Abstract
The psychrophilic model bacterium Pseudoalteromonas haloplanktis is characterized by remarkably fast growth rates under low-temperature conditions in a range from 5°C to 20°C. In this study the proteome of cellular compartments, the cytoplasm and periplasm, of P. haloplanktis strain TAC125 was analyzed under exponential growth conditions at a permissive temperature of 16°C. By means of two-dimensional protein gel electrophoresis and mass spectrometry, a first inventory of the most abundant cytoplasmic and periplasmic proteins expressed in a peptone-supplemented minimal medium was established. By this approach major enzymes of the amino acid catabolism of this marine bacterium could be functionally deduced. The cytoplasmic proteome showed a predominance of amino acid degradation pathways and tricarboxylic acid (TCA) cycle enzymes but also the protein synthesis machinery. Furthermore, high levels of cold acclimation and oxidative stress proteins could be detected at this moderate growth temperature. The periplasmic proteome was characterized by a significant abundance of transporters, especially of highly expressed putative TonB-dependent receptors. This high capacity for protein synthesis, efficient amino acid utilization, and substrate transport may contribute to the fast growth rates of the copiotrophic bacterium P. haloplanktis in its natural environments.
The Antarctic marine environment is characterized by low temperatures that very seldom exceed 0°C. Despite the permanent low temperatures, the surface water and the pack ice zones in this region harbor a surprisingly high level of microbial activity. The most abundant bacterial populations in sea ice samples from Antarctic coastal areas belong mainly to four phylogenetic groups: the Gram-positive branch, the Flexibacter-Bacteroides-Cytophaga phylum, and the alpha and gamma subdivisions of the proteobacteria (4). Up to 50% of the identified bacterial isolates could be assigned to the gammaproteobacteria (5). It is assumed that this group of heterotrophic bacteria plays a key role in carbon cycling in this extreme environment.
A prominent, cold-adapted member of this group is the obligatory marine bacterium Pseudoalteromonas haloplanktis TAC125, which was isolated from an Antarctic coastal seawater sample (32). P. haloplanktis features remarkable metabolic versatility and has become a model organism of psychrophilic bacteria. Several cold-adapted proteins of this species have been characterized, such as aspartate aminotransferase (2), polynucleotide phosphorylase (18), DNA ligase (25), cytochrome c (15), superoxide dismutase (33), lipase (14), esterases (46), xylanase (10), cellulase (47), and alpha-amylase (21, 43). As expected, most of these enzymes possess strong catalytic activities at lower temperatures and need less activation energy than do their mesophilic or thermophilic counterparts (31). It is worth noting that, quite unexpectedly and presumably associated with the cold salt-washing process, P. haloplanktis has also been found to develop routinely on the rind of cheese (34).
The genome sequencing of P. haloplanktis strain TAC125 (32) revealed nine rRNA gene clusters and a large number of tRNA genes (106 genes). It was speculated that the large number of rRNA and tRNA genes may contribute to psychrophilic adaptation and supports the fast growth of the organism in the cold. Recently, however, it was suggested that a high number of rRNA genes reflects an ecological strategy of bacteria to better respond to perturbations in nutrient resources (28).
P. haloplanktis isolates appear to be well adapted for growth under nutrient-enriched conditions (32). Several P. haloplanktis strains have been isolated from fish and mussels (16, 24, 40). Furthermore, this species has been employed as a model organism in studies of how marine copiotrophic bacteria detect and exploit nutrient pulses (27, 44).
The availability of the genome sequence now allows global analyses of the cellular physiology of this bacterium. A first proteomic analysis led to the identification of differently expressed proteins in the presence of l-malate, including a two-component regulatory system which seems to play a role in the control of malate-specific pathways in P. haloplanktis TAC125 (37). In a recent proteome study it was shown that translation, protein folding, membrane integrity, and antioxidant activities of P. haloplanktis are upregulated at 4°C (38). It was shown that the trigger factor is the main upregulated protein at low temperatures.
At 16°C the doubling time of P. haloplanktis is more than three times faster than that of Escherichia coli under similar growth conditions (38). Recently, a P. haloplanktis expression system (12, 17) was developed, which enables the use of this host for the low-temperature heterologous expression of otherwise “intractable” proteins. Furthermore, a first fed-batch cultivation strategy for this alternative expression host was established (50). However, the physiology and metabolic pathways of this bacterium, which facilitate remarkably high growth rates on complex substrates, are so far unknown. Therefore, the aim of this study was the determination of the protein inventory of P. haloplanktis under permissive growth conditions with a complex amino acid mixture as the only carbon and nitrogen source. For this purpose the cytoplasmic and periplasmic proteome was fractionated, and the most abundant proteins located in these two cellular compartments of P. haloplanktis TAC125 during exponential growth were analyzed. With this approach major enzymes and reactions relevant for the amino acid degradation pathways of P. haloplanktis could be identified, which would not have been considered for these functions from the published genome sequence alone. Finally, potential strategies relevant for the fast growth of this psychrotolerant bacterium could be detected under conditions relevant to a future biotechnological application of this species.
MATERIALS AND METHODS
Bacterial strain and growth conditions.
P. haloplanktis TAC125 is available from the Collection de l'Institut Pasteur (Paris, France) under accession number CIP 108707. Precultures were grown in ZoBell medium (52) at 16°C at 200 rpm. The main cultures were performed at 16°C at 150 rpm in 5-liter or 3-liter shake flasks with a culture volume of 600 ml or 1,000 ml of a modified SHATZ medium (45). The modified SHATZ medium contained 5 g/liter peptone N-Z-Soy BL4 (Sigma), 0.14 g/liter KH2PO4, 1 g/liter NH4NO3, 10 g/liter NaCl, 0.2 g/liter MgSO4·7H2O, 0.01 g/liter FeSO4·7H2O, and 0.01 g/liter CaCl2·2H2O. All main cultures were inoculated with exponentially growing cells to a starting optical density at 540 nm (OD540) of about 0.2.
Preparation of the cytoplasmic and periplasmic protein fractions.
P. haloplanktis TAC125 cells were harvested during exponential growth at an OD540 of 1 by centrifugation at 6,000 × g for 15 min at 4°C. Bacterial pellets were washed in Tris-EDTA (TE) buffer, and the purification of the periplasmic protein fraction was performed according to a method described previously by Watt et al. (49). In brief, the pellet was washed with 1 M NaCl at 4°C, centrifuged as described above, resuspended in sucrose buffer (20% [wt/vol] sucrose, 1 mM EDTA, and one Complete Mini protease inhibitor tablet per 10 ml of buffer), and incubated for 5 min at room temperature (RT) to disrupt the outer membrane and extract the periplasmic proteins. After a further round of centrifugation (30 min at 8,000 × g at 4°C), the supernatant containing the periplasmic proteins was decanted. By resuspension of the resulting pellet in 5 ml of double-distilled water at 4°C, the cytoplasmic proteins were extracted. The periplasmic proteins in the supernatant were precipitated overnight at −20°C with 20 ml ice-cold acetone and spun down at 8,000 × g for 45 min at 4°C. After the centrifugation step, the proteins were washed with 5 ml 96% (vol/vol) ethanol and prepared for two-dimensional (2-D) polyacrylamide gel electrophoresis (2-DE) analysis as described previously by Antelmann et al. (1).
2-D gel electrophoresis, protein identification, and image analysis.
The isoelectric focusing (IEF) of 300 μg protein per sample was performed by using the Multiphor system (Amersham Biosciences) with nonlinear immobilized pH gradient strips with a pH range of 3 to 10 (GE Healthcare) (7). The separation according to protein mass was conducted with a Protean Plus Dodeca cell (Bio-Rad) according to a method described previously by Voigt et al. (48). Protein spots were stained with Coomassie brilliant blue (CBB) G250.
All visible spots on the 2-D gels were cut either manually or with a Spot Cutter system (Bio-Rad), digested with trypsin, and analyzed by using matrix-assisted laser desorption ionization-time of flight (MALDI-ToF) mass spectrometry (MS). The measurements were carried out with a Proteome-Analyzer 4700 (Applied Biosystems) and evaluated by using the “peak-to-mascot” script of 4700 Explorer software as previously described by Eymann et al. (20). Peak lists were compared to the sequence database by using the Mascot search engine (Matrix Science). Proteins that yielded twice a Mowse score of at least 49 and a sequence coverage of at least 30% were considered positive identifications. Image analysis and color coding of the scanned 2-D gels were accomplished with DECODON Delta 2D software.
RESULTS AND DISCUSSION
The objective of this study was to determine proteomic signatures and, thus, key enzymes relevant for P. haloplanktis cells under fast growth conditions. The genome sequence of P. haloplanktis TAC125 indicated that this bacterium does not encode a cyclic AMP (cAMP)-catabolite activator protein (CAP) complex that regulates carbon availability in related organisms (32). Also, a phosphoenolpyruvate-dependent phosphotransferase system (PTS) for the transport and first metabolic step of so-called PTS sugars is missing in this marine bacterium, which is the main reason for P. haloplanktis' deficient growth on glucose. It was shown previously by several studies that P. haloplanktis grows with remarkably high growth rates in defined seawater medium with peptone as the only carbon and nitrogen source. It is assumed that these growth conditions resemble the preferred natural environment of this marine bacterium, which can easily be isolated from damaged tissues of fishes or oysters, where such complex protein substrates are available.
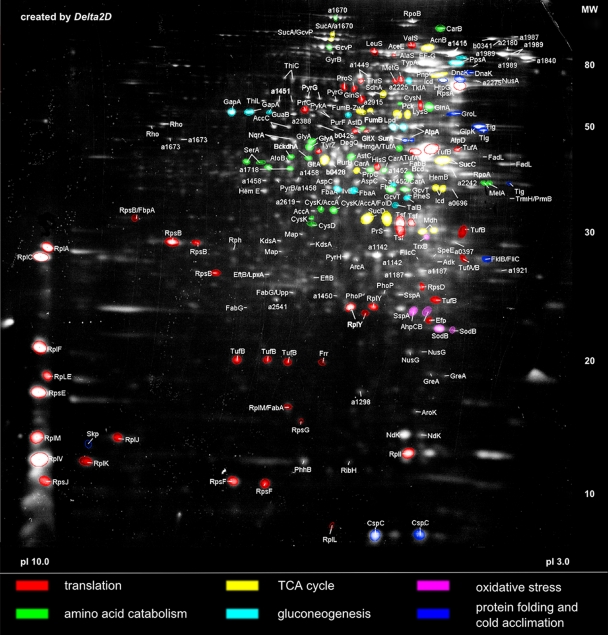
The soluble proteome of the major cellular compartment, the cytoplasm, isolated from P. haloplanktis TAC125 cells grown exponentially at 16°C with peptone as the only carbon source was separated at a pH range of 3 to 10 (Fig. 1). Of the visible protein spots in the cytoplasmic proteome, the most abundant 310 spots were excised. Two hundred seventy-seven of these proteins could be unambiguously identified by MALDI-ToF MS.
FIG. 1.
The most abundant functional categories of the cytoplasmic proteome of P. haloplanktis TAC125. The protein fraction was two-dimensionally separated at a pH range of 3 to 10. Specific functional groups are indicated by the different colors of the protein spots. MW, molecular weight (in thousands).
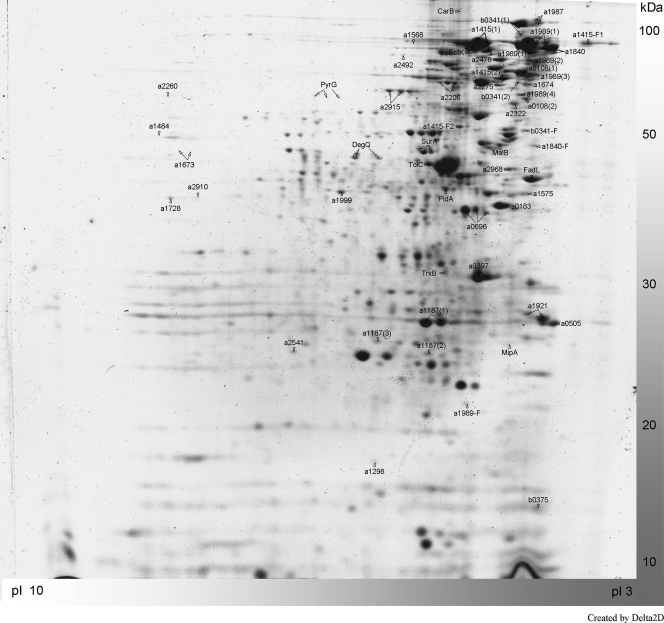
Furthermore, the periplasmic protein fraction was isolated and also separated at a pH range of 3 to 10 (Fig. 2). The image analysis of the electrophoretically separated periplasmic P. haloplanktis TAC125 proteome resulted in 381 discernible abundant protein spots, from which 293 proteins could be unequivocally identified by MALDI-ToF MS. Overall, 212 different unique proteins were detected in both fractions and assigned to a functional category based on the classification described previously by Medigue et al. (32) (see Table S1 in the supplemental material).
FIG. 2.
Two-dimensional gel electrophoresis of the periplasmic protein fraction of P. haloplanktis TAC125 at a pH range of 3 to 10. The most abundant protein spots with predicted signal sequences are labeled.
Cytoplasmic proteome.
The most abundant functional categories in the cytoplasmic proteome in terms of protein numbers were protein synthesis (44), amino acid metabolism (38), carbohydrate metabolism (28), nucleotide metabolism (13), and lipid metabolism (8) (Fig. 1, and see Table S1 in the supplemental material).
Altogether, 18 of the 50 annotated ribosomal proteins could be identified in the cytoplasmic proteome. Many of the corresponding protein spots had high densities, and the distinct alkaline predominance in the cytoplasmic 2-D gel (Fig. 1) can be attributed largely to ribosomal proteins. This functional category incurs by far the highest sum of spot quantitation values in the cytoplasmic proteome (approximately 13% of the relative spot volume [data not shown]). Furthermore, a high abundance of translation elongation and termination factors (e.g., EF-Tu, EF-Ts, EF-P, RRF, and PrfC) was shown. Moreover, half of the annotated aminoacyl-tRNA synthetases (13 out of 24 subunits) could be detected in the proteome. Viewed in the context of the strong quantitative prevalence of ribosomal proteins, the high qualitative appearance of aminoacyl-tRNA synthetases backs the assumed pivotal role of the protein synthesis apparatus for ensuring fast growth. The proteomic prominence of the translational machinery could also reflect an adaptive response in order to compensate for impaired translational efficiency under low-temperature conditions. Piette et al. (38) previously inferred that 30% of the identified cold acclimation proteins in P. haloplanktis TAC125 are related directly to protein synthesis. However, mesophilic fast-growing bacteria like Bacillus subtilis or Bacillus licheniformis also reveal similar high spot intensities of ribosomal proteins or other translation-specific proteins under optimal growth conditions (7, 48). In general, the expression of these protein functions is usually growth rate dependent (29).
Also, a relatively high abundance of all tricarboxylic acid (TCA) cycle enzymes (Fig. 1) is required for the efficient catabolism of the peptone-based amino acids. The high abundance of these housekeeping protein functions is in line with the remarkably high maximal growth rate of 0.35 h−1 (i.e., a doubling time of approximately 2 h) of P. haloplanktis TAC125 under the growth conditions investigated in this study. The doubling time of P. haloplanktis at 16°C is thus almost three times faster than that of E. coli under similar growth conditions (38). In line with this high growth rate and as reported recently, at room temperature P. haloplanktis reveals a very efficient chemotactic response, which is 10 times faster than that of E. coli and presumably allows the organism to exploit nutrient patches in the marine environment before they dissipate (44).
The fast-growth behavior of P. haloplanktis at 16°C might also be supported by the abundance of the cold shock protein CspC (PSHAa1184) and the trigger factor Tig (PSHAa2063), which was suggested to be the primary chaperone of P. haloplanktis under low-temperature conditions (38). Besides, a putative cold-active aminopeptidase (PSHAa2915) and five other aminopeptidases (PSHAa1484, Map [PSHAa2037], Dcp [PSHAa2184], PSHAa2388, and PSHAa2492) could be detected. These enzymes contain different peptidase superfamily motifs (peptidase M1, PSHAa2492 and PSHAa2915; peptidase M3-like, PSHAa2184; peptidase M17, PSHAa1484 and PSHAa2388; methionine aminopeptidase, PSHAa2037) and possess many close homologs in other members of the Alteromonadales, other proteobacteria, and several other more distant taxa.
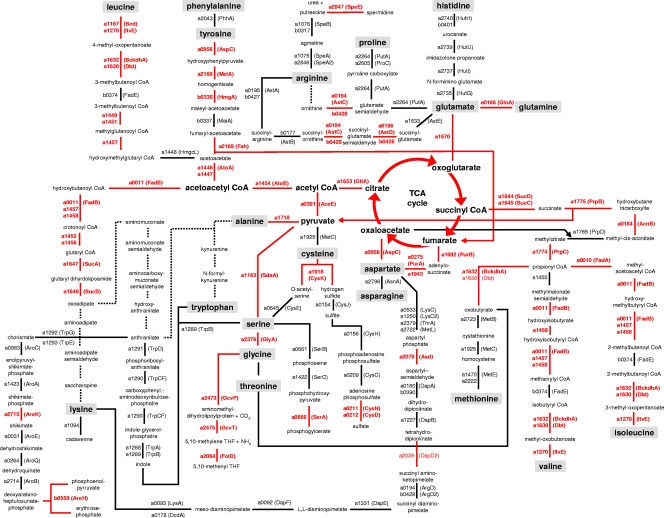
Given the complex amino acid source supplemented into the culture medium, we performed a detailed analysis of the amino acid degradation pathways based on the cytoplasmic proteome and the available genome sequence (Fig. 3). The common amino acid degradation routes are present in P. haloplanktis TAC125, except for those of tryptophan and lysine (upstream of oxoadipate). An alternative way of lysine utilization may be the decarboxylation to cadaverine by PSHAa1094 (annotated as a putative basic amino acid decarboxylase). Since the organism can synthesize both amino acids and enzymes from the biosynthetic pathways were present in the proteome, we propose that the degradation of these two amino acids proceeds via a reversion of the biosynthetic routes.
FIG. 3.
Chart of amino acid degradation pathways in P. haloplanktis TAC125. Proteins which were identified on the 2-D gels are marked by red lines and gene names; enzymatic reactions of proteins that were not identified but have a coding sequence in the genome are indicated by black lines and gene names. Dashed lines represent enzyme functions without a correlate in the P. haloplanktis TAC125 genome. The gene names were taken from the P. haloplanktis genome database of the MaGe platform (www.genoscope.cns.fr/agc/mage/psychroscope) without the organism code “pshA.” THF, tetrahydrofolate.
Only the arginase (EC 3.5.3.1) is missing in P. haloplanktis TAC125. A lack of arginase is also common in other bacterial species (11) where, like in P. haloplanktis, ornithine can be produced by an alternative route via N-acetylglutamate, N-acetylglutamate semialdehyde, and N-acetylornithine. As shown in Fig. 3, the degradation of arginine to glutamate can be accomplished in P. haloplanktis in five steps via succinylated intermediates.
Noticeably, P. haloplanktis lacks the suite of propionyl-coenzyme A (CoA) carboxylase, methylmalonyl-CoA epimerase, and methylmalonyl-CoA mutase, which converts propionyl-CoA normally derived from valine, isoleucine, methionine, and threonine degradation as well as from the degradation of odd-numbered fatty acids into succinyl-CoA. Instead, the organism utilizes the methylcitrate cycle, which converts propionyl-CoA and oxaloacetate into pyruvate and succinate in a series of reactions (Fig. 3).
Several amino acid degradation pathways were particularly prominent in the proteome, as judged by the proportion of identified components, namely, those of the branched-chain amino acids as well as of tyrosine, glycine, serine, alanine, glutamate, glutamine, and aspartate (Fig. 3). On the whole, the amino acid degradation routes feed into the TCA cycle as a central metabolic hub, which in turn supplies the precursors for gluconeogenesis. Concurrently, the P. haloplanktis TAC125 genome contains coding sequences of biosynthetic enzymes for the formation of all 20 proteinogenic amino acids. Indeed, we noticed that P. haloplanktis is able to grow on glycerol with glutamine as the only nitrogen source, indicating that this bacterium is prototrophic for all proteinogenic amino acids (data not shown).
The P. haloplanktis TAC125 genome encodes all genes required for a functional pentose phosphate pathway. In the cytoplasmic proteome the glucose-6-phosphate dehydrogenase (Zwf [PSHAa1140]), the transketolase (TktA [PSHAa0671]), and the transaldolase (TalB [PSHAa2559]) could be identified as abundant proteins. A high throughput of the pentose phosphate pathway increases the NADPH level, which provides the thioredoxin reductase with a reductive potential. The resulting elevated level of reduced thioredoxin can help to protect the P. haloplanktis proteins against the deleterious effects of an oxygen abundance. As the solubility of oxygen increases with a decline in temperature, the employment of the pentose phosphate pathway may represent a feasible means not only for carbohydrate interconversion but also for providing the organism with better oxidative stress adaptation under low-temperature conditions. In addition, a high level of the superoxide dismutase SodB (PSHAa1215), the thioredoxin reductase TrxB (PSHAa1720), and the thioredoxin-dependent peroxide reductase AhpCB (PSHAa0839) could be detected. Furthermore, a putative catalase, PSHAa1737, with extremely high-level similarity to catalases from other gamma-, beta-, and alphaproteobacteria (e.g., Psychrobacter, Mannheimia, Haemophilus, and Neisseria) was identified. These enzymatic functions seem to be crucial components for the oxidative stress resistance of P. haloplanktis TAC125.
Our proteome analysis revealed some inconsistencies for 23 detected proteins classified previously by Medigue et al. (32), which were modified accordingly (see Table S1 in the supplemental material). For instance, we amended the sole classification of the abundant ribosomal protein S1 RpsA. This protein is an essential component of the 30S ribosomal subunit but also plays a role in biosynthesis and salvage of nucleotides. This metabolic role of RpsA is derived from its association with the RNA degradosome (22) and accentuated by a conserved immediate genetic neighborhood with the cytidylate kinase Cmk (13). It is interesting that RpsA of E. coli interacts with the cold shock protein CspE and with degradosome ribonucleases.
Periplasmic proteome.
The proteome signature of the periplasmic protein fraction is shown in Fig. 2. To better visualize the differences in the proteomes of the two investigated cellular compartments, differently false-colored images of the cytoplasmic (green) and periplasmic (red) protein gels were superimposed (see Fig. S1 in the supplemental material). On the resulting dual-channel image, proteins with similar spot intensities on both gels appear yellow, while spots overrepresented on one gel retain the color of that gel. Selected highly expressed cytoplasmic proteins (such as GyrA, Fbp, and GapA) can also be found as technical impurities in the enriched periplasmic protein fraction (see also Table S1 in the supplemental material). This common technical problem of impurities of abundant cytoplasmic proteins in subcellular protein fractions was also extensively described previously by Brown et al. (6) for the periplasmic proteome of the Gram-negative bacterium Shewanella oneidensis and by Wolff et al. (51) for the membrane proteome of Staphylococcus aureus.
The periplasmic proteome revealed a clear abundance of protein spots in the acidic pH range. A similar preference of periplasmic proteins for lower isoelectric points was revealed previously by Fulda et al. for the cyanobacterial Synechocystis sp. strain PCC 6803 (23). Fifty-three of the identified proteins in the periplasmic protein fraction possess potential Sec-dependent signal sequences (Fig. 2). Three of the periplasmic proteins are cell wall related (MipA [PSHAa2838], PldA [PSHAa0175], and PSHAa0183), 5 enzymes are involved in amino acid metabolism (Dcp [PSHAa2184], PSHAa1484, PSHAa2492, PSHAa2910, and PSHAa2915), 3 are linked to protein secretion (PSHAa1869 to PSHAa1871), and 13 proteins have transport functions (FbpA [PSHAa2663], MalB [PSHAb0217], TolC [PSHAa2592], PSHAa0108, PSHAa1575, PSHAa1840, PSHAa1987, PSHAa2206, PSHAa2275, PSHAa2478, PSHAa2541, PSHAa2567, and PSHAb0341).
Interestingly, 8 of the 13 transport-related proteins belong to TonB-dependent transport systems (TBDTs). The relative spot intensities of these transporters account for approximately 13% of the periplasmic proteome (by far the highest value compared to those for other functional categories) (data not shown). The enrichment of this group of transporters indicates its probable important role in the metabolism of P. haloplanktis and could be a further prerequisite for the fast growth of this psychrophilic bacterium under nutrient-rich conditions. TBDTs form a transport channel in the outer membrane and allow the passage of their specific substrate(s) into the periplasm after energy transduction mediated by cytoplasmic membrane-anchored TonB has resulted in structural changes of the pore. Originally demonstrated to be crucial for iron transport (30), TBDTs have been linked to a multitude of other substrates, such as nickel (41), disaccharides like sucrose (3) or maltose (36), oligosaccharides (8), polysaccharides (39), and protein degradation products (35). Apart from the TBDTs, other detected putative substrate transport-related proteins are 3 ABC transporters, 4 porins, and the transporter TolB. The high proportion of TBDTs in the P. haloplanktis TAC125 genome (32) and their enrichment in the periplasmic proteome support the notion that these transporters allow the cells to efficiently utilize and scavenge a broad array of different substrates from the marine environment. Thus, the dominance of TBDTs in P. haloplanktis TAC125 could be an important component of the specialized copiotrophic strategy of this cold-adapted bacterium.
Extracellular proteome.
To assess whether P. haloplanktis secretes extracellular proteins during exponential growth into the medium, we also performed 2-D gel electrophoreses with precipitated proteins from supernatants of 1-liter cultures. However, no genuinely extracellular proteins could be detected in the precipitated protein fraction of supernatants of P. haloplanktis TAC125 cell cultures under exponential growth conditions (data not shown). The only proteins detected were abundant cytoplasmic and periplasmic proteins most probably leaked into the medium owing to cell lysis, suggesting that the secretion of proteins into the medium is not advantageous during planktonic growth. In this respect, it is worth mentioning that most of the investigated bacteria show an enhanced secretion of proteins under nutrient limitation conditions but not during exponential growth. So far, only very few studies have comprehensively examined the extracellular proteomes of marine bacteria. A secretome analysis of early-stationary-phase cell cultures of Pseudoalteromonas tunicata revealed, for example, the importance of iron transport and acquisition (19). Also, an analysis of extracellular proteins of the copiotrophic strain Ruegeria pomeroyi DSS-3 led to the identification of several transport functions (9). However, an analysis of the planctomycete Rhodopirellula baltica also revealed no true extracellular proteins in the supernatant of exponentially growing cells (26).
Conclusions.
So far, only a few studies of the proteome of marine bacteria are available (42). This initial proteomic characterization of exponentially growing P. haloplanktis cells shows that the observed fast growth of this cold-adapted marine bacterium at 16°C correlates with a prevalence of the protein synthesis machinery, an efficient utilization of available amino acids, a high abundance of transport capacities, and the presence of cold acclimation and oxidative stress proteins. This study constitutes a basis for future in-depth functional proteome studies to further specify the physiological adaptation of this versatile marine psychrophilic bacterium to its natural environmental conditions. Finally, the comprehensive deduction of relevant amino acid degradation pathways will help to develop improved media and fermentation strategies for this promising alternative low-temperature expression host.
Supplementary Material
Acknowledgments
We thank the proteome facility of the ZIK FunGene for supporting this work. We are grateful to Martin Fraunholz for signal peptide analyses.
This study was financially supported by the Ministry of Economy of Mecklenburg-Vorpommern (grant number 230-630.8-TIFA-356).
Footnotes
Published ahead of print on 23 December 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Antelmann, H., et al. 2001. A proteomic view on genome-based signal peptide predictions. Genome Res. 11:1484-1502. [DOI] [PubMed] [Google Scholar]
- 2.Birolo, L., et al. 2000. Aspartate aminotransferase from the Antarctic bacterium Pseudoalteromonas haloplanktis TAC 125. Cloning, expression, properties, and molecular modelling. Eur. J. Biochem. 267:2790-2802. [DOI] [PubMed] [Google Scholar]
- 3.Blanvillain, S., et al. 2007. Plant carbohydrate scavenging through tonB-dependent receptors: a feature shared by phytopathogenic and aquatic bacteria. PLoS One 2:e224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bowman, J. P., S. A. McCammon, M. V. Brown, D. S. Nichols, and T. A. McMeekin. 1997. Diversity and association of psychrophilic bacteria in Antarctic sea ice. Appl. Environ. Microbiol. 63:3068-3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brinkmeyer, R., et al. 2003. Diversity and structure of bacterial communities in Arctic versus Antarctic pack ice. Appl. Environ. Microbiol. 69:6610-6619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown, R. N., M. F. Romine, A. A. Schepmoes, R. D. Smith, and M. S. Lipton. Mapping the subcellular proteome of Shewanella oneidensis MR-1 using sarkosyl-based fractionation and LC-MS/MS protein identification. J. Proteome Res. 9:4454-4463. [DOI] [PubMed]
- 7.Buttner, K., et al. 2001. A comprehensive two-dimensional map of cytosolic proteins of Bacillus subtilis. Electrophoresis 22:2908-2935. [DOI] [PubMed] [Google Scholar]
- 8.Cheng, Q., M. C. Yu, A. R. Reeves, and A. A. Salyers. 1995. Identification and characterization of a Bacteroides gene, csuF, which encodes an outer membrane protein that is essential for growth on chondroitin sulfate. J. Bacteriol. 177:3721-3727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Christie-Oleza, J. A., and J. Armengaud. 2010. In-depth analysis of exoproteomes from marine bacteria by shotgun liquid chromatography-tandem mass spectrometry: the Ruegeria pomeroyi DSS-3 case-study. Mar. Drugs 8:2223-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Collins, T., et al. 2002. A novel family 8 xylanase, functional and physicochemical characterization. J. Biol. Chem. 277:35133-35139. [DOI] [PubMed] [Google Scholar]
- 11.Cunin, R., N. Glansdorff, A. Pierard, and V. Stalon. 1986. Biosynthesis and metabolism of arginine in bacteria. Microbiol. Rev. 50:314-352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cusano, A. M., E. Parrilli, G. Marino, and M. L. Tutino. 2006. A novel genetic system for recombinant protein secretion in the Antarctic Pseudoalteromonas haloplanktis TAC125. Microb. Cell Fact. 5:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Danchin, A. 1997. Comparison between the Escherichia coli and Bacillus subtilis genomes suggests that a major function of polynucleotide phosphorylase is to synthesize CDP. DNA Res. 4:9-18. [DOI] [PubMed] [Google Scholar]
- 14.de Pascale, D., et al. 2008. The cold-active Lip1 lipase from the Antarctic bacterium Pseudoalteromonas haloplanktis TAC125 is a member of a new bacterial lipolytic enzyme family. Extremophiles 12:311-323. [DOI] [PubMed] [Google Scholar]
- 15.Di Rocco, G., et al. 2008. Cloning, expression and physicochemical characterization of a di-heme cytochrome c (4) from the psychrophilic bacterium Pseudoalteromonas haloplanktis TAC 125. J. Biol. Inorg. Chem. 13:789-799. [DOI] [PubMed] [Google Scholar]
- 16.Donovan, C. J., et al. 2009. Pseudoalteromonas bacteria are capable of degrading paralytic shellfish toxins. Appl. Environ. Microbiol. 75:6919-6923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Duilio, A., M. L. Tutino, and G. Marino. 2004. Recombinant protein production in Antarctic Gram-negative bacteria. Methods Mol. Biol. 267:225-237. [DOI] [PubMed] [Google Scholar]
- 18.Evangelista, G., P. Falasca, I. Ruggiero, M. Masullo, and G. Raimo. 2009. Molecular and functional characterization of polynucleotide phosphorylase from the Antarctic eubacterium Pseudoalteromonas haloplanktis. Protein Pept. Lett. 16:999-1005. [DOI] [PubMed] [Google Scholar]
- 19.Evans, F. F., M. J. Raftery, S. Egan, and S. Kjelleberg. 2007. Profiling the secretome of the marine bacterium Pseudoalteromonas tunicata using amine-specific isobaric tagging (iTRAQ). J. Proteome Res. 6:967-975. [DOI] [PubMed] [Google Scholar]
- 20.Eymann, C., et al. 2004. A comprehensive proteome map of growing Bacillus subtilis cells. Proteomics 4:2849-2876. [DOI] [PubMed] [Google Scholar]
- 21.Feller, G., et al. 1998. Characterization of the C-terminal propeptide involved in bacterial wall spanning of alpha-amylase from the psychrophile Alteromonas haloplanctis. J. Biol. Chem. 273:12109-12115. [DOI] [PubMed] [Google Scholar]
- 22.Feng, Y., H. Huang, J. Liao, and S. N. Cohen. 2001. Escherichia coli poly(A)-binding proteins that interact with components of degradosomes or impede RNA decay mediated by polynucleotide phosphorylase and RNase E. J. Biol. Chem. 276:31651-31656. [DOI] [PubMed] [Google Scholar]
- 23.Fulda, S., F. Huang, F. Nilsson, M. Hagemann, and B. Norling. 2000. Proteomics of Synechocystis sp. strain PCC 6803. Identification of periplasmic proteins in cells grown at low and high salt concentrations. Eur. J. Biochem. 267:5900-5907. [DOI] [PubMed] [Google Scholar]
- 24.Gauthier, G., M. Gauthier, and R. Christen. 1995. Phylogenetic analysis of the genera Alteromonas, Shewanella, and Moritella using genes coding for small-subunit rRNA sequences and division of the genus Alteromonas into two genera, Alteromonas (emended) and Pseudoalteromonas gen. nov., and proposal of twelve new species combinations. Int. J. Syst. Bacteriol. 45:755-761. [DOI] [PubMed] [Google Scholar]
- 25.Georlette, D., et al. 2000. A DNA ligase from the psychrophile Pseudoalteromonas haloplanktis gives insights into the adaptation of proteins to low temperatures. Eur. J. Biochem. 267:3502-3512. [DOI] [PubMed] [Google Scholar]
- 26.Hieu, C. X., et al. 2008. Detailed proteome analysis of growing cells of the planctomycete Rhodopirellula baltica SH1T. Proteomics 8:1608-1623. [DOI] [PubMed] [Google Scholar]
- 27.Jackson, G. A. 1989. Simulation of bacterial attraction and adhesion to falling particles in an aquatic environment. Limnol. Oceanogr. 34:514-530. [Google Scholar]
- 28.Klappenbach, J. A., J. M. Dunbar, and T. M. Schmidt. 2000. rRNA operon copy number reflects ecological strategies of bacteria. Appl. Environ. Microbiol. 66:1328-1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Klumpp, S., Z. Zhang, and T. Hwa. 2009. Growth rate-dependent global effects on gene expression in bacteria. Cell 139:1366-1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee, B. C. 1995. Quelling the red menace: haem capture by bacteria. Mol. Microbiol. 18:383-390. [DOI] [PubMed] [Google Scholar]
- 31.Marx, J. C., T. Collins, S. D'Amico, G. Feller, and C. Gerday. 2007. Cold-adapted enzymes from marine Antarctic microorganisms. Mar. Biotechnol. (NY) 9:293-304. [DOI] [PubMed] [Google Scholar]
- 32.Medigue, C., et al. 2005. Coping with cold: the genome of the versatile marine Antarctica bacterium Pseudoalteromonas haloplanktis TAC125. Genome Res. 15:1325-1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Merlino, A., et al. 2008. Crystallization and preliminary X-ray diffraction studies of a psychrophilic iron superoxide dismutase from Pseudoalteromonas haloplanktis. Protein Pept. Lett. 15:415-418. [DOI] [PubMed] [Google Scholar]
- 34.Mounier, J., C. Monnet, N. Jacques, A. Antoinette, and F. Irlinger. 2009. Assessment of the microbial diversity at the surface of Livarot cheese using culture-dependent and independent approaches. Int. J. Food Microbiol. 133:31-37. [DOI] [PubMed] [Google Scholar]
- 35.Nagano, K., et al. 2007. Characterization of RagA and RagB in Porphyromonas gingivalis: study using gene-deletion mutants. J. Med. Microbiol. 56:1536-1548. [DOI] [PubMed] [Google Scholar]
- 36.Neugebauer, H., et al. 2005. ExbBD-dependent transport of maltodextrins through the novel MalA protein across the outer membrane of Caulobacter crescentus. J. Bacteriol. 187:8300-8311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Papa, R., et al. 2006. Proteomic identification of a two-component regulatory system in Pseudoalteromonas haloplanktis TAC125. Extremophiles 10:483-491. [DOI] [PubMed] [Google Scholar]
- 38.Piette, F., et al. 2010. Proteomics of life at low temperatures: trigger factor is the primary chaperone in the Antarctic bacterium Pseudoalteromonas haloplanktis TAC125. Mol. Microbiol. 76:120-132. [DOI] [PubMed] [Google Scholar]
- 39.Reeves, A. R., J. N. D'Elia, J. Frias, and A. A. Salyers. 1996. A Bacteroides thetaiotaomicron outer membrane protein that is essential for utilization of maltooligosaccharides and starch. J. Bacteriol. 178:823-830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reichelt, J. L., and P. Baumann. 1973. Change of the name Alteromonas marinopraesens (ZoBell and Upham) Baumann et al. to Alteromonas haloplanktis (ZoBell and Upham) comb. nov. and assignment of strain ATCC 23821 (Pseudomonas enalia) and strain c-A1 of De Voe and Oginsky to this species. Int. J. Syst. Bacteriol. 23:438-441. [Google Scholar]
- 41.Schauer, K., B. Gouget, M. Carriere, A. Labigne, and H. de Reuse. 2007. Novel nickel transport mechanism across the bacterial outer membrane energized by the TonB/ExbB/ExbD machinery. Mol. Microbiol. 63:1054-1068. [DOI] [PubMed] [Google Scholar]
- 42.Schweder, T., S. Markert, and M. Hecker. 2008. Proteomics of marine bacteria. Electrophoresis 29:2603-2616. [DOI] [PubMed] [Google Scholar]
- 43.Srimathi, S., G. Jayaraman, G. Feller, B. Danielsson, and P. R. Narayanan. 2007. Intrinsic halotolerance of the psychrophilic alpha-amylase from Pseudoalteromonas haloplanktis. Extremophiles 11:505-515. [DOI] [PubMed] [Google Scholar]
- 44.Stocker, R., J. R. Seymour, A. Samadani, D. E. Hunt, and M. F. Polz. 2008. Rapid chemotactic response enables marine bacteria to exploit ephemeral microscale nutrient patches. Proc. Natl. Acad. Sci. U. S. A. 105:4209-4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tosco, A., et al. 2003. GroEL from the psychrophilic bacterium Pseudoalteromonas haloplanktis TAC 125: molecular characterization and gene cloning. Extremophiles 7:17-28. [DOI] [PubMed] [Google Scholar]
- 46.Tutino, M. L., G. di Prisco, G. Marino, and D. de Pascale. 2009. Cold-adapted esterases and lipases: from fundamentals to application. Protein Pept. Lett. 16:1172-1180. [DOI] [PubMed] [Google Scholar]
- 47.Violot, S., et al. 2003. Expression, purification, crystallization and preliminary X-ray crystallographic studies of a psychrophilic cellulase from Pseudoalteromonas haloplanktis. Acta Crystallogr. D Biol. Crystallogr. 59:1256-1258. [DOI] [PubMed] [Google Scholar]
- 48.Voigt, B., et al. 2004. A proteomic view of cell physiology of Bacillus licheniformis. Proteomics 4:1465-1490. [DOI] [PubMed] [Google Scholar]
- 49.Watt, S. A., A. Wilke, T. Patschkowski, and K. Niehaus. 2005. Comprehensive analysis of the extracellular proteins from Xanthomonas campestris pv. campestris B100. Proteomics 5:153-167. [DOI] [PubMed] [Google Scholar]
- 50.Wilmes, B., A. Hartung, M. Lalk, M. Liebeke, T. Schweder, and P. Neubauer. 2010. Fed-batch process for the psychrotolerant marine bacterium Pseudoalteromonas haloplanktis. Microb. Cell Fact. 9:72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wolff, S., H. Hahne, M. Hecker, and D. Becher. 2008. Complementary analysis of the vegetative membrane proteome of the human pathogen Staphylococcus aureus. Mol. Cell. Proteomics 7:1460-1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zobell, C. E. 1941. Studies on marine bacteria. The cultural requirements of heterotrophic aerobes. J. Mar. Res. 4:41-75. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.