Abstract
The fungus Aspergillus niger forms (sub)millimeter microcolonies within a liquid shaken culture. Here, we show that such microcolonies are heterogeneous with respect to size and gene expression. Microcolonies of strains expressing green fluorescent protein (GFP) from the promoter of the glucoamlyase gene glaA or the ferulic acid esterase gene faeA were sorted on the basis of diameter and fluorescence using the Complex Object Parametric Analyzer and Sorter (COPAS) technology. Statistical analysis revealed that the liquid shaken culture consisted of two populations of microcolonies that differ by 90 μm in diameter. The population of small microcolonies of strains expressing GFP from the glaA or faeA promoter comprised 39% and 25% of the culture, respectively. Two populations of microcolonies could also be distinguished when the expression of GFP in these strains was analyzed. The population expressing a low level of GFP consisted of 68% and 44% of the culture, respectively. We also show that mRNA accumulation is heterogeneous within microcolonies of A. niger. Central and peripheral parts of the mycelium were isolated with laser microdissection and pressure catapulting (LMPC), and RNA from these samples was used for quantitative PCR analysis. This analysis showed that the RNA content per hypha was about 45 times higher at the periphery than in the center of the microcolony. Our data imply that the protein production of A. niger can be improved in industrial fermentations by reducing the heterogeneity within the culture.
The germination of fungal spores results in the formation of hyphae that grow at their apices and that branch subapically. As a result, a network of interconnected cells is formed, which is called a mycelium. Such a mycelium can be small (microcolonies with a diameter in the [sub]millimeter range) or large (macrocolonies with a diameter in the centimeter-to-meter range). The cytoplasm within these mycelia is believed to be continuous. This is due to the fact that the septa that divide hyphae into compartments have large pores that allow intercompartmental and intercellular streaming of molecules and even organelles (9).
Filamentous fungi secrete large amounts of enzymes into the environment. These enzymes degrade the substrate into small molecules that can be taken up by the fungus to serve as nutrients. Aspergillus niger is an example of a fungus with an enormous secretion capacity. Some strains secrete up to 30 g of protein per liter (6), which makes this fungus an important production platform for industrial and pharmaceutical proteins (3, 16). A. niger forms macrocolonies on a solid substrate. Using such colonies, it was shown previously that proteins are secreted only by a limited number of growing hyphae within the mycelium (20). Over and above this, not every growing hypha secretes a particular protein. For instance, glucoamylase is secreted by growing hyphae at the periphery of a macrocolony of A. niger but not by the growing hyphae in the central zone (20). In contrast, lignin peroxidase is secreted in the central growth zone but not at the periphery of macrocolonies of Phanerochaete chrysosporium (14). Taken together, it can be concluded that a fungal macrocolony is not a mass of identical hyphae. Indeed, RNA profiles of concentric zones of macrocolonies of A. niger are distinct (12). For instance, 9% of the genes that are active in a macrocolony are expressed in only one of five concentric zones. Moreover, more than 25% of the active genes show at least a 2-fold difference in expression between the outer and innermost zones of the colony. For example, the level of expression of the glucoamylase gene glaA was more than 3-fold higher at the periphery of maltose-grown colonies than at the center of the mycelium. Similarly, the level of expression of the ferulic acid esterase gene faeA was five times higher at the periphery of xylose-grown colonies. The differences in gene expression within a macrocolony of A. niger can be explained by the availability of the carbon source and, to a similar extent, by medium-independent mechanisms (12). Differences in zonal expression have also been found for macrocolonies of Neurospora crassa (10) and Aspergillus oryzae (13), suggesting that this is a widespread phenomenon in the fungal kingdom.
In industry, A. niger is grown in bioreactors. Under this condition, A. niger forms microcolonies instead of macrocolonies. Here, we addressed whether microcolonies within a liquid shaken culture of A. niger are heterogeneous with respect to size and gene expression. Moreover, we assessed whether zones within individual microcolonies are heterogeneous with respect to gene expression. To this end, the laser microdissection and pressure catapulting (LMPC) technique was used, as was the Complex Object Parametric Analyzer and Sorter (COPAS) technique. In fact, the COPAS technique was used for the first time in microbiology.
MATERIALS AND METHODS
Strains.
A. niger strains N593 (pyrA6 cspA1) (8), AR9#2 (17), and UU-A005.4 (18) were used in this study. These strains are derivatives of A. niger N402 (2). Strains AR9#2 and UU-A005.4 express green fluorescent protein (GFP) from the glaA and faeA promoters, respectively. Strain AR9#2 contains 7 copies of the reporter construct, which have integrated at 4 different sites in the genome. Strain UU-A005.4 contains about 20 copies of the reporter construct, which have integrated at 13 different positions in the chromosome.
Media and culture conditions.
Spores of A. niger strain AR9#2 were isolated from minimal medium plates containing 3% xylose, whereas spores of UU-A005.4 and N593 were isolated from plates containing 2% glucose. Xylose represses the expression of glaA (1), while glucose represses the expression of faeA (4). Liquid shaken cultures were inoculated with 7 × 108 spores ml−1 and incubated for 16 h at 30°C and 250 rpm in 1-liter Erlenmeyer flasks with 250 ml transformation medium (TM) (11). Maltose (50 mM) or xylose (50 mM) was used as a carbon source. N593 cells were grown in the presence of uridine (0.2 g liter−1), while nicotinamide (1 mg liter−1), leucine (0.2 g liter−1), and arginine (0.2 g liter−1) were added in the case of UU-A005.4.
Flow cytometry using COPAS Plus.
Microcolonies were harvested by filtration over a Büchner funnel with nylon gauze, washed with 50 ml phosphate-buffered saline (PBS), and fixed for 15 min at room temperature with 4% formaldehyde in PBS. The fixative was removed by washing 2 times in excess PBS. Microcolonies were simultaneously sorted based on size (time of flight [TOF]) and fluorescence using a COPAS Plus profiler equipped with a 1-mm nozzle (Union Biometrica, Holliston, MA). Sorting parameters were set in such a way that clusters of colonies or debris were excluded from the analysis. GFP fluorescence was detected with a 488-nm solid-state laser combined with a green PMT 514/523-nm optical emission filter. To relate the microcolony diameter to TOF, the diameters of 20 sorted microcolonies were determined by microscopy. These measurements showed that the relationship between colony diameter and TOF can be described as follows: diameter (μm) = 0.46 × TOF + 250.
Laser microdissection and laser pressure catapulting.
The laser microdissection and laser pressure catapulting (LMPC) technique was performed with the Palm CombiSystem (Carl Zeiss MicroImaging, Munich, Germany) equipped with an Axiovert 200 M Zeiss inverted microscope (Carl Zeiss AG, Oberkochen, Germany) and a 3-chip charge-coupled-device (3CCD) color camera (HV-D30; Hitachi Kokusai Electric Inc., Tokyo, Japan). The 10× objective was routinely used. The 40× objective was used when it was impossible to catapult the mycelium all at one time.
Microcolonies were harvested by filtration over a Büchner funnel with nylon gauze, fixed with 70% ethanol on a 1-mm polyethylene naphthalate (PEN) membrane-covered microscope glass slide (Carl Zeiss MicroImaging, Munich, Germany), and subsequently air dried. Microcolonies of approximately 750 μm were selected and cut by using LMPC. Larger colonies cannot be cut by the laser because they exceed the depth of field of the optical system and are therefore beyond the focus of the laser. Four zones within the microcolonies were assigned by using Palm RoboSoftware (v4.0; Carl Zeiss MicroImaging, Munich, Germany), after which the laser dissected and catapulted these zones from the center (zone 1) to the periphery (zone 4). The laser intensity was set at 80% to cut the inner three zones, whereas a 40% laser intensity was used to cut the PEN membrane in front of the outer hyphae of zone 4. The inner zone 1 could not be catapulted at once because some hyphae were still attached to the hyphae in zone 2. These hyphae were disconnected by using laser pressure catapulting. The catapulting energy was set automatically. To this end, the laser focal point was defined and set at a minimum power still enabling the cutting of the membrane. The microdissected material was catapulted into the cap of a sample tube placed above the section. The cap contained 50 μl RNAlater (Qiagen, Hilden, Germany) to enable RNA extraction.
RNA isolation.
Individual microcolonies or parts thereof were soaked in 50 μl RNAlater (Qiagen, Hilden, Germany), after which the material was snap-frozen in liquid nitrogen in a 2-ml Eppendorf tube to which 2 metal bullets (4.76 mm in diameter) were added. Subsequently, samples were ground in a Micro-Dismembrator U instrument (B. Braun Biotech Int., Melsungen, Germany) in a chilled container at 1,500 rpm for 60 s. The frozen material was taken up in 250 μl Trizol reagent (Invitrogen, Carlsbad, CA) by vortexing. After removing the metal bullets, 200 μl chloroform was added. After mixing well, samples were centrifuged at 10,000 × g for 10 min. The water phase (usually around 200 μl) was mixed with 700 μl RLT from the RNeasy MinElute cleanup kit (Qiagen, Hilden, Germany), to which 143 mM β-mercaptoethanol was added. RNA was purified according to the instructions provided with the kit. The purified RNA was eluted by using 14 μl RNase-free water.
cDNA synthesis and quantitative PCR analysis.
cDNA was synthesized from purified total RNA by using the QuantiTect reverse transcription kit (Qiagen, Hilden, Germany). Quantitative PCR (QPCR) was performed by using ABI Prism 7900HT SDS and SYBR green chemistry (Applied Biosystems, Carlsbad, CA). Cycle threshold (CT) levels were measured for the 18S rRNA gene, the actin gene, glaA, and faeA. Primers were designed according the recommendations of the manufacturer of the PCR master mix (Applied Biosystems, Carlsbad, CA). Levels of actin gene mRNA were determined with primers QPCRactFW1 and QPCRactRV1, and those of the 18S rRNA gene were determined with primers QPCR18SFW1 and QPCR18SRV1 (Table 1). These products had an amplification efficiency of 2. cDNAs of glaA and faeA were amplified by using primers QPCRglaAFW3 and QPCRglaARV3 and primers QPCRfaeAFW4 and QPCRfaeARV4, respectively (Table 1). The glaA cDNA was amplified with an efficiency of 1.98, and the faeA cDNA was amplified with an efficiency of 1.92.
TABLE 1.
Primers used in this study
| Primer | Sequence |
|---|---|
| QPCRactFW1 | 5′-GTTGCTGCTCTCGTCATT-3′ |
| QPCRactRV1 | 5′-AACCGGCCTTGCACATA-3′ |
| QPCR18SFW1 | 5′-GGCTCCTTGGTGAATCATAAT-3′ |
| QPCR18SRV1 | 5′-CTCCGGAATCGAACCCTAAT-3′ |
| QPCRglaAFW3 | 5′-GCACCAGTACGTCATCAA-3′ |
| QPCRglaARV3 | 5′-GTAGCTGTCAGATCGAAAGT-3′ |
| QPCRfaeAFW4 | 5′-GACGGCATCCCAAACTT-3′ |
| QPCRfaeARV4 | 5′-CTCACAGCACTGTACTTCAT-3′ |
Acridine orange staining and fluorescence microscopy.
A. niger microcolonies were stained with acridine orange as described previously (7). Microcolonies were fixed in 70% ethanol, after which a 1/10 volume of 4 mM acridine orange in PBS (Becton Dickinson, Franklin Lakes, NJ) was added. After staining for 5 min, the microcolonies were washed twice with PBS. The fluorescence of acridine orange was monitored by using a Zeiss Axioscope 2 Plus instrument (Carl Zeiss AG, Oberkochen, Germany) equipped with an HBO 100-W mercury lamp and a Leica LFC 420C camera (2,592 by 1,944 pixels) using standard fluorescein isothiocyanate (FITC) and tetramethyl rhodamine isocyanate (TRITC) filters.
Statistical analysis.
The statistical significance of differences in QPCR data was tested by using a t test. Flow cytometry data were subjected to descriptive statistics using SPSS software. The normality of the data was tested by using the Kolmogorov-Smirnov (KS) test with the Lilliefors correction. To assess whether distributions in size and fluorescence can be explained by a mixture of two normally distributed components, the data were modeled in the following probability distribution (19): ϕ:ϕ(x) = pN(x; μ1, σ1) + (1 − p)N(x; μ2, σ2), where 0 < p < 1 and where x ↛ N(x; μ, σ) is the probability density of the normal distribution with parameters μ and σ. This model describes a mixture of an N(μ1; σ1) distribution and an N(μ2; σ2) distribution in which the degree of participation of the component N(μ1; σ1) is p. The confidence interval for the participation frequency of component 1 was set to 95%. In the statistical analysis the five parameters in the model (p, μ1, σ1, μ2, and σ2) were fit to the empirical data by means of the maximum likelihood principle. Bootstrapping (500 replicates) was used to obtain interval estimates for the parameters. The fit procedures were implemented with Scilab software. The scripts of the Scilab functions are available at http://www.bio.uu.nl/∼microbio/Microbiology/Tools.htm.
RESULTS
Fluorescence and volumes of microcolonies are not normally distributed.
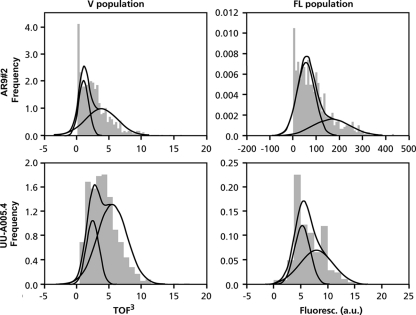
A. niger strains AR9#2 and UU-A005.4 express GFP from the promoter of the glucoamlyase gene glaA and the feruoyl esterase gene faeA, respectively. These strains were grown as liquid shaken cultures in medium containing 50 mM maltose and 50 mM xylose, respectively. Maltose induces the AmyR-regulated gene glaA (1, 15), whereas xylose induces the XlnR-regulated gene faeA (4). After 16 h, microcolonies were fixed, and their size and fluorescence were monitored by using the COPAS technique. The KS test showed that the size and fluorescence of the microcolonies in the cultures of both strains did not follow a normal distribution (P < 0.05) (data not shown). Mathematical modeling showed that the distribution of the volume of the microcolonies within the liquid shaken cultures of strains AR9#2 and UU-A005.4 can be explained by two normally distributed populations (Fig. 1 and Table 2, and see Table S1 in the supplemental material). The cultures of strains AR9#2 and UU-A005.4 consisted of 61% and 75%, respectively, of microcolonies with an average diameter of 595 μm, whereas 39% and 25% of the cultures, respectively, measured 505 μm on average. Mathematical modeling also showed that the fluorescence distribution of GFP of microcolonies within liquid shaken cultures of strains AR9#2 and UU-A005.4 can be explained by two normally distributed populations (Fig. 1, Table 2, and Table S2). Highly fluorescent microcolonies comprised 56% of the culture of strain UU-A005.4, whereas 44% of the microcolonies made up the lowly fluorescent population. The highly fluorescent population of strain AR9#2 consisted of 32% of the microcolonies, while the lowly fluorescent population comprised 68% of the microcolonies (Fig. 1, Table 2, and Table S2). The volume and GFP expression of microcolonies were also heterogeneous for other strains tested (Tables S3 and S4). These data show that heterogeneity is not the result of the copy number and the site of integration.
FIG. 1.
Heterogeneity of microcolonies of A. niger in liquid shaken cultures with respect to size and gene expression. The dark gray areas represent the distribution of volume (V population) and glaA and faeA expression (FL population) of microcolonies within liquid cultures. These distributions can be explained by assuming the existence of two populations of microcolonies (indicated by the curves). Strains AR9#2 and UU-A005.4 express GFP from the glaA and the faeA promoters, respectively. a.u., arbitrary units.
TABLE 2.
Volume and fluorescence distribution of microcolonies in liquid shaken cultures of A. niger can be explained by the existence of two populations of microcoloniesa
| Strain | Reporter construct | No. of analyzed microcolonies | Participation level of microcolonies with: |
|||
|---|---|---|---|---|---|---|
| Low V | High V | Low FL | High FL | |||
| AR9#2 | PglaA::GFP | 662 | 0.39 | 0.61 | 0.68 | 0.32 |
| UU-A005.4 | PfaeA::GFP | 522 | 0.25 | 0.75 | 0.44 | 0.56 |
FL, fluorescence; V, microcolony volume (TOF).
Heterogeneous RNA distribution within microcolonies.
QPCR was used to determine glaA and faeA expression levels in individual microcolonies of strain AR9#2. The expression levels of these genes was related to the expression levels of 18S rRNA gene and the mRNA of the actin gene. RNA was isolated from individual microcolonies with a diameter of 750 to 800 μm that had been formed in liquid shaken cultures containing either 50 mM maltose, 50 mM xylose, or 110 mM glucose as a carbon source. QPCR showed that the accumulation of faeA mRNA was 3,000-fold higher in xylose-grown microcolonies than in maltose- and glucose-grown microcolonies. The glaA gene had a high level of expression on maltose, whereas expression levels were 2.4 and 20 times lower on glucose and xylose, respectively. These data agree with data from previously reported Northern analyses of RNA from whole liquid shaken cultures (1, 4) and show that the RNA extraction and QPCR procedures that were developed to assess gene expression in individual microcolonies (see Materials and Methods for details) are reliable and reproducible.
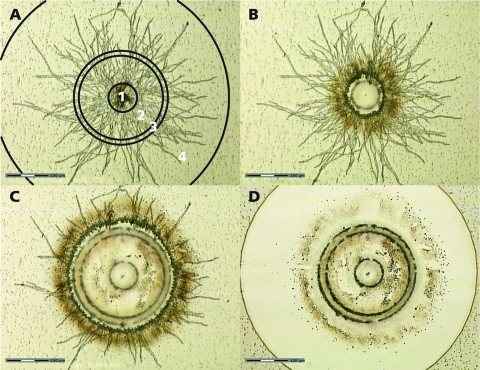
In the next step, glaA and faeA RNA levels in the center and periphery of individual microcolonies were determined. To this end, ethanol-fixed microcolonies of approximately 750 μm were cut by using laser microdissection and pressure catapulting (LMPC). Four zones were distinguished in the microcolonies. Zone 1 represents the inner 100 μm and consists of aggregated spores. The central zone 2 represents the hyphae that are present between 100 and 300 μm. This zone is separated from the peripheral zone 4 by zone 3, which extends to 350 μm (Fig. 2A). To isolate RNA from zones 2 and 4, the central zone 1 was first dissected and catapulted to remove it from the rest of the microcolony (Fig. 2B). This was followed by cutting zone 2 and by catapulting the hyphal material of this zone into a cap containing 50 μl of RNAlater (Fig. 2C). After removing zone 3 with the laser, zone 4 was cut. Hyphae of this zone were also catapulted in RNAlater (Fig. 2D). Expression levels of the 18S rRNA, actin, glaA, and faeA genes in zones 2 and 4 of individual microcolonies were determined by QPCR. Similar CT values were obtained in the case of the central zone 2 and the peripheral zone 4 (P ≥ 0.95) (Table 3). However, the RNA from the central zone 2 was derived from 45-times-more hyphal material than was the RNA from the peripheral zone 4, which contained approximately 100 hyphae only. This finding indicates that the RNA levels of these genes per unit mass of hypha are much higher in zone 4 than in zone 2. In agreement with this, acridine orange staining showed that the periphery of microcolonies of A. niger is rich in RNA, while the center is not (5). Acridine orange binds to single- and double-stranded nucleic acids, resulting in red and green fluorescence, respectively. We repeated this experiment in our experimental setup. As expected, red fluorescence (indicative of RNA) was more pronounced in the peripheral zone 4 than was green fluorescence (indicative of DNA), while the reverse was observed for the central zone 2 (Fig. 3).
FIG. 2.
Laser dissection and pressure catapulting of microcolonies of A. niger grown in a liquid shaken culture. (A) Four zones are distinguished in a microcolony. (B) First, the central zone 1 was cut. (C) This was followed by the cutting of zone 2. (D) After the cutting of zone 3, the outer zone 4 was dissected. Hyphal material of each of the zones can be isolated by catapulting.
TABLE 3.
Accumulation of transcripts in the central zone 2 and the peripheral zone 4 of microcolonies of A. nigera
| Zone (no. of hyphae) | Avg CT value (±SD) |
|||
|---|---|---|---|---|
| 18S rRNA gene | Actin gene | glaA | faeA | |
| Center (4,500) | 16.31 (±1.79) | 29.73 (±1.61) | 28.81 (±2.42) | 33.86 (±2.25) |
| Periphery (100) | 16.42 (±1.13) | 28.02 (±2.33) | 27.14 (±1.75) | 31.46 (±2.30) |
CT values were obtained with QPCR. QPCR was performed on the total RNA extracted from the 4,500 hyphae of zone 2 and the 100 hyphae of zone 4. Biological triplicates were used to calculate the average CT values and the standard deviations. glaA encodes glucoamylase, and faeA encodes ferulic acid esterase.
FIG. 3.
Heterogeneous distribution of RNA in microcolonies of a liquid shaken culture of A. niger. (A and B) Acridine orange staining shows that double-stranded DNA is present mainly in the center of a microcolony (A), whereas single-stranded mRNA is observed predominantly at the periphery (B). (C) Overlay of A and B. Zones within the colony are indicated by vertical lines.
DISCUSSION
Previously, it was shown that zones of centimeter-scale macrocolonies of A. niger are heterogeneous with respect to gene expression and protein secretion (12, 20). However, in liquid cultures, such as those of industrial bioreactors, (sub)millimeter-scale microcolonies are formed. These microcolonies are exposed to a more homogeneous medium than are macrocolonies that are grown on a static solid medium. This fact and the enormous size difference between micro- and macrocolonies raised the question of whether zones in microcolonies of A. niger are also heterogeneous in RNA abundance and composition. Moreover, we addressed for the first time whether a liquid shaken culture of A. niger consists of a homogeneous population of microcolonies or whether distinct populations of microcolonies can be distinguished that differ in size and gene expression.
Microcolonies of 800 μm were subjected to laser microdissection and laser pressure catapulting, which enabled us to isolate RNA from the central zone 2 and the peripheral zone 4. QPCR could not show differences in the levels of 18S rRNA and of RNAs of the actin gene, glaA, and faeA between the zones. However, the RNA from the central zone 2 originated from 45-times-more hyphae than did the RNA from the peripheral zone 4. Acridine orange staining confirmed that the hyphae in zone 2 contained less RNA, while staining with propidium iodide showed that ≥99% of the hyphae were alive (our unpublished results). The huge difference in RNA abundances between the periphery and the center was not observed for macrocolonies grown on a solid medium (our unpublished results). We have no explanation for this finding. The size and open structure of the microcolonies suggest that the center was not affected in the uptake of nutrients and transfer of gases.
The COPAS technique was used to assess whether microcolonies in a liquid shaken culture of A. niger are heterogeneous with respect to volume and the expression of glaA and faeA. Statistical analyses showed that 16-h-old cultures consist of two populations of pellets that can be distinguished on the basis of their volume. The small pellets generally consist of about 25% of the population. Two populations of microcolonies could also be distinguished in the case of the expression of glaA and faeA. Interestingly, the population of microcolonies with low glaA and faeA expression levels was larger than the population of small microcolonies. This indicates that heterogeneity in glaA and faeA expression in a liquid shaken culture of A. niger is only partially caused by the heterogeneity in the size of the microcolonies and, thus, also depends on an unknown other factor. This also seems to be the case for the expression of the acid amylase gene aamA, which, like glaA, is regulated by AmyR (see Table S4 in the supplemental material).
The fact that microcolonies within liquid cultures are heterogeneous with respect to size and gene expression has implications for how data from analyses of RNA, proteins, and metabolites from whole cultures should be interpreted. By using the whole culture, an average composition or activity of the microcolonies is determined. This average may by far not reflect the composition or activity of each of the populations within the liquid culture. Therefore, individual populations should be studied to understand the mechanisms underlying biological processes in a liquid culture. The COPAS technique enables the rapid sorting of populations of living or fixed microcolonies. We envision that the level of protein production in liquid cultures can be increased by reducing the heterogeneity of the microcolonies.
Supplementary Material
Acknowledgments
This research was supported by the IOP Genomics program of the Dutch Ministry of Economics Affairs and by the Dutch Technology Foundation STW, the Applied Science Division of NWO, and the Technology Program of the Ministry of Economic Affairs.
Footnotes
Published ahead of print on 17 December 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Boel, E., et al. 1984. Glucoamylases G1 and G2 from Aspergillus niger are synthesized from two different but closely related mRNAs. EMBO J. 3:1097-1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bos, C. J., et al. 1988. Genetic analysis and the construction of master strains for assignment of genes to six linkage groups in Aspergillus niger. Curr. Genet. 14:437-443. [DOI] [PubMed] [Google Scholar]
- 3.Conesa, A., P. J. Punt, N. van Luijk, and C. A. M. J. J. van den Hondel. 2001. The secretion pathway in filamentous fungi: a biotechnological view. Fungal Genet. Biol. 33:155-171. [DOI] [PubMed] [Google Scholar]
- 4.de Vries, R. P., and J. Visser. 1999. Regulation of the feruloyl esterase (faeA) gene from Aspergillus niger. Appl. Environ. Microbiol. 65:5500-5503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.El-Enshasy, H., J. Kleine, and U. Rinas. 2006. Agitation effects on morphology and protein productive fractions of filamentous and pelleted growth forms of recombinant Aspergillus niger. Process Biochem. 41:2103-2112. [Google Scholar]
- 6.Finkelstein, D. B., et al. 1989. Protein secretion in Aspergillus niger, p. 295-300. In C. L. Hershberger, S. W. Queener, and G. Hegeman (ed.), Genetics and molecular biology of industrial microorganisms. American Society for Microbiology, Washington, DC.
- 7.Freudenberg, S., K. Fasold, S. R. Müller, D. Siedenberg, G. Kretzmer, K. Schügerl, and M. Giuseppin. 1996. Fluorescence microscopic investigation of Aspergillus awamori growing on synthetic and complex media and producing xylanase. J. Biotechnol. 46:265-273. [Google Scholar]
- 8.Goosen, T., et al. 1987. Transformation of Aspergillus niger using the homologous orotidine-5′-phosphate-decarboxylase gene. Curr. Genet. 11:499-503. [DOI] [PubMed] [Google Scholar]
- 9.Jennings, D. H., J. D. Thornton, M. F. Galpin, and C. R. Coggins. 1974. Translocation in fungi. Symp. Soc. Exp. Biol. 28:139-156. [PubMed] [Google Scholar]
- 10.Kasuga, T., and N. L. Glass. 2008. Dissecting colony development of Neurospora crassa using mRNA profiling and comparative genomics approaches. Eukaryot. Cell 7:1549-1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kusters-van Someren, M. A., J. A. M. Harmsen, H. C. M. Kester, and J. Visser. 1991. Structure of the Aspergillus niger pelA gene and its expression in Aspergillus niger and Aspergillus nidulans. Curr. Genet. 20:293-299. [DOI] [PubMed] [Google Scholar]
- 12.Levin, A. M., et al. 2007. Spatial differentiation in the vegetative mycelium of Aspergillus niger. Eukaryot. Cell 6:2311-2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Masai, K., et al. 2006. Square-plate culture method allows detection of differential gene expression and screening of novel, region-specific genes in Aspergillus oryzae. Appl. Microbiol. Biotechnol. 71:881-891. [DOI] [PubMed] [Google Scholar]
- 14.Moukha, S. M., H. A. B. Wösten, E. Mylius, M. Asther, and J. G. H. Wessels. 1993. Spatial and temporal accumulation of mRNAs encoding two common lignin peroxidases in Phanerochaete chrysosporium. J. Bacteriol. 175:3672-3678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Petersen, K. L., J. Lehmbeck, and T. Christensen. 1999. A new transcriptional activator for amylase genes in Aspergillus. Mol. Gen. Genet. 262:668-676. [DOI] [PubMed] [Google Scholar]
- 16.Punt, P. J., et al. 2002. Filamentous fungi as cell factories for heterologous protein production. Trends Biotechnol. 20:200-206. [DOI] [PubMed] [Google Scholar]
- 17.Siedenberg, D., et al. 1999. GlaA promoter controlled production of a mutant green fluorescent protein (S65T) by recombinant Aspergillus niger during growth on defined medium in batch and fed-batch cultures. Biotechnol. Prog. 15:43-50. [DOI] [PubMed] [Google Scholar]
- 18.Vinck, A., et al. 16 August 2010, posting date. Heterogenic expression of genes encoding secreted proteins at the periphery of Aspergillus niger colonies. Environ. Microbiol. doi: 10.1111/j.1462-2920.2010.02322.x. [DOI] [PubMed]
- 19.Vinck, A., et al. 2005. Hyphal differentiation in the exploring mycelium of Aspergillus niger. Mol. Microbiol. 58:693-699. [DOI] [PubMed] [Google Scholar]
- 20.Wösten, H. A. B., S. M. Moukha, J. H. Sietsma, and J. G. H. Wessels. 1991. Localization of growth and secretion of proteins in Aspergillus niger. J. Gen. Microbiol. 137:2017-2023. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.