Abstract
Helicobacter pylori is considered one of the major risk factors underlying the development of gastritis and gastric and duodenal ulcers. Moreover, 50% of the population carries this bacterium, and consequently, when it is detected, eradication of H. pylori is strongly recommended. Regarding the use of probiotics as functional agents, several studies have shown that there is a direct relationship between the addition of certain probiotic bacteria and in vitro inhibition of H. pylori; however, in vivo studies showing bifidobacterial activity against H. pylori remain scarce. In this study, a Bifidobacterium bifidum strain which proved active in vitro against H. pylori has been isolated, with inhibition levels reaching 81.94% in the case of the supernatant and even 94.77% inhibition for supernatant purified by cationic exchange followed by an inverse phase. In vivo studies using a BALB/c mouse model have proved that this strain partially relieves damage to gastric tissues caused by the pathogen and also decreases the H. pylori pathogenicity ratio. This novel strain fulfills the main properties required of a probiotic (resistance to gastrointestinal juices, biliary salts, NaCl, and low pH; adhesion to intestinal mucus; and sensitivity to antibiotics). Furthermore, the absence of undesirable metabolites has been demonstrated, and its food safety status has been confirmed by acute ingestion studies in mice. In summary, the results presented here demonstrate that Bifidobacterium bifidum CECT 7366 can be considered a probiotic able to inhibit H. pylori both in vitro and in vivo.
Helicobacter pylori is a Gram-negative, spiral to pleomorphic, rod bacterium that is considered one of the major risk factors underlying the development of gastritis and gastric and duodenal ulcers (20), gastric cancer (15, 23), and gastric mucosa-associated lymphoid tissue lymphomas (41) and associated with gastric diffuse large B cell lymphomas (DLBCL) (52). In fact, in 1994 the International Agency for Research on Cancer declared H. pylori a class I carcinogen (24). The eradication of H. pylori is recognized to be a valuable treatment against the aforementioned diseases (5, 37, 48, 53).
H. pylori is typically acquired via the fecal-oral or gastrointestinal-oral route within families, normally during early childhood, and persists thereafter (44), although transmission mechanisms remain unclear (11, 12, 31, 32). H. pylori is able to colonize and persists in a unique biological niche within the gastric lumen, causing gastric inflammation, disruption of the gastric mucosal barrier, and alteration of gastric physiology (12). Regarding the prevalence of this bacterium, it is widely accepted that 50% of the population carries H. pylori and that developing countries have the highest prevalence rates, reaching up to 80% (47). Consequently, when it is detected, eradication of H. pylori is strongly recommended (20).
Currently, antibiotic-based treatment of H. pylori infection is neither sufficient nor satisfactory, with the most successful treatments reaching 75 to 90% eradication rates (51). Improved treatments are still needed, and therapies alternative or complementary to antibiotic therapy are being assessed. In this respect, the use of probiotics is a potentially promising tool to prevent H. pylori infections.
According to an expert consultation conducted by the Food and Agriculture Organization (FAO) and the World Health Organization (WHO), probiotics are “live microorganisms which when administered in adequate amounts confer a health benefit on the host” (18). The regular intake of probiotic microorganisms has been demonstrated to prevent several infectious, allergic disorders, diarrhea, and inflammatory diseases such as inflammatory bowel disease (36). Moreover, probiotics or their metabolites have been suggested to play an important role in the formation or establishment of a well-balanced, indigenous, intestinal microbiota in newborn children and adults (22, 45). Among probiotics, Bifidobacterium is one of the favorite genera in studies focused on the prevention of gastrointestinal infection and is often used in fermented dairy products or food supplements.
Several studies have demonstrated that a direct relationship exists between the addition of potential probiotic strains and the in vitro inhibition of H. pylori growth. Lactobacillus acidophilus (2, 3, 30), Lactobacillus casei strain Shirota (46), Bacillus subtilis (42), and even Weissella confusa (38) have rendered an antagonistic effect against the bacterium. Some in vitro studies showing bifidobacterial activity against H. pylori have been reported, with different levels of success (34, 35), but they remain scarce. Regarding the metabolites involved in growth inhibition, lactic acid and other organic acids are considered to play a major role (2, 35). However, other studies have pointed to the existence of other metabolites that may exert an antagonistic effect. Accordingly, in a study on L. acidophilus supernatant, Coconnier et al. (6) observed an in vitro and in vivo reduction in H. pylori viability and urease activity, independent of pH and lactic acid levels.
Bacteriocins with anti-H. pylori activity have chiefly been studied in probiotic strains of Bacillus subtilis (42), Enterococcus faecium (49), and Bifidobacterium (7); and other probiotic metabolites such as lysines have been proposed to have an antagonistic effect (30). Moreover, in vitro studies based on competition for cell-binding sites have demonstrated that there is a decrease in interleukin-8 secretion when lactobacilli are added to the medium (27, 50).
Positive results showing the inhibitory activity of probiotic bacteria in vitro have been ratified with preclinical studies, using germfree mice, for Lactobacillus salivarius (1), Lactobacillus gasseri OLL2716 (50), L. casei (46), and a mixture of L. acidophilus R0052 and Lactobacillus rhamnosus R0011 (26). Furthermore, in clinical trials with human volunteers, the incidence of gastrointestinal disorders declined after ingestion of lactobacilli, and in some cases, H. pylori levels decreased, although H. pylori infection was hardly ever fully eradicated (3). With respect to Bifidobacterium, Miki et al. (35) reported that ingestion of B. bifidum in fermented milk by H. pylori-positive subjects affected their urea breath test values, serum pepsinogen levels, and upper gastrointestinal symptoms. The aforementioned results indicate that even though positive results have been obtained with probiotics pertaining to the genus Lactobacillus, few studies have focused on Bifidobacterium activity against H. pylori. Furthermore, the mechanisms underlying the activity against this bacterium are strain dependent, and most of them remain unknown.
Hence, the aim of this study was to select a potential probiotic strain that hampers H. pylori growth in vitro and then go on to identify the nature of the metabolites responsible for this inhibitory activity. Furthermore, we report an in vivo demonstration of its capacity using a murine model as well as further toxicological assays to ensure that the selected strain is innocuous.
MATERIALS AND METHODS
Isolation of Bifidobacterium strains and growth conditions.
Feces of breast-fed infants (1 g) were dissolved in phosphate-buffered saline buffer (phosphate buffer, pH 7.0) and homogenized for 3 min in a stomacher (Bagmixer 400P; Interscience Worldwide, Saint Nom la Bretèche, France). After homogenization, 1 ml was incubated in commercial de Man, Rogosa, and Sharpe (MRS) broth medium (Oxoid, Basingstoke, United Kingdom) supplemented with 0.05% (wt/vol) cysteine (Sigma-Aldrich, St. Louis, MO) (MRS-C medium) and Bifidobacterium sp. medium (casein peptone, 10 g/liter; meat extract, 5 g/liter; yeast extract, 5 g/liter; glucose, 10 g/liter; K2HPO4, 3 g/liter; Tween 80, 1 ml/liter; filtered-sterilized sodium ascorbate, 10 g/liter; cysteine, 0.5 g/liter; pH 6.8) for 17 h at 37°C in an anaerobic atmosphere generated by means of an AnaeroGen system (Oxoid). Aliquot of 100 μl of these preincubated homogenates and dilutions were spread on plates of MRS-C broth medium and Bifidobacterium sp. medium, and the plates were incubated anaerobically at 37°C for 36 to 72 h. Thirty Gram-positive, catalase-negative colonies with typical bifidobacterial or lactobacillus cell morphology were stored at −80°C in glycerol for further analyses. Six Bifidobacterium sp. strains (numbered CP1 to CP6) were randomly selected for further assays of activity against Helicobacter pylori. The B. bifidum strain selected in this study was deposited at the Spanish Type Culture Collection (CECT; Burjassot, Spain) under accession number CECT 7366.
Bacterial strains and growth conditions.
Bifidobacterium isolates CP1 to CP6 were grown on MRS medium supplemented with 0.05% (wt/vol) cysteine and incubated anaerobically at 37°C for 36 to 72 h. The reference bacterial strain used in this study, L. rhamnosus GG ATCC 53103 (referred to here as L. rhamnosus GG), was supplied by the American Type Culture Collection (ATCC). Strain L. rhamnosus GG, was grown on MRS medium and incubated anaerobically at 37°C for 17 to 24 h.
Identification and taxonomic characterization of isolates by sequencing.
In order to identify the bacterial strains, an almost full sequence of the 16S rRNA was amplified and sequenced using an ABI Prism BigDye Terminator cycle sequencing ready reaction kit (Applied Biosystems Inc., Foster City, CA). DNA from pure culture was extracted using a High Pure PCR kit (Roche), spectrophotometrically quantified, and adjusted to a final concentration of 40 ng/μl in ultrapure water (Sigma-Aldrich). The DNA was checked for purity, using standard methods. DNA templates were amplified by PCR on a thermocycler TC-5000 instrument (Bibby Scientific, Stone, United Kingdom), using universal primers amplifying a 1,500-bp region of the 16S rRNA gene: primer 616V (5′-AGAGTTTGATYMTGGCTCAG-3′) and primer 630R (5′-CAKAAAGGAGGTGATCC-3′). The amplification mixture (100 μl) comprised 2 μl (50 pmol/μl) each of primers 616V and 630R (Thermo Fisher Scientific, Waltham, MA); 0.5 μl (2 U/μl) of Taq DNA polymerase (Finnzymes, Espoo, Finland); 10 μl of 10× reaction buffer (Finnzymes); 10 μl of a deoxynucleoside triphosphate mixture containing 1 mM (each) dATP, dGTP, dCTP, and dTTP (Roche Diagnostics GmbH, Penzberg, Germany); 70 μl of sterile filtered water (Milli-Q purification system; Millipore, Billerica, MA); and 5.5 μl of DNA template. The DNA template was amplified by initial denaturation at 94°C for 10 min, followed by 40 cycles of denaturation at 94°C for 1 min, annealing at 55°C for 1 min, and extension at 72°C for 1 min and a final extension at 72°C for 10 min. Controls devoid of DNA were simultaneously included in the amplification process. The integrity of the PCR products was assayed by detection of single bands following electrophoresis for 1 h at 100 V in 2% (wt/vol) agarose gels in Tris-borate-EDTA buffer. Amplicons were purified using a commercial kit, the QIAquick PCR purification kit (Qiagen Inc., Valencia, CA), and subsequent sequencing reactions were performed using the Big Dye Terminator (version 3.1) cycle sequencing kit (Applied Biosystems) in a premixed format. The resulting sequences were automatically aligned and inspected by eye and compared by use of the online tool BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi). The strain was identified on the basis of highest scores.
Assays of activity against H. pylori.
Six bifidobacteria (strains CP1 to CP6) were randomly selected for assays of inhibition of eight H. pylori strains (reference strains NCTC 11637 and NCTC 11638 and human isolates A1 to A6). Inhibition was tested by both agar diffusion assays and broth inhibition assays. In both cases, in order to obtain supernatants of bifidobacteria, strains CP1 to CP6 were grown anaerobically at 37°C in MRS-C medium. Supernatants were obtained by centrifugation at 12,000 × g for 10 min, neutralized to pH 6.5 with NaOH (1 N), and 10× concentrated by freeze-drying. Supernatants were sterilized by filtering through a 0.22-μm-pore-size filter (Minisart hydrophilic syringe filter; Sartorius Stedim Biotech GmbH, Goettingen, Germany) and storage at −20°C until use.
(i) Well diffusion agar assays.
Strains of H. pylori were grown in brain heart infusion (BHI) broth (Oxoid) supplemented with 5% (vol/vol) horse blood (Oxoid) in a microaerobic environment by means of a CampyGen system (Oxoid) at 37°C for 3 days. Cultures were centrifuged at 4,000 × g for 10 min, and cells were washed and suspended in saline solution (0.09%, wt/vol). Concentrated H. pylori solutions were spread onto the surface of BHI agar supplemented with 10% (vol/vol) horse blood. Wells (diameter, 5 mm) were made in the agar, and 50 μl of neutralized supernatant was loaded into the wells. The inhibition zones were read after incubation at 37°C for 3 days. As a control, MRS-C medium adjusted to pH 6.5 was included. Assays were performed in two independent experiments.
(ii) Broth liquid growth inhibition assays.
Concentrated H. pylori cell solutions were obtained as explained above. Assays were carried out in polystyrene 96-well (volume, 200 μl/well) multiwell plates (Maxisorp; Nunc, Roskilde, Denmark). Brain heart infusion broth supplemented with 10% (vol/vol) fetal bovine serum (FBS; Gibco Invitrogen, Paisley, United Kingdom) was inoculated with 1% (vol/vol) concentrated H. pylori cell solution, and 25 μl neutralized supernatant was added. Plates were incubated for 3 to 4 days at 37°C microaerobically, and the final optical density (OD) at 600 nm (OD600) was measured in order to obtain the H. pylori inhibition percentage for each supernatant. Assays were performed in three independent experiments.
Inhibition of H. pylori adhesion to intestinal mucus by site competition.
Pig type III mucin (Sigma-Aldrich) was used at a concentration of 0.5 mg/ml in bacterial adhesion assays. The mucus was dissolved in HEPES-HanKs buffer (HH; 10 mM HEPES, pH 7.4) at the desired concentration and immobilized in polystyrene multiwell plates (Maxisorp; Nunc, Roskilde, Denmark), adding 100 μl of this solution to each well and incubating the plates at 4°C overnight. The nonimmobilized mucus was removed by two successive washings with 200 μl of HH buffer. After the last washing, 100 μl of HH buffer was added to each well. H. pylori strains were grown in BHI broth (Oxoid) supplemented with 5% (vol/vol) horse blood (Oxoid) microaerobically at 37°C for 3 days. Cultures were centrifuged at 4,000 × g for 10 min, and cells were washed and suspended in HH buffer. Cells were adjusted to an OD of 0.25, fluorescently labeled by adding to the culture medium 10 μl/ml of carboxyfluorescein diacetate (CFDA; 100 mM; Calbiochem, Merck KGaA, Darmstadt, Germany), and incubated at 37°C for 45 min in darkness. Subsequently, 100 μl of nonlabeled bifidobacteria was added to wells with immobilized mucus and the plates were incubated at 37°C for 1 h. Nonadhered bifidobacteria were removed by two successive washings with 200 μl of HH buffer, 100 μl of CFDA-labeled H. pylori cells was added to each well, and the plates were incubated at 37°C for 1 h. Nonadhered H. pylori cells were removed by two successive washings with 200 μl of HH buffer. Adhered bacteria were released by means of scraping the immobilized mucus in each well and were later lysed with 1% (wt/vol) SDS in 0.1 N NaOH (200 μl per well), incubating the plates at 60°C for 1 h. The level of fluorescence liberated was measured at an excitation λ of 485 nm and an emission λ of 518 nm in a Fluoreskan Ascent FL apparatus (Thermo Fisher Scientific). Differences between adhesion values with and without bifidobacteria were considered the inhibition of adhesion percentage obtained with each bifidobacterium. Assays were performed in three independent experiments.
Characterization as potential probiotic. (i) Resistance to gastrointestinal juices.
The two-stage assay for resistance to gastrointestinal juices emulated the conditions and time of exposure to which bacteria are subjected during their passage through the gastrointestinal tract. In the first stage, gastric and pancreatic juices were prepared immediately before use. Pepsin of pig stomach mucous membrane (3 g/liter; Sigma-Aldrich) and pancreatin of pig pancreas (1 g/liter; Sigma-Aldrich) were resuspended in sterile saline solution (0.5%, wt/vol), and the pHs were adjusted to 3.0 and 8.0, respectively. After 24 h of growth at 37°C under anaerobic conditions, cells were washed twice in saline solution (0.09%, wt/vol) and brought to an inoculum of 108 to 109 CFU/ml.
In order to simulate passage through the gastric tract, an inoculum of 1% (vol/vol) bacteria was added to 10 ml of pepsin solution and the mixture was incubated at 37°C under anaerobic conditions for 2 h. To simulate passage through the intestinal tract, cells were neutralized by washing with phosphate buffer at pH 7.0 and were then resuspended in pancreatin solution and incubated at 37°C under anaerobic conditions for 4 h. Counts in samples taken at 0, 90, and 120 min in the first stage and at 0, 60, and 240 min in the second were done. To monitor viability, throughout the process the strain was inoculated in saline solution (0.09%, wt/vol) and incubated under the same conditions; aliquots were taken at the beginning, after 120 min, and at the end of the assay. Assays were performed in three independent experiments.
(ii) Sensitivity to antibiotics.
Strain sensitivity to antibiotics, expressed as the MIC, was determined according to European Food Safety Authority (EFSA) recommendations (16), following the method for broth dilution antimicrobial susceptibility test established by the Clinical and Laboratory Standards Institute. The MIC values of 20 antimicrobial agents were determined using the standardized LSM broth formulation previously described, which consists of a mixture of IST broth (90%, vol/vol) and Bifidobacterium sp. broth (10%, vol/vol) adjusted to pH 6.7 (28). The antibiotics were tested over a concentration range of 0.125 to 256 mg/liter. Assays were performed in three independent experiments.
(iii) Resistance to biliary salts, NaCl, and low pH.
Resistance to different concentrations of biliary salts (oxgall), NaCl, and pH was evaluated by monitoring bacterial growth on 96-well plates. To each well, 175 μl of Bifidobacterium sp. medium supplemented with 0.5%, 1%, 2%, and 3% (wt/vol) oxgall, Bifidobacterium sp. medium supplemented with 2%, 3%, 6%, 8%, and 10% NaCl (wt/vol), or Bifidobacterium sp. medium at pH 3.0, pH 2.0, and pH 1.5 was inoculated with 100 μl of an inoculum at an OD600 of 1 unit. Growth was analyzed at 600 nm in a Multiskan microplate reader (Thermo Fischer Scientific) after 24 h of incubation at 37°C under anaerobic conditions. The resistance percentage was calculated in each case by comparing the final OD600 in the supplemented medium with that of the control growth in Bifidobacterium sp. medium. Assays were performed in three independent experiments.
(iv) Adhesion to intestinal mucus.
Assays of adhesion to intestinal mucus were performed as described above for assays of inhibition of adhesion of H. pylori to intestinal mucus. In this case, 17-h cultures of strains CP5 and Lactobacillus GG were assayed in Bifidobacterium sp. medium and MRS medium, respectively. Assays were performed in three independent experiments.
Toxicological study. (i) Nondesired metabolite production.
(a) Lactic acid production. Lactic acid production was determined by using supernatant from a 24-h culture of each bacterial strain. Strain CECT 7366 was grown anaerobically at 37°C in Bifidobacterium sp. medium, and L. rhamnosus GG was grown in MRS medium. Supernatant was obtained by centrifugation at 12,000 × g for 10 min. Lactic acid isomers were quantified using a commercial kit (d-lactic acid/l-lactic acid; Roche Diagnostics, Manheim, Germany), following the manufacturer's instructions. Assays were performed in three independent experiments.
(b) Deconjugation of biliary salts. A bile salt hydrolase (BSH) activity assay was performed with glycocholate and taurocholate as substrates, following the technique of Kumar and coworkers (29). Assays were performed in three independent experiments.
(c) Formation of biogenic amines. The formation of biogenic amines (cadaverine, histamine, putrescine, and tyramine) was determined in Bifidobacterium sp. medium (CECT 7366) or MRS medium (L. rhamnosus GG) cell-free supernatant of 24-h cultures at 37°C anaerobically. Amine formation was determined following the chromatographic method described by Eerola and coworkers (13). Assays were performed in three independent experiments.
(ii) Acute ingestion study in mice.
All the procedures involving animal experiments were conducted in accordance with the regulations established by the European Community Council on the protection of animals with experimental and scientific applications (regulation 86/609/EEC).
For acute ingestion assays, a culture of the bifidobacteria growing anaerobically in 2 liters of Bifidobacterium sp. medium for 17 h at 37°C was obtained. Cells were recovered by centrifugation at 12,000 × g for 15 min and washed twice in saline solution (0.09%, wt/vol). Cells were resuspended in 200 ml of skim milk with 5% (wt/vol) sucrose, frozen at −80°C, and freeze-dried. Aliquots of 109 CFU per 100 μl of Ringer's solution were prepared as the inoculum.
Assays were carried out with 7-week-old pathogen-free male BALB/c mice. The mice had free access to water and standard rodent laboratory chow. Five days before the assay was started, animals were randomly assigned to the different experimental groups. All animals in each cage were administered the same dose. Table 1 summarizes the distribution of the experimental groups. Due to the special immunological characteristics of immune-deficient mice, the animals were maintained from the time of their arrival inside containment units provided with positive pressure. Immunosuppression was achieved by intraperitoneal administration of cyclophosphamide (150 mg/kg of body weight) 3 days before the first bifidobacterial administration and of another dose of 100 mg/kg 1 day before. Inoculum and placebo (lyophilized skim milk with sucrose [5%, wt/vol]) were administered by oral gavage for 6 days at a volume of 400 μl. Mortality and morbidity were noted twice a day throughout the study. Body weight was recorded daily. On day 7 of the study, the corresponding mice were killed, according to humane endpoints, with pentobarbital sodium salt (200 mg/kg) by intraperitoneal injection. Immediately after the mice were killed, the carcasses were disinfected by immersion in Virkon S disinfectant (DuPont Animal Health Solutions, Sudbury, United Kingdom) and dried with a gauze swab. The cardiac blood was taken with a 25-gauge needle coupled to a 1-ml syringe and collected in EDTA-coated tubes. Organs such as the liver, spleen, mesenteric lymph nodes, ileum, jejunum, cecum, and colon were extracted and weighed. The jejunum, cecum, and colon were processed for histopathological evaluation. Blood, liver, spleen, and mesenteric lymph nodes were homogenized in BFM medium (40) with a Potter-Elvehjem tissue grinder, and homogenates were stored at −80°C in 25% (vol/vol) glycerol. Bacterial counts were obtained by plate count in BFM medium. For histopathological evaluation, on the day of killing, the jejunum, cecum, and colon were collected and preserved in neutral buffered formaldehyde solution (4%, wt/wt; pH 7) for subsequent analysis. Organs collected from all animals were fixed, dehydrated, and embedded in paraffin. Sections were cut using a microtome, and slices were stained with hematoxylin-eosin and mounted. In order to investigate the mice for possible adverse effects, histopathological analyses included semiquantitative parameters such as the determination of the height of epithelial cells, the depth of crypts, and the thickness of mucosa.
TABLE 1.
Results obtained in mice during the acute ingestion studya
| Group | Immunosuppressed | Dose (no. of CFU/mouse) | Body wt gain (g)b |
Organ wt (g) |
|||
|---|---|---|---|---|---|---|---|
| Days −4 to 1 | Days 1 to 6 | Spleenc | Liver | Mesenteric lymph nodes | |||
| A | No | Placebo | 0.1 ± 0.6 | 0.4 ± 0.6 | 0.10 ± 0.04 C | 1.22 ± 0.23 | 0.0010 ± 0.0005 |
| B | No | 1 × 109 | 0.2 ± 0.6 | 0.2 ± 0.4 | 0.09 ± 0.05 D | 1.20 ± 0.15 | 0.0020 ± 0.0007 |
| C | Yes | Placebo | 0.0 ± 1.0 | 0.6 ± 0.6 | 0.23 ± 0.09 C | 1.29 ± 0.24 | 0.0020 ± 0.0005 |
| D | Yes | 1 × 109 | 0.6 ± 0.8 | 0.4 ± 0.6 | 0.20 ± 0.07 D | 1.22 ± 0.20 | 0.0020 ± 0.0007 |
Data are the means ± standard deviations of the means (n = 12).
Body weight gain is expressed as final weight minus initial weight.
Values sharing the letter C are significantly different at a P value of <0.01; those sharing the letter D are significantly different at a P value of <0.001.
Reversion effects of B. bifidum after challenge with H. pylori in mice.
Assays of reversion effects were carried out with 7-week-old pathogen-free male BALB/c mice under the same conditions used in the acute ingestion study. As in the acute ingestion assays, all the procedures involving animals were conducted in accordance with the regulations established by the European Community Council on the protection of animals with experimental and scientific applications (regulation 86/609/EEC).
In all cases, H. pylori was quantified by real-time PCR as described by Nayak and Rose (39). H. pylori was grown in BHI medium supplemented with 5% (vol/vol) FBS under microaerobic conditions at 37°C for 3 days. Cultures were centrifuged at 4,000 × g for 10 min. Cells were washed, suspended in saline solution (0.09%, wt/vol), and quantified by real-time PCR. Aliquots of 4 × 105 copies of H. pylori DNA per 100 μl Ringer's buffer were prepared and conserved at −20°C until administration. Two groups of 10 mice each were randomly formed. In experimental groups A and B, 100 μl of a suspension of H. pylori containing 4 × 105 copies of DNA was administered for three consecutive days from day 1 onwards. In group B, 400 μl of a suspension containing 5.6 × 108 CFU of strain CP5 was administered orally for 11 and 18 days after infection. In group A, 400 μl of a placebo (lyophilized skim milk with sucrose [5%, wt/vol]) was administered. On day 14, five mice from each group were killed and analyzed, and on day 21, the remaining five mice from each group were killed and analyzed.
Animal mortality was checked daily throughout the study. From the first study day on, body weight was recorded every 2 days and the mice were examined for clinical signs. On days 14 and 21, mice were killed as described above, followed by macroscopic necropsy. On the day of killing, the main organs were examined macroscopically, taking special note of the presence and quantification of gastric ulcers. Furthermore, the stomach was extracted, weighed, and homogenized in BHI medium using a Potter-Elvehjem tissue grinder at a ratio of 0.1 g/ml. The homogenized stomach sample was then used for quantitative PCR to obtain the number of copies of H. pylori DNA, and the results were corrected according to stomach weight in order to calculate the total number of DNA copies per stomach.
Purification of the substance(s) of interest by cationic-exchange chromatography followed by inverse-phase chromatography.
Supernatant was obtained by centrifugation at 12,000 × g for 15 min under refrigerated conditions of a 10-liter culture of strain CP5 in Bifidobacterium sp. medium under the standard conditions described above. Supernatant was stored at −20°C until use. An aliquot of 200 ml was added to 250 ml sodium phosphate buffer (20 mM, pH 5.8) and applied to a cationic-exchange column (HiPrep 16/10 SP FF; GE Healthcare) by means of a chromatography system (ÄKTA Explorer; Amersham Pharmacia Biotech). All those proteins of a cationic nature which were adsorbed to the resin of the column were eluted with sodium phosphate buffer (20 mM, pH 5.8) equilibrated with NaCl (1 M) without a gradient. The final peak fractions and all solutions that could contain the target protein were eluted simultaneously in a few fractions and in a minimum volume (10 ml) in order to concentrate the substance of interest. Each fraction collected by cationic exchange was subjected to an ultrafiltration process using 5,000-Da-molecular-mass-cutoff filters (Extreme Amicon; Millipore). Thus, the filtered volume contained those proteins with a molecular mass below 5,000 Da, and the retained volume kept those proteins with a higher molecular mass. The filtered volumes obtained from cationic exchange were subjected to another stage of inverse-phase chromatography to purify the substance(s) responsible for the inhibition of H. pylori. For the inverse-phase chromatography, a Resource RPC 3-ml column (GE Healthcare) was used, working in gradient with eluent A (5% 2-propanol, 0.1% trifluoroacetic acid [TFA] in Milli-Q-filtered water) and eluent B (0.1% TFA in 2-propanol). This chromatography gave rise to 2-ml fractions. An aliquot of these 2-ml fractions was taken to dryness to eliminate the dissolvent, and the product was stored at −20°C until it was screened for H. pylori inhibition. Throughout the purification process, several parameters, including UV absorbance (280, 254, and 214 nm), pressure, flow rate, pH, and ionic strength, were monitored.
Statistical analysis.
The results obtained were analyzed using Statgraphics plus (version 5.1) software (Manugistiscs, Rockville, MD). Mouse and organ weights were compared between the groups using Tukey's multiple-comparison test. The other data were subjected to a one-way and a multifactor analysis of variance (ANOVA). The least-significant-difference test was used for comparison of means.
Nucleotide sequence accession number.
The 16S rRNA sequence of B. bifidum CECT 7366 has been deposited in the GenBank nucleotide sequence database under accession number HM590860.
RESULTS
Activities of isolates against H. pylori.
Agar well diffusion assays were performed with two reference strains and six H. pylori isolates (Table 2). The results indicated that all supernatants exerted inhibitory activity against H. pylori. The greatest inhibition values corresponded to isolates CP4 and CP5, with average inhibition diameters of 6.05 and 6.59 mm, respectively. Regarding liquid broth growth inhibition assays of H. pylori NCTC 11637, strain CP5 rendered the highest growth inhibition (81.94%). The other strains rendered lower inhibition values (about 70%).
TABLE 2.
Activities of supernatants against H. pylori strains in well diffusion agara
| Strain | Diam of H. pylori inhibition (mm) |
|||||||
|---|---|---|---|---|---|---|---|---|
| NCTC 11637 | NCTC 11638 | A1 | A2 | A3 | A4 | A5 | A6 | |
| CP1 | 1.00 ± 0.23 | 5.50 ± 1.10 | 5.20 ± 0.69 | 6.70 ± 2.01 | 2.50 ± 0.42 | 5.70 ± 1.08 | 2.70 ± 0.71 | 4.50 ± 0.37 |
| CP2 | 7.25 ± 0.12 | 5.00 ± 0.63 | 6.00 ± 1.14 | 6.50 ± 0.34 | 4.50 ± 1.21 | 5.00 ± 0.98 | 6.20 ± 1.32 | 5.50 ± 0.68 |
| CP3 | 7.25 ± 1.20 | 6.00 ± 0.52 | 6.60 ± 1.22 | 6.30 ± 0.79 | 5.00 ± 0.13 | 5.00 ± 0.62 | 5.00 ± 1.05 | 4.50 ± 0.22 |
| CP4 | 6.25 ± 1.54 | 7.50 ± 1.14 | 6.00 ± 0.28 | 6.00 ± 1.05 | 4.70 ± 0.36 | 6.00 ± 1.17 | 6.00 ± 0.85 | 6.00 ± 0.33 |
| CP5 | 7.25 ± 0.61 | 7.00 ± 0.87 | 6.70 ± 0.76 | 7.80 ± 0.52 | 5.00 ± 0.47 | 5.80 ± 1.66 | 6.70 ± 0.41 | 6.50 ± 0.54 |
| CP6 | 6.00 ± 0.45 | 5.25 ± 0.15 | 5.00 ± 1.78 | 7.20 ± 1.33 | 2.50 ± 1.49 | 5.00 ± 0.92 | 6.00 ± 0.29 | 4.00 ± 0.91 |
Data are the means ± standard deviations of the means (n = 2).
Inhibition of H. pylori adhesion to intestinal mucus by site competition.
Reference strain NCTC 11637 and human isolates A1 and A2 were included in the adhesion to intestinal mucus assays. Results are summarized in Table 3. CP5 was the only strain that hindered the adhesion to intestinal mucus of all the H. pylori strains tested. In some cases, the presence of Bifidobacterium strains increased H. pylori adhesion (represented as negative values in Table 3). Thus, taking inhibition and adhesion results into account, strain CP5 was selected for further studies.
TABLE 3.
Inhibition of adhesion of H. pylori to intestinal mucus by site competitiona
| Strain | Inhibition of adhesion of H. pylori to intestinal mucus (%) |
||
|---|---|---|---|
| A1 | A2 | NCTC 11637 | |
| CP1 | 4.00 ± 0.38 | 2.11 ± 0.99 | −0.52 ± 1.65 |
| CP2 | −3.66 ± 0.47 | 6.72 ± 0.23 | 4.71 ± 1.85 |
| CP3 | −4.00 ± 5.61 | 3.80 ± 1.15 | 4.84 ± 0.54 |
| CP4 | −6.83 ± 1.62 | 1.74 ± 1.87 | 4.97 ± 0.76 |
| CP5 | 5.42 ± 5.48 | 3.59 ± 0.75 | 8.17 ± 2.60 |
| CP6 | 3.89 ± 4.82 | 1.27 ± 6.07 | −7.46 ± 0.08 |
Data are the means ± standard deviations of the means (n = 3).
Identification of CECT 7366 strain.
Using primers 616V and 630R, a 1,412-bp genomic DNA fragment from strain CP5 was obtained by PCR. On the basis of the sequence obtained, strain CP5 was identified as Bifidobacterium bifidum by comparing its sequence with the sequences included in the GenBank database, an annotated collection of all publicly available DNA sequences, using the online tool BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi).
Characterization of strain CECT 7366 as a potential probiotic.
All experiments characterizing strain CECT 7366 as a potential probiotic were done using well-characterized commercial probiotic strain L. rhamnosus GG as a control. The in vitro resistance of the CP5 and L. rhamnosus GG strains to gastrointestinal juices was calculated as the relationship between the number of viable cells obtained at each stage of the assay and the number of viable cells at time zero. The viabilities of both strains after the exposure to pepsin treatment were similar (9%), while for pancreatin treatment, strain CP5 displayed the highest survival rates (7% versus 2.9% for CECT 7366). Resistance to oxgall, NaCl, and low pH values was also assayed, and the results are shown in Table 4. Taking into account resistance to biliary salts, results showed that both the CP5 and L. rhamnosus GG strains were able to grow in the presence of biliary salts, although CP5 was slightly more resistant. When resistance to different NaCl concentrations is considered (Table 4), both strains tolerated 6% (wt/vol) NaCl in MRS-C medium. At higher concentrations (8 to 10% NaCl), growth was apparently inhibited. With respect to commercial strain L. rhamnosus GG, it was able to continue growing in MRS-C medium containing 8% NaCl. Results obtained by growing the two strains in the respective media at different pH values (Table 4) showed that the L. rhamnosus GG strain was less resistant to low pH values. Nevertheless, in subsequent assays, strain CP5 showed 84.67% survival in Bifidobacterium sp. medium at pH 4 and 100% survival in Bifidobacterium sp. medium at pH 7 (data not shown). Concerning sensitivities to antibiotics (Table 5), CECT 7366 did not show MIC values higher than the EFSA breakpoints. Finally, taking into consideration results for adhesion to the intestinal mucus, strain CP5 showed 12% adhesion, whereas L. rhamnosus GG showed 10% adhesion.
TABLE 4.
Characterization of resistance of CP5 and L. rhamnosus GG strains to biliary salts, NaCl, and low pHa
| Strain | % survivalb |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Biliary salts |
NaCl |
pH |
||||||||||
| 0.5% oxgall | 1% oxgall | 2% oxgall | 3% oxgall | 2% | 3% | 6% | 8% | 10% | 1.5 | 2.0 | 3.0 | |
| CECT 7366 | 49.02 ± 6.85 | 66.04 ± 3.74 C | 63.19 ± 1.69 B | 62.78 ± 0.90 C | 110.01 ± 0.35 | 93.04 ± 1.56 | 80.37 ± 0.42 B | 21.97 ± 0.63 B | 12.17 ± 0.98 B | 11.46 ± 0.01 D | 12.90 ± 0.07 | 13.26 ± 0.70 |
| L. rhamnosus GG | 33.2 ± 3.10 | 34.51 ± 0.60 C | 42.77 ± 2.42 B | 48.94 ± 3.01 C | 99.25 ± 1.71 | 99.50 ± 1.29 | 84.31 ± 1.57 B | 31.17 ± 4.71 B | 9.50 ± 0.14 B | 2.02 ± 0.24 D | 11.26 ± 0.40 | 11.11 ± 0.24 |
Data are the means ± standard deviations of the means (n = 3).
Pairs of values sharing the letter B, C, or D in the same column are significantly different at a P value of <0.05, <0.01, or <0.001, respectively.
TABLE 5.
Sensitivities to antibiotics of strain CECT 7366 and commercial probiotic strain L. rhamnosus GG
| Antibiotic | MIC (μg/ml) |
|
|---|---|---|
| CECT 7366 | L. rhamnosus GG | |
| Amoxicillin | 2 | 4 |
| Ampicillin | 2 | 4 |
| Carbenicillin | 16 | 16 |
| Clarithromycin | 16 | 16 |
| Clindamycin | <0.125 | 1 |
| Chloramphenicol | 4 | 16 |
| Erythromycin | 2 | 2 |
| Gentamicin | 32 | 16 |
| Kanamycin | 256 | 64 |
| Metronidazole | 4 | >256 |
| Nalidixic acid | >256 | >256 |
| Oxytetracycline | 2 | 4 |
| Penicillin | 0.25 | 1 |
| Polymyxin B | 64 | >256 |
| Rifampin | 0.25 | 1 |
| Streptomycin | 64 | 32 |
| Sulfonamide | >256 | >256 |
| Tetracycline | 8 | 4 |
| Trimethoprim | >256 | >256 |
| Vancomycin | 0.5 | >256 |
Purification of substance(s) of interest by cationic-exchange chromatography followed by inverse-phase chromatography.
The fractions with larger amounts of protein obtained by cationic exchange (fractions 3, 4, and 5) were tested for activity against H. pylori NCTC 11637 by means of liquid broth growth assays. Fractions 3, 4, and 5 rendered 50.32%, 81.29%, and 1.90% inhibition, respectively. Thus, cationic-exchange fraction 4 was selected for ultrafiltration through a 5,000-Da-molecular-mass-cutoff filter and further purification. From fraction 4 of cationic exchange, 28 fractions of inverse phase were obtained, and of these, fraction 4.11 rendered the highest inhibition value in liquid broth growth assays with strain H. pylori NCTC 11637 (94.77%).
Ex vivo toxicological studies.
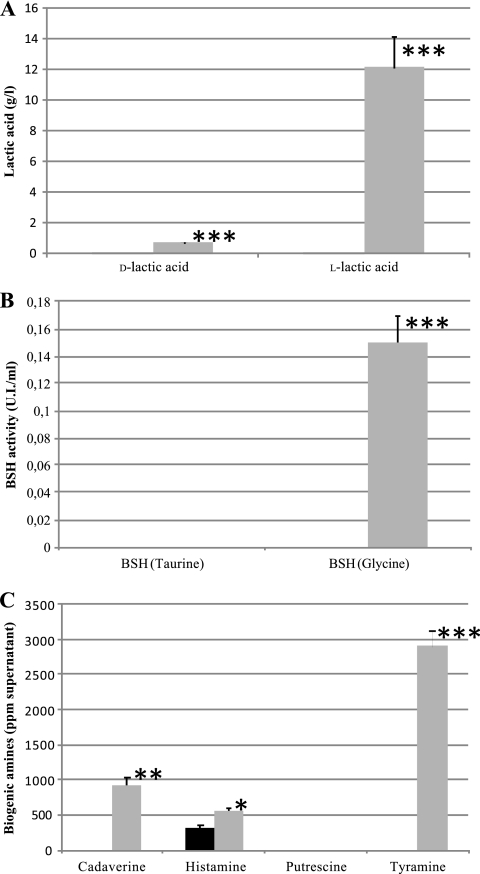
The results obtained in lactic acid production assays are shown in Fig. 1 A. Compared with strain L. rhamnosus GG, results for strain CECT 7366 show lower levels of lactic acid in the supernatant for both d- and l-lactic acid. There was no BSH activity in supernatants from 24-h cultures in either taurocholate or glycocholate, and in the case of L. rhamnosus GG, activity was obtained only with glycocholate (Fig. 1B). Figure 1C shows the results obtained for cadaverine, histamine, putrescine, and tyramine quantification in cell-free supernatants of strains CECT 7366 and L. rhamnosus GG strains grown for 24 h. Putrescine was not detected in any case. For the other amines, values ranged from 0.25 to 321.00 for CECT 7366 and 560.00 to 2,900.00 for L. rhamnosus GG. In all the analyzed amines, the values obtained for the strain L. rhamnosus GG supernatant were always higher than those obtained for the CECT 7366 supernatant.
FIG. 1.
Nondesired metabolite detection in supernatant obtained for strains CECT 7366 (solid bars) and L. rhamnosus GG (shaded bars). (A) Lactic acid; (B) BSH activity; (C) biogenic amines. Data are the means ± standard deviations of the means (n = 3). *, **, and ***, significant at P < 0.05, P < 0.01, and P < 0.001, respectively.
In vivo toxicological studies.
Neither mortality nor clinical signs were observed during the study. Regarding body weight gain (Table 1), immunosuppressed mice lost weight after the immunosuppressor was administered but recovered it after ingestion of both probiotic and vehicle. In all cases, there were no statistically significant differences between groups. Taking into account organ weights, differences in spleen weight were statistically significant between immunosuppressed and immunocompetent mice in the groups administered bifidobacteria (P < 0.001) and vehicle (P < 0.01), being heavier in immunosuppressed mice (Table 1). In the case of microbiological counts, bifidobacterial loads were not found in blood or in liver, spleen, or mesenteric lymph nodes, under the present study conditions.
Results obtained in this analysis showed no biologically significant intergroup differences between immune-competent and immune-deficient mice, with or without bifidobacterial administration (data not shown).
Reversion effects of B. bifidum after challenge with H. pylori in mice.
During the study, neither mortality nor relevantly altered clinical signs were observed in any group. Regarding body weight, no significant differences were observed between groups (Table 6). Stomach weight did not show significant differences between vehicle and bifidobacterial treatments on day 14 and on day 21. However, a significant decrease (P < 0.05, two-way ANOVA) in stomach weight was found for animals killed on day 21 compared with those for mice killed on day 14. These results indicate that there is a direct relationship between H. pylori infection and stomach weight. Taking into account the results obtained in the histomorphology analysis (Table 6), differences were observed between groups treated with bifidobacteria and vehicle at 14 and 21 days. An increase in Peyer's patches was recorded in 90% of the animals treated with vehicle and in 40% of mice treated with bifidobacteria. Signs of infection did not appear in the spleen or mesenteric ganglion in the bifidobacteria-treated group, whereas some animals treated with vehicle showed increased spleen (10%) or mesenteric ganglion (20%). Results obtained for the stomach are summarized in Table 6. Higher numbers of ulcers appeared in the vehicle-treated group than in the bifidobacteria-treated group, with larger differences being found for animals killed on day 21 of the study. Regarding the number of copies of H. pylori DNA in stomach homogenates, average results ranged from 2.1 × 106 to 4.9 × 107. The two-way ANOVA analysis showed significant differences between the group killed on day 21 of the study and the one killed on day 14 (P < 0.05), whereas no statistically significant differences were found when the results for the bifidobacteria-treated and vehicle-treated groups were compared. However, when the pathogenicity ratio of H. pylori was evaluated, results for animals killed on day 14 were significantly higher (P < 0.05, two-way ANOVA) than those for mice killed on day 21. Moreover, treatment with bifidobacteria also gave rise to a decrease in the H. pylori pathogenicity ratio in comparison with that for mice treated with the vehicle alone, although statistical significance was not found.
TABLE 6.
Results obtained in reversion studya
| Group | Dose (no. of CFU/mouse) | Study day | Wt gain (g)b | Stomach wt (g)d | No. of animals with increase in following characteristic/total no. animals per group: |
No. of DNA copies of H. pylori/stomachd | Total no. of ulcers/stomach | H. pylori pathogenicity ratioc,d | ||
|---|---|---|---|---|---|---|---|---|---|---|
| Peyer's patches | Spleen wt | Mesenteric ganglion wt | ||||||||
| A | Placebo | 14 | −0.90 ± 0.60 | 0.174 ± 0.035 D | 4/5 | 1/5 | 1/5 | 2.1 × 106 ± 1.9 × 106 F | 1.00 ± 0.71 | 0.85 ± 0.90 H |
| 21 | 0.06 ± 0.45 | 0.132 ± 0.032 D | 5/5 | 0/5 | 1/5 | 3.6 × 107 ± 6.0 × 107 F | 0.60 ± 0.89 | 0.02 ± 0.03 H | ||
| B | 1 × 109 | 14 | −0.36 ± 0.76 | 0.166 ± 0.038 E | 2/5 | 0/5 | 0/5 | 4.4 × 106 ± 3.5 × 106 G | 0.60 ± 0.55 | 0.13 ± 0.14 I |
| 21 | −0.20 ± 0.37 | 0.132 ± 0.013 E | 2/5 | 0/5 | 0/5 | 4.9 × 107 ± 3.0 × 107 G | 0.00 ± 0.00 | 0.00 ± 0.00 I | ||
Data are the means ± standard deviations of the means (n = 5).
Body weight gain is expressed as final weight minus initial weight.
H. pylori pathogenicity ratio is expressed as total number of ulcers per million copies of H. pylori DNA in the stomach.
Pairs of values sharing the same capital letter are significantly different (P < 0.05).
DISCUSSION
Nowadays, H. pylori is considered the major cause of gastric and duodenal ulcers as well as a carcinogenic agent in humans, and it is currently one of the most widely studied bacterial pathogens affecting humans (21). Within this context, studies suggest that gastroduodenal microbiota like lactobacilli may act as an active protector against this bacterium (14, 25). Following these observations, the use of probiotics as agents active against H. pylori has been evaluated in numerous preclinical and clinical studies with different degrees of success. The majority of published studies focus on in vitro and in vivo activities of probiotics; however, they do not shed light on which compounds produced by probiotics are directly related to inhibition or the underlying mechanisms involved. On the basis of these considerations, the present work has focused on the selection of a probiotic Bifidobacterium strain that shows inhibitory activity against H. pylori both in vitro and in vivo. To achieve this goal, an extensive study has been carried out and includes in vitro and in vivo analyses of the activity against H. pylori, as well as a detailed analysis of the potential use of the selected strain as a probiotic.
As a result of preliminary antimicrobial screening in both agar and liquid growth media using six Bifidobacterium isolates, strain CP5 (identified as Bifidobacterium bifidum) obtained promising H. pylori inhibition (81.94%) and decreased adhesion of the pathogen to intestinal mucus by site competition. Therefore, this strain was selected for further assays in vivo.
Some in vivo studies have demonstrated the positive effects of probiotics against H. pylori in animal models (26) and humans; however, specific studies focusing on the in vivo activity of bifidobacteria against H. pylori are still very scarce and are normally associated with other probiotics. In our study, once strain B. bifidum CECT 7366 had demonstrated its antagonistic activity against H. pylori in vitro, an in vivo reversion test was carried out using a murine model. Results indicated there were no signs of infection in spleen or in the mesenteric ganglion of those mice treated with strain B. bifidum CECT 7366. Furthermore, the group treated with vehicle developed more ulcers than the group treated with the probiotic. These results, together with the diminished H. pylori pathogenicity ratio in animals treated with bifidobacteria, indicate that strain B. bifidum CECT 7366 is a potential probiotic, exerting in vivo antagonistic activity against H. pylori. Furthermore, in vivo assays have demonstrated that it partially relieves damage to gastric tissues caused by the pathogen.
In order to identify the mechanism underlying strain CP5 inhibition of H. pylori, a purification based on cationic-exchange chromatography followed by inverse-phase chromatography was performed. This purified fraction containing those proteins with a molecular mass below 5,000 Da and the findings of in vitro functional assays pointed to the peptidic nature of the strain CP5 metabolites linked to H. pylori inhibition. In this respect, diverse studies have focused on the metabolites underlying the antagonistic activities of probiotics against human pathogens, including H. pylori. Organic acids, such as lactic acid, are the most-documented kind of metabolites directly related to the activity of lactic acid bacteria against H. pylori (46); however, there are also reports of antimicrobial compounds of proteinaceous nature with antagonistic activity against H. pylori. Of these, peptides produced by bifidobacteria with activity against H. pylori (7) have been reported but are as yet uncharacterized. In fact, to our knowledge, there is only one characterized bacteriocin from bifidobacteria (bifidocin B from B. bifidum NCFB 1454 [54]). Our study ruled out the potential inhibitory effect of pH against H. pylori because assays were carried out with neutralized extracts. Moreover, purification by cationic exchange followed by inverse phase increased the inhibition level, reaching up to 94.77%. Taken together, these results suggest that the antagonistic compounds underlying the inhibitory activity of strain B. bifidum CECT 7366 against H. pylori are of a proteinaceous nature.
The overall results confirmed strain CECT 7366 to be a potential probiotic bacterium; therefore, a screening for strain-specific probiotic properties was performed. Extensive analyses included its characterization as a potential probiotic and ex vivo and in vivo toxicological assays. Within this context, resistance to gastric conditions must be assessed. Assays evaluating survival after treatment with gastrointestinal juices, biliary salts, large amounts of NaCl, and low pH values were carried out. In all cases, results were very similar to those obtained for the well-established commercial probiotic L. rhamnosus GG. Regarding resistance to simulated gastric and pancreatic conditions, both strains exhibited limited tolerance, with less than 10% survival occurring in the presence of pepsin and pancreatin. Similar to our results, Charteris et al. (4) reported a 1-log-unit decrease in B. bifidum and L. rhamnosus GG viability after 90- and 180-min treatments with pancreatin, whereas in our case, no reduction in survival after 4 h of treatment with pancreatin appeared, even with L. rhamnosus GG and by following a very similar methodology. In the case of biliary salts, our results coincide with others previously reported for B. bifidum strains (10, 33, 34). Media with large amounts of NaCl, above 8% (wt/vol) NaCl, caused a drastic decrease in survival, as previously reported for bifidobacteria (8). Regarding pH resistance, strain CP5 showed higher survival values than those obtained for L. rhamnosus GG, reaching 11% survival at the lowest pH value tested (pH 1.5). However, differences between gastrointestinal conditions simulated in vivo and in vitro should be considered. Particularly, the low-pH transit through the stomach normally takes less than 2 h, but probiotics are ingested together with other compounds which buffer the effect of the low gastric pH (43). Finally, the capacity to adhere to intestinal mucus is considered a critical property of probiotics (25), and thus, it was assessed in the present study. Results have shown that strain CP5 has a capacity to adhere to intestinal mucus similar to that of L. rhamnosus GG and that the adherence capacity is comparable to that obtained for other bifidobacteria (25).
Once the functionality and probiotic characterization of B. bifidum CECT 7366 were demonstrated, it must be assessed for safety. Accordingly, although the B. bifidum species is included in the list of taxonomic units proposed for qualified presumption of safety (QPS) status (17) and is thus considered safe, in order to ensure strain safety, a detailed toxicological study was performed following the FAO/WHO recommendations (19). Results obtained for resistance to antibiotics were similar to those previously reported for other bifidobacteria (9, 10), and strain CECT 7366 did not show any MIC values above those recommended by EFSA (16). Regarding the production of undesired metabolites, such as lactic acid isomers, products of BSH activity, and biogenic amines, results showed that the level of production of both d- and l-lactic acid isomers was very low in comparison with that by strain L. rhamnosus GG. Concerning BSH activity, strain CECT 7366 did not display this activity with either taurocholate or glycocholate. Taking into account the biogenic amine levels obtained in supernatants, putrescine was not detected in any case, and cadaverine, histamine, and tyramine levels were very low in comparison with those for strain L. rhamnosus GG. Results obtained in the in vivo acute ingestion study showed neither mortality nor morbidity, even in immunosuppressed mice. Moreover, probiotic administration did not lead to bifidobacterial loads in organs or changes in histomorphology. Taking these results as a whole, strain B. bifidum CECT 7366 can be considered safe for human consumption.
In summary, the results presented here confer to strain B. bifidum CECT 7366 the status of a probiotic bacterium with functional activity against H. pylori in vitro and even in vivo in murine models. In this study, a fraction containing peptides with molecular masses below 5,000 Da underlying its functionality has been purified, and further studies will be carried out to identify the peptides directly related to the antagonistic activity and to study in greater depth the molecular basis of the capacity of probiotic strain B. bifidum CECT 7366 to inhibit H. pylori growth. Human clinical trials must be performed before commercialization of this strain can be approved.
Acknowledgments
This work has been cofunded by the European Social Fund, grants PTQ06-2-0642 and PTQ05-01-01208.
Footnotes
Published ahead of print on 17 December 2010.
REFERENCES
- 1.Aiba, Y., N. Suzuki, A. M. Kabir, A. Takagi, and Y. Koga. 1998. Lactic acid-mediated suppression of Helicobacter pylori by the oral administration of Lactobacillus salivarius as a probiotic in a gnotobiotic murine model. Am. J. Gastroenterol. 93:2097-2101. [DOI] [PubMed] [Google Scholar]
- 2.Bhatia, S. J., N. Kochar, P. Abraham, N. G. Nair, and A. P. Mehta. 1989. Lactobacillus acidophilus inhibits growth of Campylobacter pylori in vitro. J. Clin. Microbiol. 27:2328-2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Canducci, F., et al. 2002. Probiotics and Helicobacter pylori eradication. Digest. Liver Dis. 34(Suppl. 2):S81-S83. [DOI] [PubMed] [Google Scholar]
- 4.Charteris, W. P., P. M. Kelly, L. Morelli, and J. K. Collins. 1998. Development and application of an in vitro methodology to determine the transit tolerance of potentially probiotic Lactobacillus and Bifidobacterium species in the upper human gastrointestinal tract. J. Appl. Microbiol. 84:759-768. [DOI] [PubMed] [Google Scholar]
- 5.Chen, L. T., et al. 2005. Long-term results of anti-Helicobacter pylori therapy in early-stage gastric high-grade transformed MALT lymphoma. J. Natl. Cancer Inst. 97:1345-1353. [DOI] [PubMed] [Google Scholar]
- 6.Coconnier, M. H., V. Lievin, E. Hemery, and A. L. Servin. 1998. Antagonistic activity against Helicobacter infection in vitro and in vivo by the human Lactobacillus acidophilus strain LB. Appl. Environ. Microbiol. 64:4573-4580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Collado, M. C., et al. 2005. Antimicrobial peptides are among the antagonistic metabolites produced by Bifidobacterium against Helicobacter pylori. Int. J. Antimicrob. Agents 25:389-391. [DOI] [PubMed] [Google Scholar]
- 8.Collado, M. C., and Y. Sanz. 2007. Induction of acid resistance in Bifidobacterium: a mechanism for improving desirable traits of potentially probiotic strains. J. Appl. Microbiol. 103:1147-1157. [DOI] [PubMed] [Google Scholar]
- 9.D'Aimmo, M. R., M. Modesto, and B. Biavati. 2007. Antibiotic resistance of lactic acid bacteria and Bifidobacterium spp. isolated from dairy and pharmaceutical products. Int. J. Food Microbiol. 115:35-42. [DOI] [PubMed] [Google Scholar]
- 10.Delgado, S., E. O'Sullivan, G. Fitzgerald, and B. Mayo. 2008. In vitro evaluation of the probiotic properties of human intestinal Bifidobacterium species and selection of new probiotic candidates. J. Appl. Microbiol. 104:1119-1127. [DOI] [PubMed] [Google Scholar]
- 11.Drumm, B., G. I. Pérez-Pérez, M. J. Blaser, and P. M. Sherman. 1990. Intrafamilial clustering of Helicobacter pylori infection. N. Engl. J. Med. 322:359-363. [DOI] [PubMed] [Google Scholar]
- 12.Dunn, B. E., H. Cohen, and M. J. M. J. Blaser. 1997. Helicobacter pylori. Clin. Microbiol. Rev. 10:720-741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eerola, S., R. Hinkkanen, E. Lindfors, and T. Hirvi. 1993. Liquid chromatographic determination of biogenic amines in dry sausages. J. AOAC Int. 76:575-577. [PubMed] [Google Scholar]
- 14.Elliot, S. N., A. Buret, W. McKnight, M. J. Miller, and J. L. Wallace. 1998. Bacteria rapidly colonize and modulate healing of gastric ulcers in rats. Am. J. Physiol. 275:G425-G432. [DOI] [PubMed] [Google Scholar]
- 15.Eurogast Study Group. 1993. An international association between Helicobacter pylori infection and gastric cancer. Lancet 341:1359-1362. [PubMed] [Google Scholar]
- 16.European Food Safety Authority. 2008. Update of the criteria used in the assessment of bacterial resistance to antibiotics of human or veterinary importance. EFSA J. 732:1-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.European Food Safety Authority. 2008. Scientific opinion of the Panel on Biological Hazards on the maintenance of the list of QPS microorganisms intentionally added to food or feed. EFSA J. 923:30-48. [Google Scholar]
- 18.FAO/WHO. 2001. Health and nutritional properties of probiotics in food including powder milk with live lactic acid bacteria. Report of a joint FAO/WHO expert consultation on evaluation of health and nutritional properties of probiotics in food including powder milk with live lactic acid bacteria. FAO/WHO, Córdoba, Argentina.
- 19.FAO/WHO. 2002. Drafting guidelines for the evaluation of probiotics in food. Report of a joint FAO/WHO working group. FAO/WHO, London, Ontario, Canada.
- 20.Fox, J. G., and T. C. Wang. 2001. Helicobacter pylori—not a good bug after all. N. Engl. J. Med. 345:829-832. [DOI] [PubMed] [Google Scholar]
- 21.Gotteland, M., O. Brunser, and S. Cruchet. 2006. Systematic review: are probiotics useful in controlling gastric colonization by Helicobacter pylori? Aliment. Pharmacol. Ther. 23:1077-1086. [DOI] [PubMed] [Google Scholar]
- 22.He, T., et al. 2008. Effects of yogurt and bifidobacteria supplementation on the colonic microbiota in lactose-intolerant subjects. J. Appl. Microbiol. 104:595-604. [DOI] [PubMed] [Google Scholar]
- 23.Herrera, V., and J. Parsonnet. 2009. Helicobacter pylori and adenocarcinoma. Clin. Microbiol. Infect. 15:971-976. [DOI] [PubMed] [Google Scholar]
- 24.IARC Working Group. 1994. IARC working group on the evaluation of carcinogenic risks to humans: some industrial chemicals Lyon, 15-22 February 1994. IARC Monogr. Eval. Carcinog. Risks Hum. 60:1-560.7869568 [Google Scholar]
- 25.Izquierdo, I., M. Medina, S. Ennahar, E. Marchioni, and Y. Sanz. 2008. Resistance to simulated gastrointestinal conditions and adhesion to mucus as probiotic criteria for Bifidobacterium longum strains. Curr. Microbiol. 56:613-618. [DOI] [PubMed] [Google Scholar]
- 26.Johnson-Henry, K. C., et al. 2004. Probiotics reduce bacterial colonization and gastric inflammation in H. pylori-infected mice Dig. Dis. Sci. 49:1095-1102. [DOI] [PubMed] [Google Scholar]
- 27.Kabir, A. M., et al. 1997. Prevention of Helicobacter pylori infection by lactobacilli in a gnotobiotic murine model. Gut 41:49-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Klare, I., et al. 2005. Evaluation of new broth media for microdilution antibiotic susceptibility testing of lactobacilli, pediococci, lactococci, and bifidobacteria. Appl. Environ. Microbiol. 71:8982-8986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kumar, R. S., et al. 2006. Structural and functional analysis of a conjugated bile salt hydrolase from Bifidobacterium longum reveals an evolutionary relationship with penicillin V acylase. J. Biol. Chem. 281:32516-32525. [DOI] [PubMed] [Google Scholar]
- 30.Lorca, G. L., T. Wadstrom, G. F. Valdez, and A. Ljungh. 2001. Lactobacillus acidophilus autolysins inhibit Helicobacter pylori in vitro. Curr. Microbiol. 42:39-44. [DOI] [PubMed] [Google Scholar]
- 31.Lu, Y., T. E. Redlinger, R. Avitia, A. S. Galindo, and K. Goodman. 2002. Isolation and genotyping of Helicobacter pylori from untreated municipal wastewater. Appl. Environ. Microbiol. 68:1436-1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Malaty, H., et al. 1991. Transmission of Helicobacter pylori infection. Studies in families of healthy individuals. Scand. J. Gastroenterol. 30:876-879. [DOI] [PubMed] [Google Scholar]
- 33.Margolles, A., L. García, B. Sánchez, M. Gueimonde, and C. G. de los Reyes-Gavilán. 2003. Characterisation of a Bifidobacterium strain with acquired resistance to cholate—a preliminary study. Int. J. Food Microbiol. 82:191-198. [DOI] [PubMed] [Google Scholar]
- 34.Midolo, P. D., J. R. Lambert, R. Hull, F. Luo, and M. L. Grayson. 1995. In vitro inhibition of Helicobacter pylori NC NCTC 11637 by organic acids and lactic acid bacteria. J. Appl. Bacteriol. 79:475-479. [DOI] [PubMed] [Google Scholar]
- 35.Miki, K., et al. 2007. Effect of Bifidobacterium bifidum fermented milk on Helicobacter pylori and serum pepsinogen levels in humans. J. Dairy Sci. 90:2630-2640. [DOI] [PubMed] [Google Scholar]
- 36.Minocha, A. 2009. Probiotics for preventive health. Nutr. Clin. Pract. 24:227-241. [DOI] [PubMed] [Google Scholar]
- 37.Montalban, C., A. Santon, D. Boixeda, and C. Bellas. 2001. Regression of gastric high grade mucosa associated lymphoid tissue (MALT) lymphoma after Helicobacter pylori eradication. Gut 49:584-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nam, H., M. Ha, O. Bae, and Y. Y. Lee. 2002. Effect of Weissella confusa strain PL9001 on the adherence and growth of Helicobacter pylori. Appl. Environ. Microbiol. 68:4642-4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nayak, A. K., and J. B. Rose. 2007. Detection of Helicobacter pylori in sewage and water using a new quantitative PCR method with SYBR green. J. Appl. Microbiol. 103:1931-1941. [DOI] [PubMed] [Google Scholar]
- 40.Nebra, Y., and A. R. Blanch. 1999. A new selective medium for Bifidobacterium spp. Appl. Environ. Microbiol. 65:5173-5176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Parsonnet, J., et al. 1994. Helicobacter pylori infection and gastric lymphoma. N. Engl. J. Med. 330:1267-1271. [DOI] [PubMed] [Google Scholar]
- 42.Pinchuk, I. V., et al. 2001. In vitro anti-Helicobacter pylori activity of the probiotic strain Bacillus subtilis 3 is due to secretion of antibiotics. Antimicrob. Agents Chemother. 45:3156-3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ritter, P., C. Kohler, and U. von Ah. 2009. Evaluation of the passage of Lactobacillus gasseri K7 and bifidobacteria from the stomach to intestines using a single reactor model. BMC Microbiol. 9:87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rowland, M., et al. 2006. Age-specific incidence of Helicobacter pylori. Gastroenterology 130:65-72. [DOI] [PubMed] [Google Scholar]
- 45.Salazar, N., et al. 2009. Exopolysaccharides produced by Bifidobacterium longum IPLA E44 and Bifidobacterium animalis subsp. lactis IPLA R1 modify the composition and metabolic activity of human faecal microbiota in pH-controlled batch cultures. Int. J. Food Microbiol. 135:260-267. [DOI] [PubMed] [Google Scholar]
- 46.Sgouras, D., et al. 2004. In vitro and in vivo inhibition of Helicobacter pylori by Lactobacillus casei strain Shirota. Appl. Environ. Microbiol. 70:518-526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Taylor, D. N., and M. J. Blazer. 1991. The epidemiology of Helicobacter pylori infection. Epidemiol. Rev. 13:42-59. [DOI] [PubMed] [Google Scholar]
- 48.Thiede, C., et al. 2000. Eradication of Helicobacter pylori and stability of remissions in low-grade gastric B-cell lymphomas of the mucosa-associated lymphoid tissue: results of an ongoing multicenter trial. Recent Results Cancer Res. 156:125-133. [DOI] [PubMed] [Google Scholar]
- 49.Tsai, C. C., L. F. Huang, C. C. Lin, and H. Y. Tsen. 2004. Antagonistic activity against Helicobacter pylori infection in vitro by a strain of Enterococcus faecium TM39. Int. J. Food Microbiol. 96:1-12. [DOI] [PubMed] [Google Scholar]
- 50.Ushiyama, A., et al. 2003. Lactobacillus gasseri OLL2716 as a probiotic in clarithromycin-resistant Helicobacter pylori infection. J. Gastroenterol. Hepatol. 18:986-991. [DOI] [PubMed] [Google Scholar]
- 51.Vergara, M., M. Vallve, J. P. Gisbert, and X. Calvet. 2003. Meta-analysis: comparative efficacy of different proton-pump inhibitors in triple therapy for Helicobacter pylori eradication. Aliment. Pharmacol. Ther. 18:647-654. [DOI] [PubMed] [Google Scholar]
- 52.Wotherspoon, A. C., C. Ortiz-Hidalgo, M. R. Falzon, and P. G. Isaacson. 1991. Helicobacter pylori-associated gastritis and primary B-cell gastric lymphoma. Lancet 338:1175-1176. [DOI] [PubMed] [Google Scholar]
- 53.Wotherspoon, A. C., et al. 1993. Regression of primary low-grade B-cell gastric lymphoma of mucosa-associated lymphoid tissue type after eradication of Helicobacter pylori. Lancet 342:575-577. [DOI] [PubMed] [Google Scholar]
- 54.Yildirim, Z., D. K. Winters, and M. G. M. G. Johnson. 1999. Purification, amino acid sequence and mode of action of bifidocin B produced by Bifidobacterium bifidum NCFB 1454. J. Appl. Microbiol. 86:45-54. [DOI] [PubMed] [Google Scholar]