|---|
| Genogroup I | |||
| Capsid (241) | SRI-1/reverse | CCAACCCARCCATTRTACAT | 5652-5671 |
| SRI-2/forward | AAATGATGATGGCGTCTA | 5356-5373 | |
| SRI-3/reverse | AAAAYRTCACCGGGKGTAT | 5578-5596 | |
| Capsid (313) | NV-GIF1M/forward | CTGCCCGAATTYGTAAATGATGAT | 5342-5365 |
| NV-GIF2/forward | ATGATGATGGCGTCTAAGGACGC | 5358-5380 | |
| NV-GIR1M/reverse | CCAACCCARCCATTRTACATYTG | 5649-5671 | |
| Genogroup II | |||
| RdRp (203) | SRII-1/reverse | CGCCATCTTCATTCACAAA | 5078-5096 |
| SRII-2/forward | TWCTCYTTYTATGGTGATGATGA | 4583-4605 | |
| SRII-3/reverse | TTWCCAAACCAACCWGCTG | 4767-4785 | |
| Capsid (310) | NV-GIIF1M/forward | GGGAGGGCGATCGCAATCT | 5049-5067 |
| NV-GIIF3M/forward | TTGTGAATGAAGATGGCGTCGART | 5079-5102 | |
| NV-GIIR1M/reverse | CCRCCIGCATRICCRTTRTACAT | 5367-5389 |
The primer sets and RT-PCR conditions described by Shieh et al. (17) were applied for the detection of pan-enterovirus.
Statistical analysis.
The correlations between the NoV detection rate and water quality (physical-chemical parameters), bacterial indicators, and viral indicators were analyzed with the chi-square test. A P value of <0.05 was considered significant. SPSS software (SPSS, Inc., Chicago, IL) was used for all analyses.
Phylogenetic analyses.
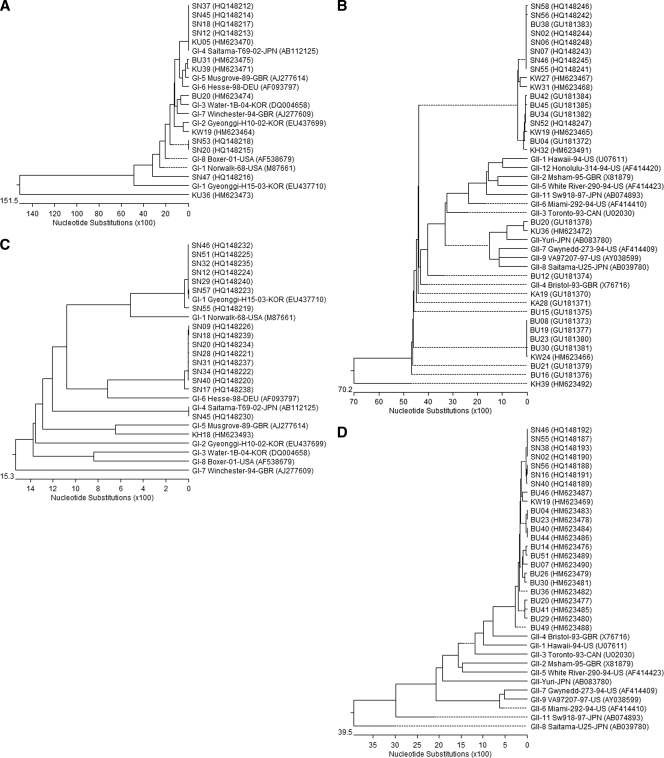
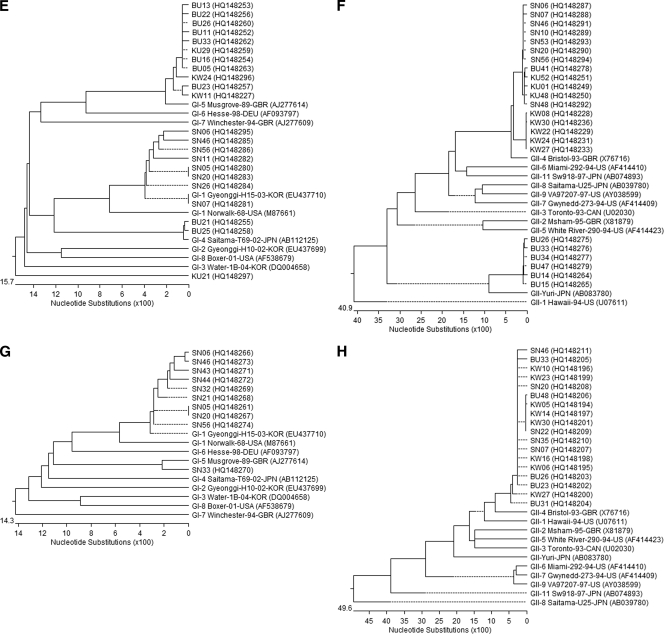
DNAStar version 5.07 software package was applied for the phylogenetic analyses. The CLUSTAL W method and the neighbor-joining method were used for the DNA sequence alignments and dendrograms (Fig. 2).
FIG. 2.
Phylogenetic tree constructed with the partial nucleotide sequences of capsid or RdRp gene of NoV isolates of summer (A to D) and winter seasons (E to H). The data were analyzed separately depending on the primer set used for the detection (see Table 1). Panels: A and E, GI; B and F, GII; C and G, SRI; D and H, SRII.
Nucleotide sequence accession numbers.
The nucleotide sequences of the environmental NoV isolates were submitted to GenBank database under the following accession numbers: GU181370 to GU181385, HM623464 to HM623493, and HQ148187 to HQ148297.
RESULTS
Location and seasonal pattern of NoV detection.
In the present study, NoVs were detected in 65 of the total 300 sites in summer (21.7%), and 52 of the total 300 sites in winter (17.3%); in all, NoVs were detected in 117 of the 600 total sites (19.5%). This nationwide inspection showed different regional NoV detection rates. In particular, the summer rates were distinctly high (>30%) in several regions, including Seoul, Gyeongsangnam-do, Ulsan, Gyeonggi-do, and Gyeongsangbuk-do. Two metropolises, Seoul and Deajeon, showed high NoV-positive rates in winter as well (>30%).
Two metropolises, Daegu and Gwangju, had no NoV-positive sites in summer or winter. In addition, the southwestern provinces Jeollanam-do, Jeollabuk-do, Chungcheongnam-do, and Chungcheongbuk-do showed relatively low virus detection rates compared to the southeastern provinces (Gyeongsangnam-do and Gyeongsangbuk-do). Three regions—Gyeongsangnam-do, Jeollabuk-do, and Daejeon—showed considerable changes in the NoV detection rates between summer and winter (69.2 to 23.1%, 15.0 to 40.0%, and 0 to 33.3%, respectively) (Table 2).
TABLE 2.
Detection rates and genotypes of NoV in South Korea in 2008
| Region | No. of samples tested per season | No. of NoV-positive detections (%) |
NoV genotype(s) |
|||
|---|---|---|---|---|---|---|
| Summer | Winter | Totala | Summer | Winter | ||
| Gangwon-do | 32 | 2 (6.3) | 0 (0) | 2 (3.1) | GII-4 | |
| Gyeonggi-do | 38 | 16 (42.1) | 9 (23.7) | 25 (32.9) | GI-1, -4, -6, -8; GII-4 | GI-1, 5; GII-4 |
| Gyeongsangnam-do | 13 | 9 (69.2) | 3 (23.1) | 12 (46.2) | GII-4 | GI-5; GII-Yuri |
| Gyeongsangbuk-do | 37 | 12 (32.4) | 11 (29.7) | 23 (31.1) | GI-3, -4, -5; GII-4, -Yuri | GI-4, -5; GII-4, -Yuri |
| Jeollanam-do | 20 | 1 (5.0) | 0 (0) | 1 (2.5) | GII-4 | |
| Jeollabuk-do | 20 | 3 (15.0) | 8 (40.0) | 11 (27.5) | GI-2; GII-4 | GI-5; GII-4 |
| Chungcheongnam-do | 15 | 2 (13.3) | 0 (0) | 2 (6.7) | GI-5; GII-4 | |
| Chungcheongbuk-do | 23 | 2 (8.7) | 2 (8.7) | 4 (8.7) | GI-5, -8; GII-Yuri | GII-4 |
| Jeju-do | 4 | 1 (25.0) | 1 (25.0) | 2 (25.0) | GII-4 | GII-4 |
| Seoul | 7 | 5 (71.4) | 4 (57.1) | 9 (64.3) | GI-1, -4,- 6; GII-4 | GI-1; GII-4 |
| Busan | 15 | 4 (26.7) | 3 (20.0) | 7 (23.3) | GII-4 | GI-5; GII-4 |
| Incheon | 36 | 6 (16.7) | 7 (19.4) | 13 (18.1) | GI-1, -4, -8; GII-4 | GI-1, -3, -5; GII-4 |
| Daejeon | 9 | 0 (0) | 3 (33.3) | 3 (16.6) | GII-4 | |
| Daegu | 15 | 0 (0) | 0 (0) | 0 (0) | ||
| Gwangju | 12 | 0 (0) | 0 (0) | 0 (0) | ||
| Ulsan | 4 | 2 (50.0) | 1 (25.0) | 3 (37.5) | GII-4 | GII-Yuri |
| Nationwide | 300 | 65 (21.7) | 52 (17.3) | 117 (19.5) | ||
The total virus is the sum of those detected in both summer and winter. The total percentage was calculated as follows: total virus detected/total samples tested.
The highest variety of NoV genotypes was found in the area within and surrounding the capital—including Seoul, Gyeonggi-do, and Incheon—which contains nearly half the South Korean population. A positive rate of at least 30% of various NoV genotypes was detected in each of these areas. Another vast area, the Southeastern provinces (Gyeongsangnam-do and Gyeongsangbuk-do) and Busan, along with the area around the capital city (Seoul, Incheon, and Gyeonggi-do), accounted for a large portion of the NoV detection rate, at 76.1% of the total detected NoV (89 positive of 117 total NoV-positive samples) (Table 2).
A total of 18 sites (KW24, KW27, BU14, BU15, BU16, BU21, BU23, BU26, BU31, BU34, BU41, SN06, SN07, SN20, SN32, SN46, SN53, and SN56) located in Jeollabuk-do, Gyeongsangnam-do, Gyeongsangbuk-do, Gyeonggi-do, Ulsan, Incheon, and Seoul showed positive results in both summer and winter (Table 3).
TABLE 3.
Water quality (physical-chemical parameters) and microorganism analyses data of NoV detected samples
| Season and sample no. | Sampling date (mo/day), codea | Sampling site/setting | Drinking or nondrinkingb | pH | Temp (°C) | Turbidity (NTU) | Residual chlorine (ppm) | Nitrite nitrogen (ppm) | Bacteria (CFU/ml) | Total coliforms/ 100 ml | E. coli/100 ml | Somatic phage (PFU/ 100 ml) | Male-specific phage (PFU/ 100 ml) | Pan- enterovirus | NoV genotype(s) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Summer | |||||||||||||||
| 1 | 7/16, KA19 | Gangwon-do/elementary school | Nd | 6.2 | 18.8 | 0.24 | 0 | 4.42 | 0 | Pc | Nd | 0 | 0 | N | II-4 |
| 2 | 7/28, KA28 | Gangwon-do/resort | Dr | 5.35 | 18.7 | 0.44 | 0 | 1.74 | 0 | P | P | 0 | 0 | N | II-4 |
| 3 | 6/30, KU05 | Gyeongsangbuk-do/village | Dr | 8.23 | 20.4 | 0 | 0 | 1.08 | 4 | P | P | 100 | 0 | N | I-4 |
| 4 | 7/16, KU36 | Chungcheongbuk-do/troops | Dr | 6.67 | 20.1 | 0 | 0 | 2.45 | 203 | P | P | 8 | 2 | N | I-8, II-Yuri |
| 5 | 7/02, KU39 | Chungcheongbuk-do/youth hostel | Dr | 7.29 | 18.2 | 0 | 0 | 0.78 | 0 | P | N | 20 | 7 | P | I-5 |
| 6 | 8/11, KH18 | Chungcheongnam-do/youth hostel | Dr | 7.65 | 17.8 | 0.16 | 0.07 | 1.10 | 14 | P | P | 0 | 0 | N | I-5 |
| 7 | 8/01, KH32 | Jeollanam-do/house | Nd | 7.82 | 17.2 | 0.08 | 0.08 | 15.12 | 144 | P | N | 0 | 0 | N | II-4 |
| 8 | 6/24, KH39 | Chungcheongnam-do/golf course | Dr | 6.65 | 17.0 | 0 | 0.11 | 5.04 | 28 | P | P | 0 | 0 | N | II-4 |
| 9 | 8/07, KW19 | Jeollabuk-do/youth hostel | Nd | 6.50 | 22.1 | 0.22 | 0 | 29.10 | 81 | P | P | 0 | 0 | P | I-2, II-4 |
| 10 | 7/31, KW24 | Jeollabuk-do/middle school | Nd | 6.50 | 19.8 | 0.35 | 0 | 1.46 | 0 | N | N | 0 | 0 | N | II-4 |
| 11 | 8/01, KW27 | Jeollabuk-do/house | Nd | 6.10 | 18.3 | 0.64 | 0 | 0 | 2 | P | N | 0 | 0 | N | II-4 |
| 12 | 8/26, KW31 | Jeju-do/golf course | Nd | 7.77 | 15.6 | 0.51 | 0 | 0.10 | 0 | N | N | 0 | 0 | N | II-4 |
| 13 | 7/01, BU04 | Busan/high school | Dr | 6.70 | 17.0 | 0.17 | 0 | 3.70 | 4 | N | N | 0 | 0 | N | II-4 |
| 14 | 7/02, BU07 | Busan/high school | Nd | 6.67 | 18.3 | 0.16 | 0 | 7.00 | 10 | N | N | 0 | 0 | P | II-4 |
| 15 | 7/02, BU08 | Busan/apartment | Dr | 6.91 | 19.9 | 0.94 | 0 | 3.60 | 14 | P | P | 0 | 0 | N | II-4 |
| 16 | 7/04, BU12 | Gyeongsangbuk-do/water supply facilities | Dr | 6.80 | 18.2 | 0.26 | 0 | 2.60 | 2 | N | N | 0 | 0 | N | II-4 |
| 17 | 7/04, BU14 | Gyeongsangbuk-do/office | Dr | 7.20 | 14.0 | 1.56 | 0 | 0 | 210 | P | N | 0 | 0 | N | II-4 |
| 18 | 7/08, BU15 | Gyeongsangnam-do/special school | Dr | 7.50 | 25.0 | 0.12 | 0 | 0.40 | 3 | N | N | 0 | 0 | N | II-4 |
| 19 | 7/08, BU16 | Gyeongsangnam-do/youth hostel | Nd | 7.80 | 23.8 | 0.39 | 0 | 0.10 | 9 | P | N | 0 | 0 | N | II-4 |
| 20 | 7/09, BU19 | Gyeongsangbuk-do/house | Nd | 7.50 | 22.0 | 0.32 | 0 | 1.50 | 40 | P | N | 1 | 0 | N | II-4 |
| 21 | 7/09, BU20 | Gyeongsangbuk-do/office | Nd | 6.50 | 24.0 | 0.33 | 0 | 0.10 | 2,700 | P | P | 1,000 | 27 | N | I-3, II-4, II-Yuri |
| 22 | 7/11, BU21 | Gyeongsangbuk-do/office | Nd | 7.40 | 24.2 | 0.35 | 0 | 0.10 | 8 | N | N | 0 | 0 | N | II-4 |
| 23 | 7/11, BU23 | Gyeongsangbuk-do/office | Dr | 7.14 | 26.7 | 0.18 | 0 | 0.60 | 50 | N | N | 0 | 0 | N | II-4 |
| 24 | 7/11, BU26 | Gyeongsangbuk-do/village | Dr | 7.35 | 29.1 | 0.68 | 0 | 0.30 | 43 | N | N | 0 | 0 | P | II-4 |
| 27 | 7/15, BU29 | Ulsan/public bath | Nd | 6.90 | 25.0 | 0.25 | 0 | 2.20 | 166 | P | N | 0 | 0 | N | II-4 |
| 25 | 7/14, BU30 | Gyeongsangbuk-do/youth hostel | Dr | 8.91 | 25.6 | 0.16 | 0 | 0.40 | 2 | N | N | 0 | 0 | N | II-4 |
| 26 | 7/14, BU31 | Gyeongsangbuk-do/youth hostel | Nd | 8.54 | 25.5 | 0.21 | 0 | 0.10 | 4 | P | N | 0 | 0 | N | I-5 |
| 28 | 7/15, BU34 | Ulsan/apartment | Dr | 6.10 | 20.0 | 0.64 | 0 | 7.40 | 4 | N | N | 0 | 0 | N | II-4 |
| 29 | 7/16, BU36 | Gyeongsangnam-do/middle school | Dr | 6.43 | 23.3 | 0.10 | 0 | 7.50 | 2 | N | N | 0 | 0 | N | II-4 |
| 30 | 7/16, BU38 | Gyeongsangnam-do/middle school | Nd | 6.79 | 22.4 | 0.08 | 0 | 0.4 | 3 | N | N | 0 | 0 | N | II-4 |
| 31 | 7/17, BU40 | Gyeongsangbuk-do/high school | Nd | 6.75 | 18.0 | 0.20 | 0 | 1.4 | 2 | N | N | 0 | 0 | N | II-4 |
| 32 | 7/17, BU41 | Gyeongsangbuk-do/museum | Dr | 6.25 | 28.3 | 0.31 | 0 | 1.60 | 14 | P | P | 1 | 0 | N | II-4 |
| 33 | 7/17, BU42 | Gyeongsangnam-do/youth hostel | Dr | 7.38 | 17.3 | 0.46 | 0 | 0.30 | 88 | N | N | 0 | 0 | N | II-4 |
| 34 | 7/21, BU44 | Gyeongsangnam-do/high school | Nd | 6.40 | 21.4 | 2.87 | 0 | 6.20 | 131 | P | P | 0 | 0 | N | II-4 |
| 35 | 7/17, BU45 | Gyeongsangnam-do/youth hostel | Dr | 7.60 | 25.0 | 0.11 | 0 | 1.40 | 26 | N | N | 0 | 0 | N | II-4 |
| 36 | 7/17, BU46 | Gyeongsangnam-do/youth hostel | Dr | 6.20 | 24.5 | 0.10 | 0 | 3.40 | 12 | P | N | 0 | 0 | N | II-4 |
| 37 | 7/23, BU49 | Busan/apartment | Dr | 6.4 | 21.0 | 0.21 | 0 | 4.20 | 17 | P | N | 0 | 0 | N | II-4 |
| 38 | 7/23, BU51 | Gyeongsangnam-do/high school | Dr | 7.01 | 23.0 | 0.15 | 0 | 0 | 7 | P | N | 0 | 0 | N | II-4 |
| 39 | 7/02, SN02 | Incheon/high school | Dr | 7.30 | 17.9 | 0.17 | 0 | 0.50 | 0 | N | N | 0 | 0 | N | II-4 |
| 40 | 7/31, SN06 | Incheon/youth hostel | Dr | 6.10 | 20.0 | 1.47 | 0 | 1.37 | 7 | P | N | 0 | Te | N | II-4 |
| 41 | 7/02, SN07 | Incheon/elementary school | Dr | 6.19 | 19.8 | 0.06 | 0 | 0.96 | 0 | P | N | 0 | 0 | N | II-4 |
| 42 | 7/10, SN09 | Incheon/elementary school | Dr | 6.93 | 20.2 | 0.23 | 0 | 5.03 | 0 | P | N | 0 | 0 | N | I-6 |
| 43 | 7/03, SN12 | Incheon/youth hostel | Dr | 5.89 | 16.0 | 0.18 | 0 | 6.50 | 17 | P | P | 3 | 0 | N | I-1, -4 |
| 44 | 7/08, SN16 | Gyeonggi-do/gas station | Nd | 7.63 | 19.9 | 0.09 | 0 | 13.08 | 9 | N | N | 0 | 0 | N | II-4 |
| 45 | 8/04, SN17 | Gyeonggi-do/factory | Nd | 6.65 | 19.5 | 0.45 | 0 | 8.93 | 16 | P | N | 0 | 0 | N | I-6 |
| 46 | 8/04, SN18 | Gyeonggi-do/house | Nd | 6.61 | 17.5 | 1.13 | 0 | 2.15 | 34 | P | P | 0 | 1 | N | I-4, -6 |
| 47 | 7/23, SN20 | Gyeonggi-do/spring | Dr | 6.18 | 19.6 | 0.11 | 0 | 4.13 | 1 | P | N | 0 | 0 | N | I-6, -8 |
| 48 | 6/23, SN28 | Gyeonggi-do/restaurant | Dr | 6.53 | 17.7 | 0.12 | 0 | 5.03 | 114 | P | P | 0 | 12 | N | I-6 |
| 49 | 8/05, SN29 | Gyeonggi-do/museum | Dr | 6.79 | 23.2 | 1.83 | 0 | 0.05 | 273 | N | N | 474 | 0 | N | I-1 |
| 50 | 8/04, SN31 | Gyeonggi-do/youth hostel | Dr | 6.26 | 18.4 | 0.17 | 0 | 4.46 | 17 | N | N | 0 | 0 | N | I-6 |
| 51 | 7/31, SN32 | Gyeonggi-do/troops | Dr | 6.54 | 17.9 | 1.79 | 0 | 0.02 | 127 | P | P | 0 | T | N | I-1 |
| 52 | 6/26, SN34 | Gyeonggi-do/spring | Dr | 7.09 | 13.7 | 0.09 | 0 | 2.99 | 1,090 | P | N | 0 | 0 | N | I-6 |
| 53 | 6/19, SN37 | Seoul/building | Nd | 8.20 | 18.3 | 0.13 | 0 | 1.02 | 14 | P | P | 0 | 0 | P | I-4 |
| 54 | 7/28, SN38 | Seoul/building | Nd | 8.46 | 29.6 | 0.41 | 0 | 0 | 270 | P | N | 0 | 0 | N | II-4 |
| 55 | 6/17, SN40 | Seoul/building | Nd | 7.49 | 22.3 | 0.71 | 0 | 11.76 | 45 | N | N | 0 | 0 | N | I-6, II-4 |
| 56 | 7/15, SN45 | Gyeonggi-do/troops | Dr | 6.87 | 16.8 | 0.40 | 0 | 1.19 | 17 | P | P | 0 | 0 | P | I-4 |
| 57 | 7/22, SN46 | Gyeonggi-do/youth hostel | Dr | 6.79 | 18.5 | 0.34 | 0 | 2.50 | 17 | P | P | 0 | 0 | N | I-1, II-4 |
| 58 | 7/24, SN47 | Gyeonggi-do/youth hostel | Dr | 7.09 | 16.3 | 0.23 | 0 | 1.43 | 0 | N | N | 0 | 0 | N | I-1 |
| 59 | 7/09, SN51 | Gyeonggi-do/community center | Dr | 6.94 | 17.4 | 0.09 | 0 | 17.56 | 8 | P | N | 0 | 0 | N | I-1 |
| 60 | 7/30, SN52 | Gyeonggi-do/Troops | Dr | 6.67 | 17.0 | 0.14 | 0 | 2.61 | 12 | N | N | 6 | T | N | II-4 |
| 61 | 8/05, SN53 | Incheon/farm | Nd | 7.25 | 16.4 | 0.52 | 0 | 10.46 | 10 | N | N | 0 | 3 | N | I-8 |
| 62 | 6/11, SN55 | Seoul/church | Nd | 7.02 | 21.3 | 0.66 | 0 | 14.17 | 5 | P | N | 0 | 0 | N | I-1, II-4 |
| 63 | 6/11, SN56 | Seoul/bus terminal | Nd | 6.92 | 19.3 | 0.16 | 0 | 12.81 | 4 | P | N | 0 | 0 | N | II-4 |
| 64 | 6/26, SN57 | Gyeonggi-do/elementary school | Dr | 6.81 | 15.5 | 0.24 | 0 | 7.00 | 280 | N | N | 0 | 0 | N | I-1 |
| 65 | 7/28, SN58 | Gyeonggi-do/troops | Dr | 6.62 | 16.1 | 0.06 | 0 | 13.04 | 10 | N | N | 0 | 0 | N | II-4 |
| Winter | |||||||||||||||
| 1 | 11/03, KU01 | Gyeongsangbuk-do/village | Nd | 7.66 | 16.6 | 1.10 | 0 | 0.01 | 47 | P | N | 0 | 0 | N | II-4 |
| 2 | 12/01, KU21 | Incheon/office | Nd | 7.19 | 15.8 | 2.45 | 0.06 | 0.97 | 1,560 | P | P | 100 | 0 | N | I-3 |
| 3 | 11/25, KU29 | Incheon/elementary school | Dr | 7.91 | 13.0 | 0 | 0 | 2.33 | 2 | N | N | 20 | 7 | P | I-5 |
| 4 | 11/12, KU48 | Chungcheongbuk-do/golf course | Dr | 7.84 | 16.4 | 0 | 0 | 0.22 | 42 | N | P | 8 | 2 | N | II-4 |
| 5 | 11/18, KU52 | Chungcheongbuk-do/park | Dr | 7.69 | 11.7 | 0 | 0 | 0 | 0 | N | P | 0 | 0 | N | II-4 |
| 6 | 11/17, KW05 | Daejeon/park | Dr | 6.07 | 15.7 | 0.13 | 0 | 6.51 | 6 | N | N | 0 | 0 | N | II-4 |
| 7 | 11/13, KW06 | Daejeon/middle school | Nd | 7.31 | 15.6 | 0.12 | 0.09 | 2.85 | 0 | N | P | 0 | 0 | P | II-4 |
| 8 | 11/17, KW08 | Daejeon/high school | Nd | 6.69 | 16.7 | 0.30 | 0 | 6.75 | 28 | P | P | 0 | 12 | N | II-4 |
| 9 | 11/27, KW10 | Jeollabuk-do/village | Dr | 7.83 | 13.3 | 0.42 | 0 | 0.51 | 114 | P | N | 0 | 0 | N | II-4 |
| 10 | 11/27, KW11 | Jeollabuk-do/village | Dr | 7.39 | 13.9 | 0.11 | 0 | 0.46 | 350 | P | N | 0 | 0 | N | I-5 |
| 11 | 11/18, KW14 | Jeollabuk-do/high school | Nd | 6.48 | 14.8 | 0.22 | 0 | 6.01 | 46 | P | N | 0 | 0 | N | II-4 |
| 12 | 11/21, KW16 | Jeollabuk-do/house | Nd | 6.31 | 10.9 | 0.10 | 0 | 4.13 | 8 | P | N | 0 | 0 | N | II-4 |
| 13 | 11/14, KW22 | Jeollabuk-do/park | Nd | 7.00 | 13.2 | 0.11 | 0 | 0.54 | 21 | P | P | 3 | 0 | N | II-4 |
| 14 | 11/11, KW23 | Jeollabuk-do/high school | Dr | 7.68 | 19.2 | 0.11 | 0 | 4.55 | 17 | N | N | 0 | 0 | N | II-4 |
| 15 | 11/12, KW24 | Jeollabuk-do/middle school | Nd | 6.39 | 16.6 | 0.10 | 0 | 8.69 | 41 | P | N | 0 | 0 | N | I-5, II-4 |
| 16 | 11/12, KW27 | Jeollabuk-do/house | Nd | 6.24 | 16.1 | 0.26 | 0 | 5.31 | 19 | N | N | 0 | 0 | N | II-4 |
| 17 | 11/26, KW30 | Jeju-do/golf course | Nd | 7.55 | 12.4 | 0.14 | 0 | 0.29 | 6 | N | P | 0 | 0 | P | II-4 |
| 18 | 11/07, BU05 | Busan/village | Dr | 6.48 | 22.8 | 0.25 | 0 | 0.79 | 0 | P | N | 0 | 0 | N | I-5 |
| 19 | 11/10, BU11 | Busan/village | Nd | 6.48 | 21.7 | 0.86 | 0 | 9.93 | 0 | P | P | 0 | 0 | N | I-5 |
| 20 | 11/11, BU13 | Gyeongsangbuk-do/village | Nd | 6.8 | 21.7 | 23.50 | 0 | 1.18 | 3 | P | N | 0 | 0 | N | I-5 |
| 21 | 11/11, BU14 | Gyeongsangbuk-do/village | Nd | 6.8 | 19.1 | 0.61 | 0 | 0.05 | 322 | P | N | 0 | 0 | N | II-Yuri |
| 22 | 11/12, BU15 | Gyeongsangnam-do/village | Dr | 6.87 | 18.9 | 0.27 | 0 | 3.25 | 0 | N | P | 0 | 0 | P | II-Yuri |
| 23 | 11/12, BU16 | Gyeongsangnam-do/village | Nd | 6.85 | 15.5 | 0.75 | 0 | 3.23 | 7 | P | N | 0 | 0 | N | I-5 |
| 24 | 11/24, BU21 | Gyeongsangbuk-do/village | Nd | 6.75 | 18.9 | 0.11 | 0 | 0.60 | 0 | N | N | 0 | 0 | N | I-4 |
| 25 | 11/24, BU22 | Gyeongsangbuk-do/village | Nd | 7.80 | 19.2 | 1.67 | 0 | 3.39 | 0 | N | N | 0 | 0 | N | I-5 |
| 26 | 11/13, BU23 | Gyeongsangbuk-do/village | Nd | 6.70 | 13.5 | 0.17 | 0 | 0.51 | 2 | N | P | 0 | 0 | N | I-5, II-4 |
| 27 | 11/13, BU25 | Gyeongsangbuk-do/village | Dr | 6.70 | 17.0 | 0.32 | 0 | 1.09 | 7 | P | N | 0 | 0 | N | I-4 |
| 28 | 11/13, BU26 | Gyeongsangbuk-do/village | Dr | 7.20 | 14.0 | 0.38 | 0 | 0.78 | 0 | P | N | 0 | 0 | N | I-5, II-4, II-Yuri |
| 29 | 11/14, BU31 | Gyeongsangbuk-do/village | Dr | 8.49 | 24.4 | 0.14 | 0 | 0.05 | 44 | P | N | 0 | 0 | N | II-4 |
| 30 | 11/24, BU33 | Gyeongsangbuk-do/village | Nd | 6.56 | 19.2 | 0.09 | 0 | 1.79 | 0 | N | N | 0 | 0 | N | I-5, II-4, II-Yuri |
| 31 | 10/31, BU34 | Ulsan/village | Dr | 6.43 | 19.6 | 0.12 | 0 | 7.24 | 0 | N | N | 0 | 0 | N | II-Yuri |
| 32 | 11/18, BU41 | Gyeongsangbuk-do/village | Dr | 6.90 | 18.7 | 0.14 | 0 | 1.70 | 12 | P | N | 1 | 0 | N | II-4 |
| 33 | 11/05, BU47 | Gyeongsangnam-do/village | Nd | 6.97 | 22.0 | 0.43 | 0 | 0.53 | 0 | N | P | 1,000 | 27 | N | II-Yuri |
| 34 | 11/10, BU48 | Busan/village | Dr | 6.62 | 16.6 | 0.74 | 0 | 4.58 | 768 | P | N | 0 | 0 | N | II-4 |
| 35 | 11/11, SN05 | Incheon/elementary school | Dr | 6.95 | 15.0 | 0.3 | 0 | 7.39 | 61 | P | N | 0 | 0 | N | I-1 |
| 36 | 12/22, SN06 | Gyeonggi-do/troops | Dr | 6.45 | 12.9 | 0.18 | 0 | 0.82 | 0 | P | N | 0 | 0 | P | I-1, II-4 |
| 37 | 11/18, SN07 | Incheon/elementary school | Dr | 7.90 | 14.4 | 0.30 | 0 | 6.68 | 0 | N | N | 0 | 0 | N | I-1, II-4 |
| 38 | 12/02, SN10 | Incheon/troops | Dr | 6.56 | 13.7 | 0.10 | 0 | 5.03 | 6 | N | N | 0 | 0 | N | II-4 |
| 39 | 11/27, SN11 | Incheon/elementary school | Dr | 6.88 | 14.0 | 0.12 | 0.75 | 11.54 | 0 | N | N | 0 | 0 | N | I-1 |
| 40 | 11/06, SN20 | Gyeonggi-do/spring | Dr | 6.38 | 15.4 | 0.37 | 0 | 4.25 | 5 | P | N | 0 | 0 | N | I-1, II-4 |
| 41 | 12/08, SN21 | Gyeonggi-do/factory | Nd | 6.68 | 16.5 | 0.09 | 0 | 5.47 | 0 | P | N | 0 | 0 | N | I-1 |
| 42 | 12/16, SN22 | Gyeonggi-do/spring | Dr | 6.94 | 12.6 | 0.14 | 0 | 1.62 | 0 | N | N | 0 | 0 | N | II-4 |
| 43 | 12/10, SN26 | Gyeonggi-do/water supply facilities | Dr | 7.36 | 13.6 | 28.90 | 0 | 1.71 | 31 | P | N | 0 | 0 | N | I-1 |
| 44 | 12/15, SN32 | Gyeonggi-do/troops | Dr | 6.80 | 11.0 | 0.75 | 0 | 0.63 | 0 | P | P | 1 | 0 | N | I-1 |
| 45 | 12/12, SN33 | Gyeonggi-do/troops | Dr | 7.25 | 15.1 | 1.45 | 0 | 1.01 | 2 | P | N | 0 | 0 | N | I-5 |
| 46 | 12/04, SN35 | Gyeonggi-do/park | Dr | 6.63 | 13.3 | 0.75 | 0 | 0.84 | 0 | N | N | 0 | 0 | N | II-4 |
| 47 | 12/20, SN43 | Seoul/park | Dr | 6.63 | 12.3 | 0.20 | 0 | 5.24 | 0 | P | N | 0 | 0 | N | I-1 |
| 48 | 11/28, SN44 | Gyeonggi-do/park | Dr | 6.88 | 15.6 | 0.15 | 0 | 9.17 | 0 | N | P | 0 | 0 | N | I-1 |
| 49 | 12/20, SN46 | Seoul/park | Dr | 6.61 | 12.3 | 0.14 | 0 | 5.24 | 0 | P | N | 0 | 0 | N | I-1, II-4 |
| 50 | 12/20, SN48 | Seoul/park | Dr | 6.43 | 14.6 | 0.11 | 0 | 7.64 | 0 | P | N | 0 | 0 | N | II-4 |
| 51 | 12/02, SN53 | Incheon/farm | Nd | 7.24 | 13.3 | 0.21 | 0 | 10.74 | 1 | P | N | 0 | 0 | N | II-4 |
| 52 | 11/06, SN56 | Seoul/bus terminal | Nd | 7.01 | 20.2 | 0.16 | 0 | 10.89 | 12 | N | N | 0 | 0 | N | I-1, II-4 |
The sampling codes were defined as the combined designation for the processing institute and the sampling location.
Dr, drinking; Nd, nondrinking.
P, present.
N, not detected.
T, too numerous to count.
Genetic analyses of NoV positive samples.
The sequence analyses of the detected NoV revealed more varieties of NoV genotypes in summer (GI-1, -2, -3, -4, -5, -6, -8 and GII-4 and -Yuri) than in winter (GI-1, -3, -4, -5 and GII-4 and -Yuri). GII-4 was the most frequent genotype in the summer (44 detections, 58.7%) and winter (30 detections, 47.6%) (Table 2). Among the GI genotypes, the most common were GI-1 and -6 in summer (8 detections each) and GI-1 and -5 in winter (12 detections each). GI-1 was the most frequently detected genotype collectively (8 and 12 detections in the summer and winter, respectively). Interestingly, GI-5 and GII-Yuri were each detected only 3 and 2 times each in the summer but showed much higher frequencies (12 and 6 detections, respectively) in the winter. In contrast, genotypes GI-6 and -8 showed dramatic reductions of 8 and 3, respectively, to none. The mixture of multiple NoV genogroups and genotypes were detected in 9 of 65 (13.8%) sites in the summer and in 9 of 52 (17.3%) sites in the winter (Table 3).
Among the samples from 18 sites in which positivity was observed in both seasons, 10 site samples (KW24, KW27, BU23, BU26, BU41, SN06, SN07, SN32, SN46, and SN56) contained identical genotypes in either season (Table 3).
Sequence and phylogenetic analyses showed that the NV-GI primer sets (NV-GIF1M, -GIF2, and -GIR1M) (Table 1) detected GI-1, -2, -3, -4, -5, and -8 genotypes with 1, 1, 1, 5, 2, and 3 cases in the summer (total 13), and GI-1, -3, -4, and -5 with 8, 1, 2, and 11 cases in winter (total of 22), respectively. In addition, the SRI primer set (SRI-1, -2, and -3) detected GI-1, -4, -5, and -6 genotypes with 7, 1, 1, and 8 cases in the summer (total of 17) and GI-1 and -5 genotypes with 9 and 1 cases in the winter (total of 10), respectively. The NV-GII primer set for GII NoV (NV-GIIF1M, -GIIF3M, and -GIIR1M) detected the GII-4 and GII-Yuri genotypes with 30 and 2 cases in summer (total of 32) and 17 and 6 cases in winter (total of 23), respectively. Finally, the GII NoV primer set and the SRII primer set (SRII-1, -2, and -3) only showed positive results for the GII-4 genotypes, with 23 cases (summer) and 18 cases (winter).
Correlation of NoV with water quality and microorganism contents.
Water quality analysis data showed that the average pH, water temperature, nitrite nitrogen, and turbidity were 6.99, 19.46°C, 0.58 nephelometric turbidity units (NTU), and 6.744 ppm, respectively, for summer and 6.94, 14.97°C, 0.62 NTU, and 4.312 ppm, respectively, for winter. A statistical analysis showed that only two independent variables—water temperature and turbidity—were correlated with NoV detection rates. For this analysis, temperature data were divided into five ranges (≤9.99, 10 to 14.99, 15 to 19.99, 20 to 24.99, and ≥25), and matched NoV detection rates showed different frequencies of 0.0% (0/22), 17.8% (24/135), 18.5% (59/319), 24.3% (25/103), and 42.9% (9/21), respectively. Turbidity was divided into three ranges (0, 0.001 to 0.5, and ≥0.501) with NoV detection rates of 5.5% (7/127), 23.4% (82/350), and 21.1% (26/123), respectively. Among the combinations of variables, water temperature and turbidity were most significantly correlated with the NoV detection rate. However, other combinations of variables were also correlated with the NoV detection rate, including average pH and nitrite nitrogen with turbidity.
Microorganism analysis indicated that the detection rates of heterotrophic bacteria, total coliforms, E. coli, somatic coliphage, and male-specific coliphage were 15.67% (47/300), 62.00% (186/300), 19.33% (58/300), 10.67% (32/300), and 16.00% (48/300), respectively, in the summer and 9.67% (29/300), 44.33% (133/300), 6.00% (18/300), 5.67% (17/300), and 11.00% (33/300), respectively, in the winter. The statistical analyses showed that the NoV detection rate was correlated with only one single variable, male-specific coliphage; no correlations were found with any other single or combinations of microorganisms. The pan-enteroviruses were detected in six sites in the summer and five sites in the winter. No further analysis of enterovirus, including sequencing, classification, and correlation analysis, was performed due to the low numbers detected (Table 3).
DISCUSSION
During the water quality control process, it is important and thus necessary for public health to monitor waterborne pathogenic microorganisms. The use of contaminated water for drinking and food preparation may be the major cause of food-borne disease outbreaks. The World Health Organization reported that diarrheal disease accounted for an estimated 4.1% of the total disability-adjusted life year (DALY) global burden of disease. It is also responsible for 1.8 million deaths every year that primarily occur in developing countries and children. Deficiencies in water quality, sanitation, and hygiene were estimated to contribute to 88% of the worldwide disease burden (21).
Numerous waterborne diseases, including diarrhea, polio, cholera, hepatitis, and malaria, are known to be caused by protozoa, viruses, or bacteria. Among the waterborne diseases, one of the most serious is gastroenteritis caused by NoVs. NoV infections are associated annually with an estimated 64,000 hospitalizations and 900,000 clinical visits in industrialized countries and up to 200,000 child deaths in developing countries (16). In South Korea, NoV-related viral gastroenteritis has been a major public health issue, as shown by several outbreaks since the virus was first reported in 2005 (9, 10).
In the present study, water samples were collected from 300 groundwater sites located in nine provinces and seven metropolises for general coverage of the national South Korean boundaries (Fig. 1). Two inspections were performed—in the summer and winter of 2008—to analyze the seasonal pattern. This nationwide study made it possible to determine the regional differences in NoV contamination. We found more NoV contaminations in the groundwater of southwestern regions than in the groundwaters of southeastern regions. The capital, Seoul, and its neighboring areas, Incheon and Gyeonggi-do, showed a potentially high risk of outbreak due to their high NoV detection rates in their highly populated environment. Seoul, Gyeongsangnam-do, Ulsan, Gyeonggi-do, and Gyeongsangbuk-do showed relatively high annual NoV detection rates (>30%). Compared to these regions, Daegu, Gwangju, Jeollanam-do, Gangwon-do, Chungcheongnam-do, and Chungcheongbuk-do had much lower detection rates (<10%).
In addition to the nationwide analyses of virus distribution, the seasonal analyses showed distinct changes in detection rates from summer to winter in some regions (Gyeongsangnam-do [69.2 to 23.1%], Jeollabuk-do [15.0 to 40.0%], and Daejeon [0 to 33.3%]). This provides important information for groundwater quality management (Table 2).
Although NoV infection has been referred to as the “winter vomiting disease,” this study showed a higher NoV detection rate in summer (21.7%) than in winter (17.3%). A recent study reported the identification of a new GII-4 variant with an atypical NoV epidemic pattern (8), which implied that such seasonal changes in NoV contamination may be due to the appearance of new variants. However, a more reasonable explanation for the seasonal changes in groundwater NoV is the specific climate conditions of South Korea, characteristically with torrential rains in summer. The basis lies in that one of the main sources of viral contamination in groundwater being the wastewater that contains discharged virus; in summer, the torrential rainfall can cause the sewage to overflow, and some wastewater may infiltrate the groundwater (20). This environmental condition may explain why the seasonal pattern is different from that of the general pattern in acute gastroenteritis outbreak cases.
We found diverse NoV genotypes distributed nationwide and mixed multiple genotypes at the same sites. These results supported the hypothesis that the contamination of groundwater was caused by infiltration of virus-containing wastewater.
As stated above, NoV has several characteristics that facilitate its spread. Of these, antigenic shift and recombination can increase the frequency of outbreaks. We found that GII-4 was the most frequent seasonal, nationwide, and regional genotype, and other rare genotypes were detected as well. These findings are valuable data for monitoring the diversity of strains and potential variant strains that arise from antigenic shift and recombination of NoV.
We applied two primer sets to each of GI and GII genotype detection and found that each primer set showed different genotype detection patterns on phylogenetic analysis with sequence data. Consequently, the application of double primer sets proved to be meaningful in covering diverse and mixed multiple genotypes in environmental samples.
The water quality, microorganism content, and correlation analysis data are important for indexing total water purity and can be used for groundwater management. These data have value in setting criteria for classifying groundwater for various purposes.
The statistical study showed that several factors were related to the NoV detection rate. Water turbidity, temperature, and male-specific coliphage were shown to be critical for the determination of the potability of groundwater for both drinking and food preparation.
As one of the main water resources in South Korea, groundwater must be analyzed and monitored for the maintenance of public health. Previous studies that measured NoV in environmental samples in South Korea were limited to a restricted sampling area and comprised a small number of samples (12). Thus, the present study is the first nationwide inspection of groundwater microorganism content, quality, and pan-enterovirus contamination in South Korea focusing on NoV location, seasonal distribution, and genetic diversity.
Acknowledgments
This research was supported by the Basic Science Research Program (2009-0083937); the Waterborne Virus Bank through the National Research Foundation of Korea funded by the Ministry of Education, Science, and Technology; and the National Institute of Environment Research (Ministry of Environment).
Footnotes
Published ahead of print on 23 December 2010.
REFERENCES
- 1.Baert, L., et al. 2009. Reported foodborne outbreaks due to noroviruses in Belgium: the link between food and patient investigations in an international context. Epidemiol. Infect. 137:316-325. [DOI] [PubMed] [Google Scholar]
- 2.Bailey, M. S., et al. 2005. Gastroenteritis outbreak in British troops, Iraq. Emerg. Infect. Dis. 11:1625-1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Centers for Disease Control and Prevention. 2002. Outbreaks of gastroenteritis associated with noroviruses on cruise ships-United States, 2002. MMWR Morb. Mortal. Wkly. Rep. 51:1112-1115. [PubMed] [Google Scholar]
- 4.Dey, S. K., et al. 2007. Molecular and epidemiological trend of norovirus associated gastroenteritis in Dhaka City, Bangladesh. J. Clin. Virol. 13:908-911. [DOI] [PubMed] [Google Scholar]
- 5.Eaton, A. D., L. S. Clesceri, and A. E. Greenberg. 1995. Standard methods for the examination of water and wastewater, 19th ed. American Public Health Association, Washington, DC.
- 6.Fout, G. S., B. C. Martinson, M. Moyer, D. R. Dahling, and R. E. Stetler. 1996. ICR microbial laboratory manual. U.S. Environmental Protection Agency, Washington, DC.
- 7.Glass, R. I., U. D. Parashar, and M. K. Estes. 2009. Norovirus gastroenteritis. N. Engl. J. Med. 361:1776-1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ho, E. C., P. K. Cheng, A. W. Lau, A. H. Wong, and W. W. Lim. 2007. Atypical norovirus epidemic in Hong Kong during summer of 2006 caused by a new genogroup II/4 variant. J. Clin. Microbiol. 45:2205-2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huh, J. W., W. H. Kim, S. G. Moon, J. B. Lee, and Y. H. Lim. 2009. Viral etiology and incidence associated with acute gastroenteritis in a 5-year survey in Gyeonggi province, South Korea. J. Clin. Virol. 44:152-156. [DOI] [PubMed] [Google Scholar]
- 10.Kim, S. H., et al. 2005. Outbreaks of gastroenteritis that occurred during school excursions in Korea were associated with several waterborne strains of norovirus. J. Clin. Microbiol. 43:4836-4839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.La Rosa, G., et al. 2007. Molecular identification and genetic analysis of norovirus genogroups I and II in water environments: comparative analysis of different reverse transcription-PCR assays. Appl. Environ. Microbiol. 73:4152-4161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee, C. H., and S. J. Kim. 2008. The genetic diversity of human noroviruses detected in river water in Korea. Water Res. 42:4477-4484. [DOI] [PubMed] [Google Scholar]
- 13.Medici, M. C., et al. 2009. An outbreak of norovirus infection in an Italian residential-care facility for the elderly. Clin. Microbiol. Infect. 15:97-100. [DOI] [PubMed] [Google Scholar]
- 14.Parshionikar, S. U., et al. 2003. Waterborne outbreak of gastroenteritis associated with a norovirus. Appl. Environ. Microbiol. 69:5263-5268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Patel, M. M., A. J. Hall, J. Vinje, and U. D. Parashar. 2009. Noroviruses: a comprehensive review. J. Clin. Virol. 44:1-8. [DOI] [PubMed] [Google Scholar]
- 16.Patel, M. M., et al. 2008. Systematic literature review of role of NoVs in sporadic gastroenteritis. Emerg. Infect. Dis. 14:1224-1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shieh, Y. C., R. S. Baric, J. W. Woods, and K. R. Calci. 2003. Molecular surveillance of enterovirus and Norwalk-like virus in oysters relocated to a municipal-sewage-impacted gulf estuary. Appl. Environ. Microbiol. 69:7130-7136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.U.S. Environmental Protection Agency. 2001. Method 1602: male-specific (F+) and somatic coliphage in water by single agar layer (SAL) procedure. EPA 821-R-01-029. U.S. Environmental Protection Agency, Washington, DC.
- 19.Widdowson, M. A., et al. 2005. Norovirus and foodborne disease, United States, 1991-2000. Emerg. Infect. Dis. 11:95-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wilhelm, S. R., S. L. Schiff, and W. D. Robertson. 1994. Chemical fate and transport in a domestic septic system: unsaturated and saturated zone geochemistry. Environ. Toxicol. Chem. 13:193-203. [Google Scholar]
- 21.World Health Organization. 2004. Burden of disease and cost-effectiveness estimates. World Health Organization, Geneva, Switzerland.
- 22.Xerry, J., C. I. Gallimore, M. Iturriza-Gómara, and J. J. Gray. 2009. Tracking the transmission routes of genogroup II noroviruses in suspected food-borne or environmental outbreaks of gastroenteritis through sequence analysis of the P2 domain. J. Med. Virol. 81:1298-1304. [DOI] [PubMed] [Google Scholar]
- 23.Yoon, J. S., et al. 2008. Molecular epidemiology of norovirus infections in children with acute gastroenteritis in South Korea in November 2005 through November 2006. J. Clin. Microbiol. 46:1474-1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zheng, D. P., et al. 2006. Norovirus classification and proposed strain nomenclature. Virology 346:312-323. [DOI] [PubMed] [Google Scholar]