Abstract
A microfluidic device-based system for the rapid and semiautomated counting of bacteria in freshwater was fabricated and examined. Bacteria in groundwater and in potable water, as well as starved Escherichia coli O157:H7 spiked in pond water, were able to be on-chip stained and enumerated within 1 h using this system.
Assessing microbiological quality assurance by monitoring bacteria in various sources of freshwater used for human consumption, recreation, food preparation, and industries is necessary (14). Determination of total bacterial numbers in freshwater is a basis for the routine evaluation of microbiological water quality, and the change in number of harmful bacteria should be determined as quickly as possible if total bacterial numbers are increasing. Thus, “real-time” and “on-site” microbiological methods are required.
Culture-independent techniques using fluorescence microscopy have been widely applied to the detection of bacteria in aquatic environments because of their rapidity. However, these methods are often labor consuming, and cell concentration steps or large sample volumes are necessary to obtain statistically relevant cell numbers when freshwater samples contain low numbers of bacteria. These factors have created demands for a simple method of bacterial quantification in freshwater that does not require complex sample preparation. Flow cytometry is an effective alternative to fluorescence microscopy as a method of bacterial detection in aquatic samples (11), as the procedure is rapid and sensitive and reliably quantifies individual cells. However, flow cytometers are often rather complex and sometimes require skilled operators for their operation and maintenance. Therefore, a simpler and smaller system should be more useful for “on-site” counting of targeted microorganisms in aquatic environment.
The present study examines the use of microfluidic devices (microchips) developed during decades of progress in microfabrication technologies. Microchip-based analyses are faster, are performed on a smaller scale, and consume less sample and reagents than conventional approaches (2). Consequently, they have great potential in environmental microbiology (10). Furthermore, microfluidic devices reduce the biohazard risk because cells are analyzed in a closed system and the devices are immediately sterilized after use. Therefore, microfluidic devices in various forms have been used to miniaturize flow cytometers (on-chip flow cytometry [4, 5, 12, 16]). However, most of these microfluidic devices were developed to entrap or analyze the characteristics of targeted particles rather than to determine total numbers of particles by a simple procedure. Previously, we quantified bacterial cells in potable water (containing 104 to 105 cells/ml of bacteria) using a simplified microfluidic device (13). This system enables rapid counting of bacterial cells in potable water samples, but it requires prestaining of bacterial cells before analysis.
In this study, we investigated the ability of a newly designed microfluidic device to determine bacterial cells at a density of 104 to 106/ml without sample preparation, such as concentration or prestaining of bacterial cells. We then evaluated the ability of this new counting system (microfluidic-based system) to determine numbers of total bacteria or targeted bacterial species in freshwater samples by “on-chip staining and counting.”
Bacterial strain and fluorescence microscopy.
Escherichia coli O157:H7 ATCC 43888 cells were cultured overnight in Luria-Bertani (LB) liquid medium (10 g tryptone, 5 g yeast extract, 10 g NaCl in 1 liter of distilled water) at 37°C. The cells were harvested by centrifugation (5,000 × g, 5 min), washed twice with phosphate-buffered saline (PBS) (130 mM NaCl, 10 mM Na2HPO4, 10 mM NaH2PO4 [pH 7.2]), and fixed in 70% (wt/vol) ethanol at 4°C for 1 h. The fixed E. coli cells were resuspended in sterile deionized water. Then, a subsample of this bacterial suspension was stained with 1 μg/ml of 4,6-diamidiono-2-phenyl indole (DAPI; Nacalai Tesque, Kyoto, Japan) for 5 min at room temperature (approximately 25°C) in the dark. DAPI-stained E. coli cells were filtered onto a black polycarbonate membrane (pore size: 0.2 μm; Advantec Toyo, Tokyo, Japan) and counted at a magnification of ×1,000 under UV excitation (excitation, 330 to 380 nm; emission, 420 nm) using a fluorescence microscope (E-400; Nikon, Tokyo, Japan). Then, E. coli O157 cells (without fluorescent staining) were spiked into sterile deionized water within the range of 104 to 106 cells/ml and were used to determine the correlation between on-chip counts and conventional fluorescence microscopic counts. In addition to DAPI, fluorescein isothiocyanate (FITC)-labeled anti-E. coli O157:H7 antibody (Kirkegaard & Perry Laboratories, Gaithersburg, MD) (15) was used for the specific detection of E. coli O157:H7 cells. Samples were stained for 30 min at room temperature (approximately 25°C) by adding 1/3 volume of staining buffer (12% [wt/vol] bovine serum albumin [BSA] in PBS) and fluorescent antibody (final concentration, 4 μg/ml). Stained cells were filtered onto a black polycarbonate membrane and counted at a magnification of ×1,000 under blue excitation (excitation, 465 to 495 nm; emission, 515 to 555 nm) using a fluorescence microscope (E-400; Nikon).
Microfluidic device designed for “on-chip” staining and counting.
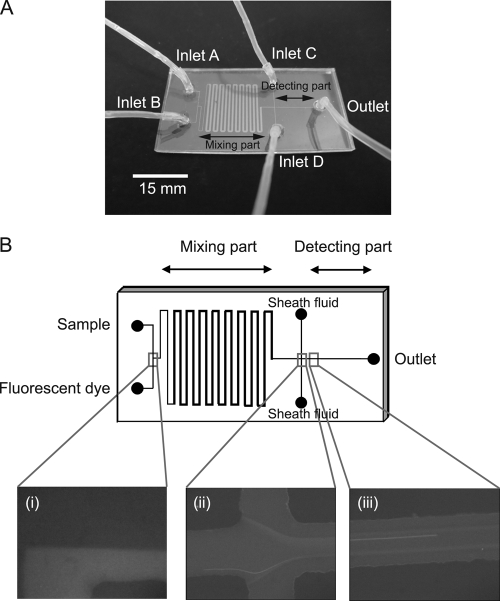
Polydimethylsiloxane (PDMS)-glass hybrid microfluidic devices were fabricated using rapid prototyping and replica molding techniques (13). The masks for channel patterns were printed on a transparent film. Ultrathick photoresist (SU-8-50; Microchem, Newton, MA) was spin-coated onto a silicon wafer and baked on a hot plate at 65°C for 2 min and 95°C for 3 min. The pattern on the mask was photolithographically transferred to the SU-8-coated silicon wafer using a mask aligner (M-1S; Mikasa, Tokyo, Japan). After development in SU-8 developer (Microchem) for 3 min, the master was washed in isopropyl alcohol and distilled water. The prepolymer of PDMS and the curing agent (Silpot 184; Toray Dow Corning, Tokyo, Japan) were mixed at a ratio of 10:1, stirred thoroughly, and then degassed under vacuum. The prepolymer mixture was poured onto the master and cured at 120°C for 40 min. After being cured, the PDMS replica was peeled off from the master. Access ports were drilled into the chip by a paper punch (diameter, 2 mm). The PDMS replica was attached to a glass substrate using a plasma reactor (SEDE/V; Meiwa Fosis, Osaka, Japan). Figure 1A shows a microfluidic device fabricated in this study (size, 50 mm by 25 mm). Samples were injected at inlet A, while fluorescent dye (2 μg/ml of DAPI or 20 μg/ml of FITC-labeled anti-E. coli O157:H7 antibody with 6% [wt/vol] bovine serum albumin) was injected at inlet B. Bacterial cells in the sample were fluorescently stained during flow through the “mixing part” (Fig. 1A). Inlets C and D were used to inject sheath fluid (PBS) for alignment of flowing bacterial cells, and fluorescently stained cells were detected in the “detecting part” (Fig. 1A). The depth of the microchannel was 20 μm, and the width was 100 μm except for the “mixing part” (500-μm width).
FIG. 1.
Detail of the microfluidic device for on-chip staining and counting. (A) Design of microfluidic device. Long and short lengths are 50 mm and 25 mm, respectively. Width of the channel is 100 μm except for the “mixing part” (500 μm). Depth of the channel is 20 μm. Samples and fluorescent dye solution were injected at inlets A and B, respectively. Sheath fluid was injected at inlets C and D. (B) On-chip staining and counting of bacterial cells. (i) Samples and fluorescent dye solution flow separately and are mixed through the “mixing part” of the microchannel. (ii) Alignment of sample flow by sheath fluid. (iii) Flow of bacterial cells in the “detecting part” of the microchannel.
We used a specified system to count bacterial cells flowing in the microchannel (13). Each sample and the fluorescent dye were placed in a 100-μl gas-tight syringe (1710LT; Hamilton Company, NV) and injected into the microchannel of the device via a Teflon tube by a syringe pump (kdS100; KD Scientific, MA; flow rate, 0.1 μl/min). Fluid in the microchip was monitored through an objective lens (UPlanApo 40×; Olympus, Tokyo, Japan) under UV excitation (excitation, 330 to 385 nm; emission, 420 nm) for DAPI-stained cells or blue excitation (excitation, 470 to 490 nm; emission, 515 to 550 nm) for antibody-stained cells with a fluorescence microscope (AX70; Olympus). Flowing cells were recorded as a movie using a color charge-coupled device (CCD) camera (ICD-878; Ikegami Electronics, Tokyo, Japan) mounted on the microscope. The frame rate of the camera was established at 30 frames per second, and the shutter speed was 33 ms. The movies were captured over 10 to 45 min and separated into bitmap images. Cells that appeared in the definite area of these frames were counted by image analysis software programmed in our laboratory (17). Bacterial numbers were calculated from the cell count at each flow volume (flow rate × measurement time). These microfluidic counts were compared with the fluorescence microscopic counts.
First, we examined the possibility of on-chip staining. Samples and fluorescent dye were separately injected at each inlet, and they produced laminar flow (Fig. 1B part i). They were mixed enough during flow through the “mixing part” of the microchannel, and fluorescently stained cells were aligned by the sheath fluid (Fig. 1B part ii). Cells were detected and counted in the “detecting part” of the microchannel (Fig. 1B part iii). Without sheath flow, the flow speed of cells flowing near the wall of the microchannel was slower than that of cells flowing in the center of the stream, and this decreased the reproducibility of counts in each sample. We used the same flow rate between the sample flow and sheath flow because the highest efficiency was obtained using this condition.
Accuracy of on-chip flow cytometric counts.
To confirm the ability of our counting system to quantify low numbers of bacterial cells, E. coli cells were diluted in sterile deionized water within the range of 1 × 104 to 1 × 106/ml and counted by our system and fluorescence microscopy (n = 5 for each bacterial concentration). The measurement time for the microfluidic-based system was adjusted to 10 to 45 min for each dilution because on-chip counts plateaued within 15 min even when bacterial numbers were low (104 cells/ml).
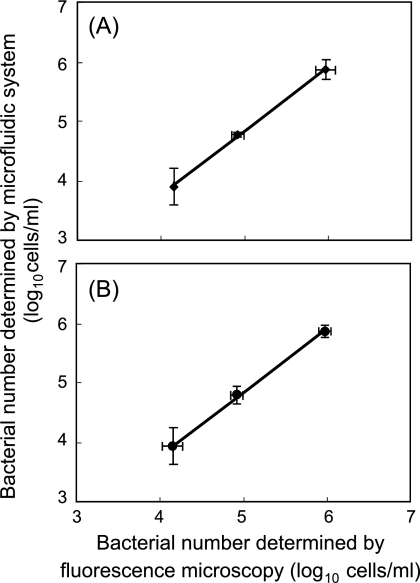
The counts of DAPI-stained cells obtained by our system and fluorescence microscopy closely correlated in the range of 1 × 104 to 1 × 106 cells/ml (Fig. 2A; r2 = 0.99). A similar correlation was obtained between on-chip counts and fluorescence microscopic counts when FITC-labeled anti-E. coli O157:H7 antibody (final concentration, 10 μg/ml) (19) was used (Fig. 2B; r2 = 0.98). That is, low numbers of bacterial cells (104 cells/ml in the sample) were rapidly determined in both on-chip DAPI staining and on-chip fluorescent antibody staining.
FIG. 2.
Correlation between microfluidic counts and conventional fluorescence microscopic counts of E. coli O157:H7 stained with DAPI (A) and FITC-labeled anti-E. coli O157:H7 antibody (B). Error bars indicate the standard deviation (n = 5).
Total direct count of bacterial cells in potable freshwater by on-chip flow cytometry.
While most potable water has been chlorinated for bacterial sterilization, natural mineral water and tap water filtered through a household water purifier have also been generally utilized for consumption. The numbers of bacterial cells in nonchlorinated water sometimes increase during storage (9). We applied our microfluidic-based system to count bacterial cells in potable freshwater, such as tap water from a household water purifier or groundwater, after confirming that our system could count bacteria at a low abundance.
Tap water samples in a household water purifier were collected at Osaka University, Suita, Japan. Groundwater samples were collected at Jurinji Temple, Hyogo, Japan. Bacterial cells in these potable freshwater samples were counted using the microfluidic-based system or fluorescence microscopy (Table 1). To determine the total bacterial number by fluorescence microscopy, bacteria in these samples were fluorescently stained with DAPI, trapped onto a polycarbonate filter, and enumerated by fluorescence microscopy as previously described. The ratio of the counts determined by our system to that by microscopy was 73% in the purified household tap water and 80% in the groundwater. The microfluidic-based system developed in this study has detected bacterial cells in potable freshwater to levels of 80% of the total cell numbers within 1 h, and this system enables detection of significant increases of bacterial number in freshwater samples rapidly, which is essential for the routine evaluation of microbiological water quality.
TABLE 1.
Bacterial numbers in potable water determined by microfluidic-based system for counting and fluorescence microscopy
| Methoda | Total bacterial no. (cells/ml)c in sample |
||
|---|---|---|---|
| Tap waterb 1 | Tap water 2 | Groundwater | |
| Microfluidics | 1.1 (± 0.33) × 105 | 3.7 (± 1.1) × 105 | 2.4 (± 0.31) × 106 |
| EFM | 1.5 (± 0.23) × 105 | 5.1 (± 1.2) × 105 | 3.0 (± 0.42) × 106 |
Microfluidics, microfluidic-based system for counting; EFM, epifluorescence microscopy.
Purified household tap water.
Numbers in parentheses indicate standard deviations (n = 5).
Selective counting of targeted bacteria in freshwater by on-chip flow cytometry.
Rapid quantification of harmful bacteria in the aquatic environment is important to ensure its safety. We therefore examined the microfluidic-based system for selective detection of targeted cells with on-chip fluorescent antibody staining. Distribution of enterohemorrhagic Escherichia coli O157 in the freshwater environment has been reported in several studies (7), and these strains sometimes cause hemorrhagic colitis (1, 6). Understanding the abundance of this pathogen in the aquatic environment may help to prevent outbreaks of related disease. Thus, starved E. coli O157:H7 cells (18) were spiked into pond water samples (collected at Osaka University) as approximately 0.2% of the total bacterial number, because E. coli O157 cells often exist at 0.2 to 0.3% of total bacterial numbers in eutrophic freshwater environments (7). The numbers of these spiked cells were determined by both the microfluidic-based system and fluorescence microscopy (Table 2). Total bacterial numbers (mean ± standard deviation) in pond water samples determined by fluorescence microscopy were as follows: pond water 1, 4.7 (± 0.72) × 106; pond water 2, 3.9 (± 0.32) × 106; pond water 3, 4.4 (± 0.71) × 106.
TABLE 2.
Selective detection of starved E. coli O157:H7 spiked into pond water by microfluidic-based system for counting and fluorescence microscopy
| Methoda | No. of E. coli O157:H7 cells/mlb in sample |
||
|---|---|---|---|
| Pond water 1 | Pond water 2 | Pond water 3 | |
| Microfluidics | 1.0 (± 0.11) × 104 | 1.4 (± 0.41) × 104 | 1.2 (± 0.42) × 104 |
| EFM | 1.1 (± 0.32) × 104 | 1.5 (± 0.23) × 104 | 1.3 (± 0.31) × 104 |
Microfluidics, microfluidic-based system for counting; EFM, epifluorescence microscopy.
Numbers in parentheses indicate standard deviations (n = 6).
On-chip counts were close to fluorescence microscopic counts (Table 2). The data suggest that the low number (0.2%) of targeted cells were selectively detected within 1 h by our newly fabricated system in the presence of high numbers and various species of indigenous bacteria.
In this study, we fabricated and examined the availability of a microfluidic-based cell-counting system for on-chip staining and counting. The system can count total bacterial cells and target bacterial species with the same microfluidic device by changing the fluorescent dye, and results can be obtained within 1 h.
For further studies counting bacteria at very low concentrations, our system probably can be integrated easily with a bacterial concentration system using one of the several forms of microfluidic devices which have been developed (3, 8).
Our system can be applied for “early warning” purposes in the microbial quality control of freshwater because of the following characteristics: (i) it is rapid (one hour to be completed); (ii) measurement is possible without complex pretreatment apparatus (filtration funnels, etc.); (iii) it is easy to use, as it uses a semiautomated system (on-chip staining and image analysis); and (iv) it could be developed as a portable system for on-site measurement by using a light-emitting diode (LED) light source. We therefore believe that this basic study using a microfluidic device will contribute to technical progress in developing systems for the quality control of freshwater.
Acknowledgments
This study was supported by the Ground-Based Research Program for Space Utilization promoted by the Japan Space Forum, by the Kurita Water and Environment Foundation, by the Kieikai Research Foundation, and by the JSPS Grant-in-Aid for Scientific Research (A) (grant no. 21256002).
Footnotes
Published ahead of print on 17 December 2010.
REFERENCES
- 1.Ackman, D., et al. 1997. Swimming-associated haemorrhagic colitis due to Escherichia coli O157:H7 infection: evidence of prolonged contamination of a fresh water lake. Epidemiol. Infect. 119:1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blankenstein, G., and U. D. Larsen. 1998. Modular concept of a laboratory on a chip for chemical and biochemical analysis. Biosens. Bioelectron. 13:427-438. [Google Scholar]
- 3.Cabrera, C. R., and P. Yager. 2001. Continuous concentration of bacteria in a microfluidic flow cell using electrokinetic techniques. Electrophoresis 22:355-362. [DOI] [PubMed] [Google Scholar]
- 4.Fu, A. Y., H. P. Chou, C. Spence, F. H. Arnold, and S. R. Quake. 2002. An integrated microfabricated cell sorter. Anal. Chem. 74:2451-2457. [DOI] [PubMed] [Google Scholar]
- 5.Gawad, S., L. Schild, and P. Renaud. 2001. Micromachined impedance spectroscopy flow cytometer for cell analysis and particle sizing. Lab Chip 1:76-82. [DOI] [PubMed] [Google Scholar]
- 6.Keene, W. E., et al. 1994. A swimming-associated outbreak of hemorrhagic colitis caused by Escherichia coli O157:H7 and Shigella sonnei. N. Engl. J. Med. 331:579-584. [DOI] [PubMed] [Google Scholar]
- 7.Kurokawa, K., K. Tani, M. Ogawa, and M. Nasu. 1999. Abundance and distribution of bacteria carrying sltII gene in natural river water. Lett. Appl. Microbiol. 28:405-410. [DOI] [PubMed] [Google Scholar]
- 8.Lapizco-Encinas, B. H., B. A. Simmons, E. B. Cummings, and Y. Fintschenko. 2004. Dielectrophoretic concentration and separation of live and dead bacteria in an array of insulators. Anal. Chem. 76:1571-1579. [DOI] [PubMed] [Google Scholar]
- 9.Leclerc, H., and A. Moreau. 2002. Microbiological safety of natural mineral water. FEMS Microbiol. Rev. 26:207-222. [DOI] [PubMed] [Google Scholar]
- 10.Liu, W. T., and L. Zhu. 2005. Environmental microbiology-on-a-chip and its future impacts. Trends Biotechnol. 23:174-179. [DOI] [PubMed] [Google Scholar]
- 11.Müller, S., and G. Nebe-von-Caron. 2010. Functional single-cell analyses: flow cytometry and cell sorting of microbial populations and communities. FEMS Microbiol. Rev. 34:554-587. [DOI] [PubMed] [Google Scholar]
- 12.Sakamoto, C., N. Yamaguchi, and M. Nasu. 2005. Rapid and simple quantification of bacterial cells by using a microfluidic device. Appl. Environ. Microbiol. 71:1117-1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sakamoto, C., et al. 2007. Rapid quantification of bacterial cells in potable water using a simplified microfluidic device. J. Microbiol. Methods 68:643-647. [DOI] [PubMed] [Google Scholar]
- 14.Sidorowicz, S. V., and T. N. Whitmore. 1995. Prospects for new techniques for rapid bacteriological monitoring of drinking water. J. Inst. Water Environ. Manag. 9:92-98. [Google Scholar]
- 15.Tanaka, Y., N. Yamaguchi, and M. Nasu. 2000. Viability of Escherichia coli O157:H7 in natural river water determined by the use of flow cytometry. J. Appl. Microbiol. 88:228-236. [DOI] [PubMed] [Google Scholar]
- 16.Wang, Z., et al. 2004. Measurements of scattered light on a microchip flow cytometer with integrated polymer based optical elements. Lab Chip 4:372-377. [DOI] [PubMed] [Google Scholar]
- 17.Yamaguchi, N., T. Ichijo, M. Ogawa, K. Tani, and M. Nasu. 2004. Multicolor excitation direct counting of bacteria by fluorescence microscopy with the automated digital image analysis software BACS II. Bioimages 12:1-7. [Google Scholar]
- 18.Yamaguchi, N., M. Sasada, and M. Nasu. 2009. Rapid detection of starved Escherichia coli with respiratory activity in potable water by signal-amplified in situ hybridization following formazan reduction. Microbes Environ. 24:286-290. [DOI] [PubMed] [Google Scholar]
- 19.Yamaguchi, N., M. Sasada, M. Yamanaka, and M. Nasu. 2003. Rapid detection of respiring Escherichia coli O157:H7 in apple juice, milk and ground beef by flow cytometry. Cytometry 54A:27-35. [DOI] [PubMed] [Google Scholar]