Abstract
The prevalence and diversity of multidrug-resistant (MDR) Salmonella enterica strains associated with cattle at harvest in the United States were examined. Hides and carcasses of cattle were sampled at processing plants (n = 6) located in four geographically distant regions from July 2005 to April 2006. The mean prevalences of Salmonella on hides, preevisceration carcasses (immediately after hide removal), and postintervention carcasses (in the chiller and after the full complement of interventions) were 89.6%, 50.2%, and 0.8%, respectively. The values for MDR Salmonella enterica strains (defined as those resistant to two or more antimicrobials) as percentages of Salmonella prevalence were 16.7% (95% confidence interval [CI], 8.3 to 25.1%; median percent prevalence, 6.9%), 11.7% (95% CI, 4.4 to 19.0%; median, 4.8%), and 0.33% (95% CI, −0.3 to 0.70%; median, 0%), respectively. In this study, 16,218 Salmonella hide and carcass isolates were screened for antimicrobial resistance. Of these, 978 (6.0%) unique MDR S. enterica isolates were identified and serotyped and their XbaI pulsed-field gel electrophoresis (PFGE) profiles determined. The predominant MDR S. enterica serotypes observed were Newport (53.1%), Typhimurium (16.6%), and Uganda (10.9%). Differences in MDR S. enterica prevalence were detected, and PFGE analysis revealed both epidemic clusters (profiles found in plants in multiple regions/seasons) and endemic clusters (profiles observed in plants in limited regions/seasons) within several of the MDR serotypes examined. Despite these differences, multiple-hurdle processing interventions employed at all plants were found to be quite effective and decreased Salmonella carcass contamination by 98.4% (95% CI, 97.6 to 99.7%).
Salmonellae are important food-borne pathogens noted for causing millions of cases of food-borne illness in the United States each year (26, 55, 66). Nontyphoidal salmonellosis is generally a self-limiting disease, and patients frequently recover without the need for medical attention. However, a small percentage of Salmonella infections result in invasive salmonellosis, a more severe form of illness requiring hospitalization and antibiotic therapy. Recent studies have found that in certain Enterobacteriaceae, including Salmonella, virulence genes may be colocalizing on transferable resistance plasmids (37, 73), a phenomenon that would lend credence to studies that have shown that antimicrobial-resistant Salmonella strains may be more invasive than Salmonella strains that are susceptible to antimicrobials (45, 71, 72). As such, there is a need to understand the complex etiology and epidemiology of these food-borne pathogens.
Extensive research aimed at characterizing antimicrobial resistance phenotypes of Salmonella enterica from a variety of food and animal sources has revealed that numerous serotypes may harbor multiple antimicrobial resistance determinants (75, 80). These multidrug-resistant (MDR) S. enterica strains (defined as strains that are resistant to two or more antimicrobial agents) may carry their resistance determinants on chromosomal locations, on resistance plasmids, or on both (3, 23, 52). Of particular importance to the medical community are resistances to the extended-spectrum cephalosporin ceftriaxone, the drug of choice for treatment of pediatric salmonellosis, and to the quinolone nalidixic acid and the fluoroquinolone ciprofloxacin, which are preferable for treatment of adults (42).
While poultry products and, more recently, contaminated fresh produce are well-established vectors for S. enterica, several food-borne disease case studies have shown undercooked ground beef and beef products to be sources of sporadic and outbreak cases of salmonellosis (43, 50, 60, 68, 72). Among the various sources or production systems that supply cattle for beef, the primary source of lean beef for the grinding industry is meat harvested from cull cattle (dairy and beef cull cows and bulls). Cull cattle, especially dairy cattle, have been implicated as a reservoir for antimicrobial-resistant S. enterica (4, 35, 41, 70). The presence of these organisms on the hides of cattle at harvest represents a risk to food safety, as they may be transferred to carcasses during the dressing process (6, 9, 15). Once on the carcass, pathogens may enter the food supply if they survive carcass-processing interventions. Thus, in order to gain a better understanding of the risk associated with processing cull cattle and the potential for introducing MDR S. enterica into the food chain, it is important to study the extent to which the hides of cattle at harvest are contaminated with these pathogens. To that end, we examined the prevalence of MDR S. enterica (here referred to as MDR Salmonella) associated with cattle at harvest in plants (n = 6) located in four geographically distant regions of the United States over the course of 10 months. The MDR Salmonella strains isolated were serotyped and their antimicrobial susceptibility phenotypes and XbaI pulsed-field gel electrophoresis (PFGE) profiles determined. Collection of these data provided a unique opportunity to observe the diversity of MDR Salmonella strains found at cattle harvest establishments over time and revealed the existence of both epidemic and endemic MDR Salmonella biotypes.
MATERIALS AND METHODS
Sample collection.
Processing plants (n = 6) that harvest cull cows, bulls, dairy cattle, and/or fed cattle, located in four geographically distant regions (here designated A to D and located within [although not respectively within] microbiological monitoring regions 2, 3, 5, and 8, as defined by the Beef Industry Food Safety Council [BIFSCo] [14]) of the United States, were sampled every 3 months (July, October, January, and April) in a 10-month period from 2005 to 2006. For cull cattle, samples were collected from four plants (three that processed both cull and fed cattle and one that processed strictly cull cattle), one in each region. Samples, including hide samples, preevisceration carcass samples (sampled after hide removal and prior to any intervention), and postintervention carcass samples (sampled after receiving the full complement of processing interventions [including steam vacuum, lactic acid hot-water wash, and spray chill carcass rinse] and having chilled for no more than 2 h), were collected (n = 95 per sample type) on each sample day, with two sample days per season, resulting in a total of 3,040 samples for each sample type. Fed-cattle samples were collected from four plants (three that processed both cull and fed cattle and one that processed strictly fed cattle), one in each region, only in the first sample season (July). Hide and preevisceration carcass samples (n = 171 to 207) were collected on one sample day per region, for a total of 755 samples for each sample type. Thus, samples were collected from six plants in all, from one or two plants in each region, on a total of 9 days. Hides and carcasses were tagged prior to sampling, such that samples were matched and collected consecutively, with approximately every fourth carcass on the processing line being sampled. All samples were shipped in coolers with ice packs and were received and processed at the U.S. Meat Animal Research Center (USMARC) within 24 h of collection.
Hide samples were obtained by swabbing approximately 1,000 cm2 with a sterile sponge (Whirl Pak; Nasco, Ft. Atkinson, WI), prewetted with 20 ml sterile Difco buffered peptone water (BPW; Becton Dickinson, Sparks, MD). Hide samples were collected from the brisket plate region of animals on the line, after stunning and exsanguination, prior to hide removal. Carcass samples were obtained by swabbing approximately 8,000 cm2 of carcass with 2 sterile sponges (Nasco), each prewetted with 10 ml BPW, as previously described (7).
Culture media and enrichment methods.
Hide as well as preevisceration and postintervention carcass sample enrichments were analyzed for the presence of Salmonella. Sponge samples were enriched, as previously described (7, 12). Briefly, Difco Trypticase soy broth (TSB; Becton Dickinson) was added to sponge samples at a 1:5 ratio, incubated at 25°C for 2 h and 42°C for 6 h, and then held at 4°C until being processed the next day. Salmonella strains were isolated from culture enrichments using immunomagnetic separation (IMS), as previously described (56). IMS bead-bacterium complexes were placed into 3 ml of Rappaport-Vassiliadis soya peptone broth (RVS; Oxoid, Basingstoke, United Kingdom) and incubated at 42°C for 18 to 20 h. These secondary enrichments were swabbed onto Difco Hektoen enteric medium (Becton Dickinson) with novobiocin at a concentration of 5 mg liter−1 (HEn) and Difco Brilliant Green agar with sulfadiazine at 80 mg liter−1 (BGS; Becton Dickinson) and then streaked for isolation and incubated at 37°C for 18 to 20 h. One to three putative Salmonella isolates were picked (depending on the number of putative isolates present on the selective medium) and further confirmed as being Salmonella by PCR for the Salmonella-specific portion of the invA gene (58, 62).
Identification and characterization of multidrug-resistant Salmonella strains.
All Salmonella isolates were initially screened for resistance to three antimicrobials (ampicillin [Ap], tetracycline [Te], and kanamycin [K]) by stamping the isolates in a 96-well block format with a Boekel microplate replicator (Boekel Scientific, Feasterville, PA) onto four tryptic soy agar (TSA) plates (150 mm) containing either no antimicrobials, Ap (32 mg liter−1), Te (32 mg liter−1), or K (64 mg liter−1) (all antimicrobials were obtained from Sigma, St. Louis, MO). These plates were incubated at 37°C for 18 to 20 h. Isolates demonstrating resistance to any of these antimicrobials were streaked for isolation onto TSA with the appropriate antimicrobial and were again incubated at 37°C for 18 to 20 h. If multiple isolates from a sample (up to three) demonstrated the same initial resistance pattern in the prescreening process, then one isolate was chosen for further characterization, as previous studies in our laboratory have shown that this frequently indicates the presence of a dominant Salmonella strain in the sample enrichment and further analysis yields identical resistance patterns. Cultures on TSA were subcultured by streaking them for isolation onto XLDtnc agar (xylose lysine desoxycholate medium; Oxoid) with 4.6 ml liter−1 tergitol (aka niaproof; Sigma), 15 mg liter−1 novobiocin, and 10 mg liter−1 cefsulodin and incubated at 37°C for 18 to 20 h. Isolates demonstrating typical Salmonella colony morphology on XLDtnc (black colonies with a clear pink outer ring) were streaked for isolation onto TSA and incubated, as described above. The resulting pure cultures were used for antimicrobial susceptibility analysis and serological identification. Antimicrobial susceptibility testing was performed using the Sensititre broth microdilution system (TREK Diagnostic Systems, Toledo, OH) and CMV1AGNF test plates, according to the manufacturer's instructions. The plates determined sensitivity to 15 antimicrobials: amikacin (Ai), Ap, amoxicillin-clavulanic acid (Am), ceftiofur (F), cefoxitin (T), ceftriaxone (Ax), chloramphenicol (C), ciprofloxacin (Cp), gentamicin (G), K, nalidixic acid (N), streptomycin (S), sulfisoxazole (Su), Te, and trimethoprim sulfamethoxazole (Sxt). Antimicrobial sensitivity was determined using a Sensititre AutoReader and the SWIN software package, which uses Clinical and Laboratory Standards Institute (CLSI)-approved MIC breakpoint guidelines for the drugs listed above (19, 75). The following organisms were used as quality control strains in the antimicrobial sensitivity assays: Pseudomonas aeruginosa ATCC 27853 (American Type Culture Collection), Escherichia coli ATCC 25922, and Staphylococcus aureus ATCC 25923. Salmonella isolates were serogrouped with a Welcollex color serogrouping kit (Remel, Lenexa, KS) and serotyped further using slide agglutination with O-factor antisera and tube agglutination with H-factor antisera (Denka Seiken Co., Ltd., Derbyshire, United Kingdom), according to the manufacturer's instructions.
PFGE analysis.
PFGE analysis was performed according to the protocol developed by the Centers for Disease Control and Prevention (CDC) (65). Agarose-embedded DNA was digested with XbaI (New England BioLabs, Beverly, MA). Salmonella enterica serotype Braenderup strain H9812 was used as a control and for standardization of gels (48). Banding patterns were inspected by visual confirmation and then further analyzed and compared using Bionumerics software (Applied Maths, Sint-Martens-Latem, Belgium), employing the Dice similarity coefficient with a 1.5% band position tolerance in conjunction with the unweighted-pair group method using arithmetic averages for clustering. For each serotype dendrogram shown, clusters were ranked first by the number of regions and seasons in which isolates within that cluster were observed and then by the percentage of isolates that were present within each cluster. Roman numerals were used for cluster designations, and in each case, Roman numeral I indicates the cluster that was found to be the most widely distributed. Clusters were designated sequentially from there, based on the percentage of isolates that fell within that cluster.
Statistics.
Cull cattle Salmonella prevalence data were analyzed by season (8 sample days per season; 2 days at each of four plants) or by region (8 sample days per plant in each region; 2 days per season). Fed-cattle prevalence values are reported as percent positive samples for one sample collection day per plant in each region. Prevalence values for cull and fed-cattle samples were calculated by dividing the number of culture-positive samples by the total number of samples collected (cull cattle n = 95 per sample type per day; fed cattle n = 171 to 207 per sample type). The percents prevalence for Salmonella and MDR Salmonella on hide and carcass samples were calculated for each sample day and are reported as the mean percent prevalence, 95% confidence interval (95% CI), and median percent prevalence values (Tables 1 and 2). For data sets that were not normally distributed, comparisons of median prevalence values were made using the Kruskal-Wallis test for nonparametric data and Dunn's multiple-comparison posttest. For data sets that were normally distributed, comparisons of mean prevalence values were made using a one-way analysis of variance (ANOVA) and Bonferroni's multiple-comparison posttest. Data were analyzed using Prism 5.0 Graph Pad software, and P values less than 0.05 were considered significant.
TABLE 1.
Mean percents Salmonella and MDR Salmonella prevalence by season
| Sample group and parameter | Value for: |
P |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Summer |
Fall |
Winter |
Spring |
Overall |
||||||||
| Not MDR | MDR | Not MDR | MDR | Not MDR | MDR | Not MDR | MDR | Not MDR | MDR | Not MDR | MDR | |
| Hide (cull) | ||||||||||||
| % Salmonella | 78.5 | 15.3 | 75.0 | 13.4 | 65.3 | 26.3 | 74.5 | 11.6 | 72.9 | 16.7 | 0.4864a | 0.8803a |
| 95% CI | 55.6-101.4 | −2.6-33.3 | 55.9-94.1 | 2.4-24.7 | 37.1-89.9 | −4.8-57.4 | 56.5-92.4 | −1.7-24.9 | 63.7-82.1 | 8.3-25.1 | ||
| Median % prevalence | 91.6 | 3.2 | 78.5 | 7.9 | 77.4 | 6.4 | 76.3 | 6.3 | 80.6 | 6.9 | ||
| Simpson's ID | 0.41 | 0.49 | 0.28 | 0.39 | 0.39 | 0.4828b | ||||||
| 95% CI | 0.20-0.61 | 0.26-0.74 | 0.02-0.53 | 0.12-0.65 | 0.29-0.49 | |||||||
| Preevisceration (cull) | ||||||||||||
| % Salmonella | 39.9 | 9.5 | 40.4 | 11.4 | 32.2 | 23.1 | 41.4 | 2.8 | 38.5 | 11.7 | 0.8928b | 0.2336a |
| 95% CI | 14.3-65.4 | 1.4-17.5 | 15.6-65.1 | 3.2-19.7 | 10.2-54.1 | −7.9-54.2 | 26.2-56.7 | 0.51-5.0 | 29.3-47.7 | 4.4-19.0 | ||
| Median % prevalence | 35.3 | 6.4 | 35.8 | 6.4 | 27.0 | 4.2 | 44.2 | 2.7 | 37.9 | 4.8 | ||
| Postintervention (cull) | ||||||||||||
| % Salmonella | 0.54 | 0.8 | 0.0 | 0.14 | 0.41 | 0.26 | 0.92 | 0.14 | 0.47 | 0.33 | 0.3284a | 0.8466a |
| 95% CI | −0.42-1.5 | −0.75-2.3 | 0.0 | −0.19-0.46 | −0.06-0.88 | −0.36-0.88 | −1.3-3.1 | −0.18-0.46 | −0.04-0.98 | −0.03-0.70 | ||
Comparisons of median prevalence values were made using the Kruskal-Wallis test for nonparametric data and Dunn's multiple-comparison posttest (P < 0.05).
Comparisons of mean prevalence or diversity values (normally distributed data sets) were made using a one-way ANOVA and Bonferroni's multiple-comparison posttest (P < 0.05).
TABLE 2.
Mean percents Salmonella and MDR Salmonella prevalence by region
| Sample group and parameter | Value for:a |
P |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A |
B |
C |
D |
Overall |
||||||||
| Not MDR | MDR | Not MDR | MDR | Not MDR | MDR | Not MDR | MDR | Not MDR | MDR | Not MDR | MDR | |
| Hide (cull) | ||||||||||||
| % Salmonella | 80.4C | 7.6AB | 80.9C | 11.2AB | 91.1C | 1.9A | 38.9D | 46.0B | 72.9 | 16.7 | 0.0004b | 0.0008b |
| 95% CI | 66.8-93.9 | 0.5-14.8 | 70.5-91.4 | 3.3-19.1 | 83.6-98.6 | -0.3-4.3 | 19.4-58.4 | 20.9-71.0 | 63.7-82.1 | 8.3-25.1 | ||
| Median % prevalence | 81.6 | 3.7 | 80.0 | 10.0 | 94.8 | 1.6 | 35.4 | 43.7 | 80.6 | 6.9 | ||
| Simpson's ID | 0.41 | 0.5 | 0.1 | 0.5 | 0.4 | 0.0785c | ||||||
| 95% CI | 0.19-0.64 | 0.26-0.74 | −0.18-0.38 | 0.33-0.59 | 0.29-0.49 | |||||||
| Hide (fed) | ||||||||||||
| % Salmonella | 81.3 | 18.7d | 79.2 | 2.9 | 98.4 | 0.0 | 69.1 | 17.6 | 82.0 | 9.8 | ||
| 95% CI | 62.6-101.4 | −5.7-25.3 | ||||||||||
| Preevisceration (cull) | ||||||||||||
| % Salmonella | 45.8CD | 6.4AB | 58.3D | 8.9A | 25.6C | 1.3B | 24.2C | 30.1A | 38.5 | 11.7 | 0.0113c | 0.0026b |
| 95% CI | 26.6-65.0 | −1.1-13.9 | 35.9-80.6 | 2.8-15.1 | 15.0-36.1 | −0.29-2.9 | 4.5-43.9 | 1.9-58.3 | 29.3-47.7 | 4.4-19.0 | ||
| Median % prevalence | 45.8 | 2.1 | 55.2 | 6.8 | 26.5 | 0.0 | 9.5 | 18.5 | 37.9 | 4.8 | ||
| Preevisceration (fed) | ||||||||||||
| % Salmonella | 53.8 | 14.6d | 34.6 | 2 | 60.2 | 0.0 | 14.4 | 4.3 | 40.8 | 5.2 | ||
| 95% CI | 7.9-73.6 | −5.1-15.6 | ||||||||||
| Postintervention (cull) | ||||||||||||
| % Salmonella | 0.54 | 0.8 | 0.27 | 0.14 | 0.14 | 0.0 | 0.9 | 0.4 | 0.5 | 0.3 | 0.8791b | 0.4525b |
| 95% CI | −0.42-1.5 | −0.75-2.3 | −0.15-0.7 | −0.19-0.46 | −0.19-0.46 | 0.0 | −1.3-3.1 | −0.26-1.1 | −0.04-0.98 | −0.03-0.70 | ||
Common uppercase superscripts (A and B, MDR Salmonella, and C and D, non-MDR Salmonella) indicate values that are not significantly different (P < 0.05).
Comparisons of median prevalence values were made using the Kruskal-Wallis test for nonparametric data and Dunn's multiple-comparison posttest.
Comparisons of mean prevalence or diversity values (normally distributed data sets) were made using a one-way ANOVA and Bonferroni's multiple-comparison posttest.
Fed cattle MDR Salmonella prevalence values for hides and carcasses in region A (measured in summer only) were reflective of cull cattle MDR Salmonella prevalence values for the same region and season. MDR Salmonella prevalence values for cull cattle hides and preevisceration carcasses in this region and season were as follows: for hide samples, 4.2% for day 1 and 20% for day 2, and for preevisceration carcass samples, 2.5% for day 1 and 47.9% for day 2. These results demonstrate the wide range in prevalence values that can be observed from one sample day or time point to the next.
The diversity of MDR Salmonella serotypes isolated from hide samples was examined by calculating Simpson's index of diversity (ID) (1 − D, where D is Σ[n(n − 1)]/N(N − 1), with n representing the number of unique isolates of each serotype of MDR Salmonella identified on a given sample day and N representing the total number of unique MDR Salmonella isolates identified on a given sample day). Simpson's index of diversity is a measure of both how many MDR serotypes are isolated in each region on a given sample day and how evenly those serotypes are represented in a given region or season. Values for 1 − D that are closer to 1 indicate high diversity, while values closer to 0 indicate low diversity. Values were averaged by region or season and reported as mean levels of diversity with the 95% CI of each calculated mean. Comparisons of values for Simpson's index of diversity were made using a one-way ANOVA and Bonferroni's multiple-comparison posttest (39, 67).
RESULTS
Prevalence of Salmonella and MDR Salmonella on cattle hide and carcass samples.
Salmonella mean prevalence values for cull cattle hide samples, preevisceration carcass samples, and postintervention carcass samples were fairly consistent across seasons and on average were 89.6, 50.2, and 0.8%, respectively, as we have reported previously (15). These values are listed in Table 1 and are separated into the percentages of samples in each season that were found to be contaminated with non-MDR Salmonella or MDR Salmonella. These data show that the proportions of hide and carcass samples that were found to contain MDR Salmonella were also fairly consistent across seasons and were on average 16.7, 11.7, and 0.33% (median values of 6.9, 4.8, and 0%) for hides, preevisceration carcass samples, and postintervention carcass samples, respectively (Table 1). In contrast with seasonal prevalence, analysis of Salmonella and MDR Salmonella prevalence by region revealed significant differences (Table 2). Specifically, while the overall prevalence of Salmonella on hides and carcasses of cull cattle sampled at harvest in region C was consistent with that found in other regions, the prevalence of MDR Salmonella detected in region C was consistently lower than (albeit not significantly different from) that observed in plants sampled in region A or B. And throughout the course of the study, MDR Salmonella prevalence values in region C were significantly lower (P = 0.0008) than those observed in region D (Table 2). No significant regional differences were observed for MDR Salmonella prevalence values associated with postintervention carcasses.
Salmonella and MDR Salmonella prevalence associated with fed cattle was examined in the summer sample season only and was found to be reflective of cull cattle prevalence in the same region (Table 2). Analysis of the MDR Salmonella strains isolated from fed cattle (for those harvested in the same plant as the cull cattle sampled in this study) revealed strong evidence of cross-contamination in the plant environment, as indistinguishable Salmonella strains (as characterized by serotype, MDR resistance phenotype, and PFGE profile) were frequently collected over consecutive sampling days. This cross-contamination effect has previously been noted by others (5, 34), and as a result, prevalence and MDR serotype/phenotype data collected from fed cattle were treated as an additional sample time point for surveying the diversity of MDR Salmonella strains entering slaughter establishments on the hides of cattle. No attempt to attribute any specific serotype or MDR phenotype to one or the other cattle type was made, as the data were not collected in such a way as to substantiate this type of analysis.
Sampling of 3,040 postintervention carcasses over the course of this study resulted in a total of 24 Salmonella isolates (10 MDR and 14 pansusceptible isolates) that were collected from 23 postintervention carcasses. Thus, despite the infrequent isolation of Salmonella from postintervention carcasses (Salmonella prevalence at this sample site was on average 0.8% [95% CI, 0.18 to 1.42%]), the mean percentage of Salmonella strains found to be MDR at this site was 43.3% (95% CI, 7.7 to 78.9%) in this study.
Salmonella serotype diversity and MDR phenotypes.
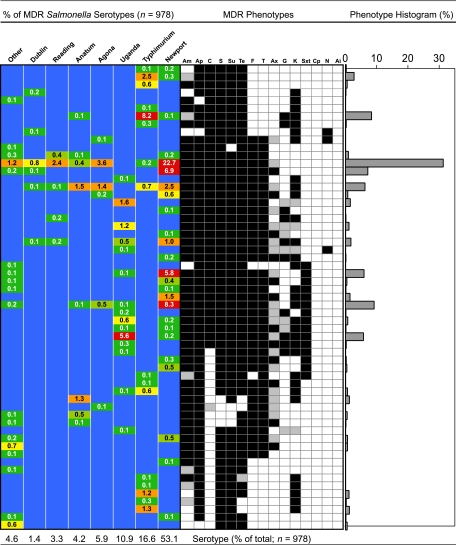
Antimicrobial sensitivity screening of 16,218 Salmonella isolates revealed 978 (6.0%) unique MDR Salmonella isolates. Unique isolates refer to a single isolate per sample except for rare instances (∼0.03%; n = 10,630 samples examined) when more than one serotype of MDR Salmonella was isolated from an enrichment. Of these isolates, 870 were obtained from cull cattle samples (n = 9,120) and 108 from fed cattle samples (n = 1,510). MDR Salmonella strains were isolated from samples collected at five of the six plants that participated in this study, and 59 different resistance phenotypes were observed (Fig. 1). Serotyping of these MDR Salmonella strains resulted in the identification of 20 serotypes. The most prevalent MDR Salmonella enterica serotype observed was Newport (53.1%), followed by Typhimurium (16.6%) and Uganda (10.9%) (Fig. 1). Other MDR Salmonella enterica serotypes identified included Agona (5.9%), Anatum (4.2%), Reading (3.3%), Dublin (1.4%), Muenster (0.8%), Ohio (0.8%), Give (0.6%), Heidelberg (0.6%), Saint Paul (0.4%), Infantis (0.3%), Derby (0.2%), Mbandaka (0.2%), Montevideo (0.2%), Cerro (0.1%), Enteritidis (0.1%), Kentucky (0.1%), and Muenchen (0.1%).
FIG. 1.
Serotype distribution heat map, antimicrobial resistance profile, and histogram depicting the percentage of each resistance phenotype observed in the 978 MDR Salmonella strains characterized. Values in the heat map indicate the percentage of each serotype that was found to exhibit the corresponding resistance phenotype. Resistance phenotypes are indicated by black (resistant), gray (intermediate), or white (susceptible) boxes. Antimicrobial abbreviations: Am, amoxicillin-clavulanic acid; Ap, ampicillin; C, chloramphenicol; S, streptomycin; Su, sulfisoxazole; Te, tetracycline; F, cefoxitin; T, ceftiofur; Ax, ceftriaxone; G, gentamicin; K, kanamycin; Sxt, sulfamethoxazole-trimethoprim; Cp, ciprofloxacin; N, nalidixic acid; and Ai, amikacin.
Analysis of MDR Salmonella serotype diversity using Simpson's index showed moderate levels of diversity in the sample periods examined. In this study, cattle were sampled consecutively (generally every fourth animal over consecutive lots), and as a result of this sampling scheme, MDR Salmonella strains present on cattle hides and carcasses in a given sample period were typically dominated by a particular serotype/MDR phenotype, likely a reflection of cattle lot effects and cross-contamination in the lairage environment (5, 15, 34). Accordingly, mean diversity values were low to moderate and ranged from 0.28 to 0.49 when analyzed by season or from 0.1 to 0.5 when analyzed by region. While not significantly different, seasonal diversity was observed to be lowest in the winter and highest in the fall (P = 0.4828), and analysis by region showed that diversity tended to be lowest in region C and highest in region B (P = 0.0785) (Tables 1 and 2).
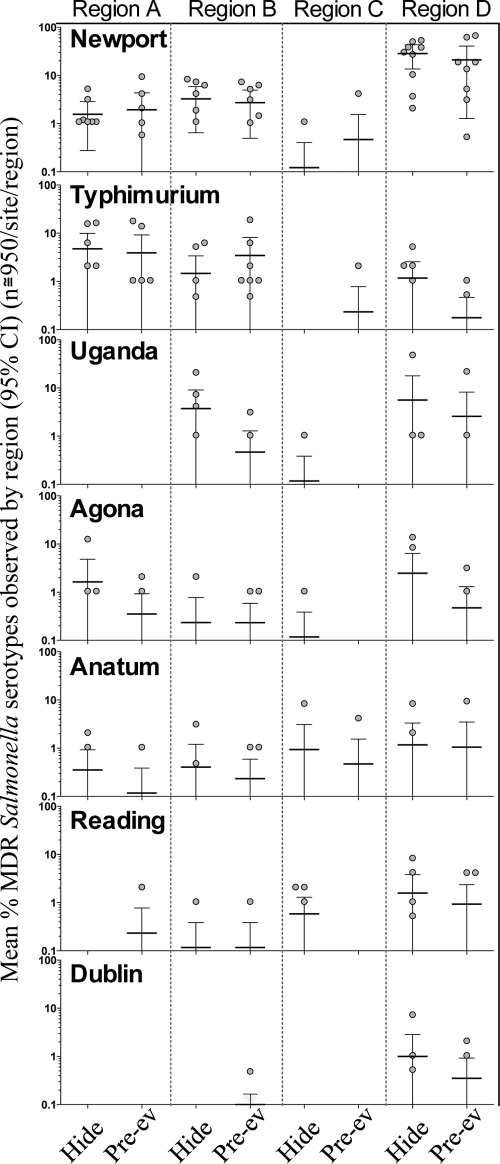
MDR Salmonella Newport was the most frequently isolated and widely distributed serotype observed in this study. It was isolated at least once from plants in all four regions and in all seasons (Fig. 2). The mean observed prevalence values per sample day ranged from 0.12% to 28.4% and from 0.47% to 21.1% for hide and preevisceration carcass samples, respectively (Fig. 2). MDR Salmonella serotypes Agona, Anatum, and Reading were also widely distributed and isolated from cattle at harvest in all four regions, albeit at lower levels than Newport (Fig. 2). While the aforementioned serotypes appeared to be widely distributed, other serotypes identified were found to have a more regional distribution. For example, although MDR Salmonella Typhimurium was isolated from cattle at harvest in plants in all four regions, considerably higher prevalence values were observed in plants in regions A and B than in region C or D. This phenomenon also was observed for MDR Salmonella serotypes Dublin and Uganda, which were predominantly isolated from cattle at harvest in plants in regions B and D (Fig. 2).
FIG. 2.
Top seven MDR Salmonella serotypes observed at processing plants in four regions of the United States over 10 months. Each point represents the percent prevalence of that serotype on a given sample day (n = 95 [cull] or 177 to 180 [fed]/sample site; values are shown for hide or preevisceration carcass [pre-ev] samples per sample day; 9 sample days per region). Also depicted are the mean (horizontal lines) and 95% confidence intervals (bars) for the percent prevalence observed for each serotype in each region.
Characterization of the antimicrobial susceptibility profiles for 978 MDR Salmonella isolates showed the most common resistance pattern to be resistance to eight antimicrobials (AmApFTCSSuTe), with decreased susceptibility to ceftriaxone (Ax) (MIC range between 16 and 32 μg ml−1) (Fig. 1). Approximately one-third of all MDR Salmonella strains isolated in this study (31.3%) demonstrated this resistance pattern. The Salmonella serotypes found exhibiting this profile included Newport, Typhimurium, Agona, Anatum, Reading, Dublin, Enteritidis, Mbandaka, Saint Paul, Heidelberg, Give, and Ohio. Resistance to Ax was detected in 12.1% of all MDR Salmonella strains examined, while decreased susceptibility to Ax [here indicated by parentheses as “(Ax)”] was detected in 66.9% of the isolates examined. Noteworthy was the low incidence of Ax resistance associated with Salmonella serotype Typhimurium (0.6% of MDR Salmonella Typhimurium strains were resistant to Ax, while 9% demonstrated decreased susceptibility). Resistance to quinolone and fluoroquinolone antimicrobials was rarely detected, with 0.3% of the MDR Salmonella strains showing resistance to nalidixic acid (Salmonella serotypes Dublin, Agona, and Uganda), and no isolates were found with resistance to ciprofloxacin. Salmonella serotype Uganda was observed to have the most extensive resistance phenotype, with resistance to 12 of the 15 antimicrobials screened (Fig. 1).
The MDR Salmonella strains isolated from postintervention carcasses (n = 10) included those of serotypes (MDR phenotypes) Typhimurium (AmApCSSuTe), Dublin [AmApFT(Ax)CGKSSuTe], Reading [AmApFT(Ax)CSSuTe], and Newport [AmApFT(Ax)CKSSuTeSxt] and a nontypeable (NT) O-group D isolate (AmCKSSuTe). The dominant MDR Salmonella serotype observed at this sample site was Typhimurium (n = 6), while the other MDR Salmonella strains were each isolated once. The pansusceptible (PS) Salmonella strains (n = 14) isolated were dominated by serotype Dublin (n = 7). Other PS Salmonella enterica serotypes identified included Muenster (n = 2), Anatum (n = 2), and Cerro, Onrieke, and Montevideo (which were each isolated once).
XbaI PFGE analysis.
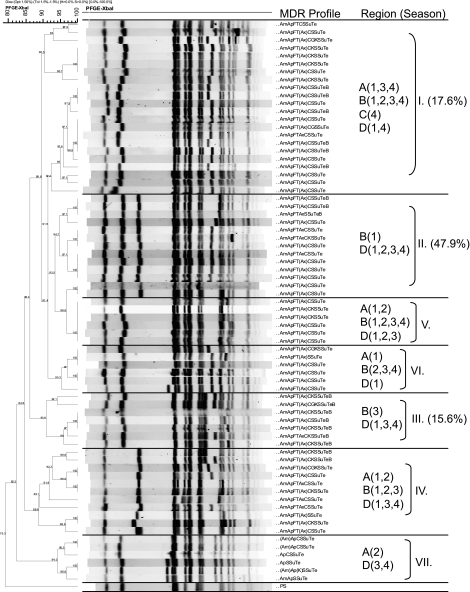
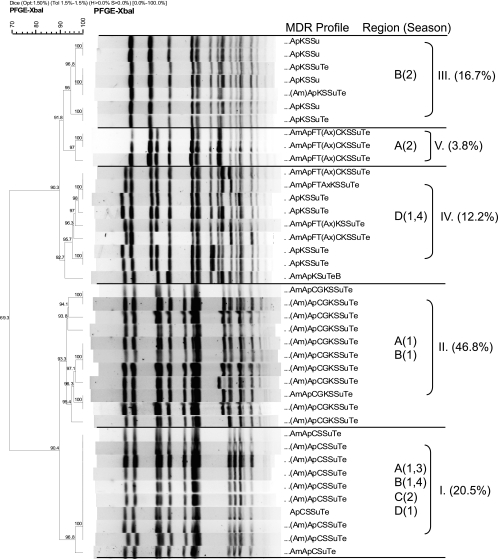
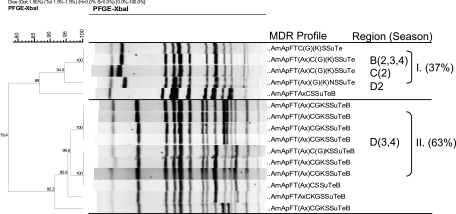
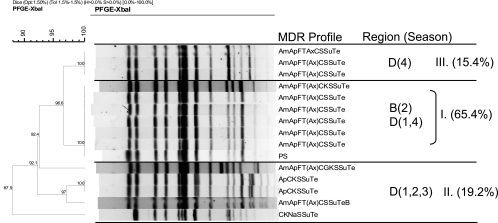
The XbaI PFGE profiles of MDR Salmonella strains collected in this study showed that isolates predominantly clustered by serotype and MDR phenotype, a phenomenon previously reported by others (44). Comparisons of XbaI profiles of the MDR Salmonella Newport isolates showed the overall similarity to be 79.3%, and the majority of isolates fell into seven PFGE clusters (Fig. 3). Certain clusters (e.g., cluster I) appeared to be widely disseminated and were isolated from cattle at harvest in plants located in multiple regions of the United States over multiple seasons. Conversely, other clusters appeared to be endemic, as they were isolated repeatedly over the course of the study, but primarily from plants in a limited number of regions (e.g., cluster II) (Fig. 3). Comparisons of the XbaI PFGE profiles of the MDR Salmonella Typhimurium isolates showed their overall similarity to be 69.3%, and these isolates fell predominantly into two major groups composed of five clusters (Fig. 4). The first group consisted of three clusters and included isolates exhibiting core resistance to ApKSSuTe (cluster III) and isolates with PFGE profiles similar to those of the recently described WA-TYP035/187 MDR clade (1, 31) (clusters IV and V), some of which demonstrated cephalosporin resistance. The second group of isolates exhibited the ApCSSuTe DT104 resistance phenotype (cluster I) or an expanded version of this phenotype with aminoglycoside resistance (cluster II). While the majority of MDR Salmonella Typhimurium isolates were obtained from plants sampled in regions A and B, isolates from cluster I (exhibiting the classic epidemic DT104 PFGE profile [27, 53]) appeared to be widely disseminated and were found in all regions sampled at various times of the year. In contrast, representatives of clusters III to V were isolated from cattle at harvest in plants only in certain regions (Fig. 4).
FIG. 3.
XbaI PFGE-based dendrogram and MDR profiles of representative Salmonella Newport isolates. Cluster analysis of banding patterns was performed using the Dice similarity coefficient and the unweighted-pair group method. Regions (A to D) and seasons (1, summer; 2, fall; 3, winter; 4, spring) where isolates were observed are indicated, in addition to the percentage of Salmonella Newport isolates found in that cluster, for the top three clusters. Antimicrobial abbreviations: Am, amoxicillin-clavulanic acid; Ap, ampicillin; C, chloramphenicol; S, streptomycin; Su, sulfisoxazole; Te, tetracycline; F, cefoxitin; T, ceftiofur; Ax, ceftriaxone; G, gentamicin; K, kanamycin; Sxt, sulfamethoxazole-trimethoprim; Cp, ciprofloxacin; N, nalidixic acid; and Ai, amikacin.
FIG. 4.
XbaI PFGE-based dendrogram and MDR profiles of representative Salmonella Typhimurium isolates. Cluster analysis of banding patterns was performed using the Dice similarity coefficient and the unweighted-pair group method. Regions (A to D) and seasons (1, summer; 2, fall; 3, winter; 4, spring) where isolates were observed are indicated, in addition to the percentage of Salmonella Typhimurium isolates found in that cluster. Antimicrobial abbreviations: Am, amoxicillin-clavulanic acid; Ap, ampicillin; C, chloramphenicol; S, streptomycin; Su, sulfisoxazole; Te, tetracycline; F, cefoxitin; T, ceftiofur; Ax, ceftriaxone; G, gentamicin; K, kanamycin; Sxt, sulfamethoxazole-trimethoprim; Cp, ciprofloxacin; N, nalidixic acid; and Ai, amikacin.
While certain MDR Salmonella Newport and Typhimurium clusters could be found widely disseminated, MDR Salmonella Uganda and Dublin strains were isolated primarily from cattle at harvest in regions B and D. The PFGE profiles of Salmonella Uganda were 79.4% similar overall and fell into two major groups. Isolates in the first group (cluster I) were found primarily in regions B and D, although one of these isolates was collected from region C in the fall (Fig. 5). In two of the four sample seasons, isolates in the second group (cluster II) were found only in region D. The Salmonella serotype demonstrating the most highly conserved PFGE profiles was MDR Dublin (87.9% similarity overall), and the isolates fell into three clusters, two of which comprised isolates found only in region D and one of which comprised isolates found in both regions B and D at different times of the year (Fig. 6).
FIG. 5.
XbaI PFGE-based dendrogram and MDR profiles of representative Salmonella Uganda isolates. Cluster analysis of banding patterns was performed using the Dice similarity coefficient and the unweighted-pair group method. Regions (B to D) and seasons (1, summer; 2, fall; 3, winter; 4, spring) where isolates were observed are indicated, in addition to the percentage of Salmonella Uganda isolates found in that cluster. Antimicrobial abbreviations: Am, amoxicillin-clavulanic acid; Ap, ampicillin; C, chloramphenicol; S, streptomycin; Su, sulfisoxazole; Te, tetracycline; F, cefoxitin; T, ceftiofur; Ax, ceftriaxone; G, gentamicin; K, kanamycin; Sxt, sulfamethoxazole-trimethoprim; Cp, ciprofloxacin; N, nalidixic acid; and Ai, amikacin.
FIG. 6.
XbaI PFGE-based dendrogram and MDR profiles of representative Salmonella Dublin isolates. Cluster analysis of banding patterns was performed using the Dice similarity coefficient and the unweighted-pair group method. Regions (B and D) and seasons (1, summer; 2, fall; 3, winter; 4, spring) where isolates were observed are indicated, in addition to the percentage of Salmonella Dublin isolates found in that cluster. Antimicrobial abbreviations: Am, amoxicillin-clavulanic acid; Ap, ampicillin; C, chloramphenicol; S, streptomycin; Su, sulfisoxazole; Te, tetracycline; F, cefoxitin; T, ceftiofur; Ax, ceftriaxone; G, gentamicin; K, kanamycin; Sxt, sulfamethoxazole-trimethoprim; Cp, ciprofloxacin; N, nalidixic acid; and Ai, amikacin.
DISCUSSION
Numerous studies have examined the prevalence of Salmonella on hides and carcasses of fed and cull cattle at slaughter (9, 11, 15, 39, 64). However, studies examining antimicrobial-resistant-Salmonella prevalence and diversity in beef production settings are fewer (9, 51, 78). Research efforts aimed at characterizing MDR Salmonella strains associated with feedlot or dairy cattle have primarily focused on fecal shedding of these pathogens from animals in farm settings (2, 29, 30, 35, 36, 74). While studies such as these add to our understanding of the epidemiology of these pathogens, surveys of MDR Salmonella prevalence on cattle hides at harvest are perhaps more directly related to addressing the food safety concerns of beef. The reasons for this are 2-fold. First, the ability to detect Salmonella in hide samples is likely enhanced in comparison with that ability for fecal samples of asymptomatic cattle. This is probably due to increased target bacterial concentration in the less restrictive, generally aerobic hide environment. Second, and perhaps more important, cattle hides are a major source of carcass contamination in beef processing environments (54, 57). Previous analysis of the aerobic bacterial load (aerobic plate count [APC]) of hide and preevisceration carcass samples collected in this study (n = 3,040 for each sample site) showed that on average ∼1.7% (95% CI, 0.96 to 3.13%) of hide contamination was observed to be transferred to carcasses during the dressing process (15). Also, numerous studies have shown that cattle hide hygiene is significantly affected by transportation and lairage prior to slaughter (5, 24, 49, 63). Accordingly, measurements of hide pathogen prevalence at harvest are an essential aspect of both understanding the magnitude of risk and determining what control measures may be necessary to mitigate those risks.
In this study, we found that MDR salmonellae were a consistently measurable subpopulation of the salmonellae present on hides and carcasses of cattle at slaughter. While the majority of Salmonella strains isolated were found to be sensitive to ampicillin, tetracycline, and kanamycin (and, in keeping with previous studies in our laboratory, were thus likely susceptible to the other 15 antimicrobials evaluated) or were found to be resistant to 1 antimicrobial (commonly tetracycline), MDR Salmonella strains were isolated from 16.7% (95% CI, 8.3 to 25.1%) of hide samples, 11.7% (95% CI, 4.4 to 19.0%) of preevisceration carcass samples, and 0.33% (95% CI, −0.03 to 0.7%) of postintervention carcass samples on average. The mean prevalence values for hide, preevisceration carcass, and postintervention carcass samples were somewhat greater than the median prevalence values (6.9%, 4.8%, and 0.0%, respectively), likely a reflection of the large differences in MDR Salmonella prevalence observed between plants in different regions. Analysis of MDR Salmonella prevalence by season showed no significant differences, although analysis by region did. Specifically, MDR Salmonella prevalence in plants in region C was consistently (although not significantly) lower than that observed in plants in region A or B. And throughout the course of the study, MDR Salmonella prevalence values for region C were significantly lower (P = 0.0008) than those observed for region D (Table 2). Given that plants that slaughter cull cows and bulls typically obtain animals from a broad geographic area, our observation here of regional differences in MDR Salmonella prevalence is puzzling. It is possible that these data reflect the impact of plant lairage environments on cattle hide hygiene at harvest. However, these environments are constantly being seeded with new microorganisms by animals that often come from many parts of the country. Undoubtedly, the factors that influence bacterial competition and persistence in lairage environments (or in upstream environments such as auction markets or buying stations) represent important areas of further study, so that the impact of these factors on food safety can be assessed. However, it is also important to emphasize the limited nature of the study described herein, given that only one or two plants per region were sampled, for a total of nine sample days per region. Accordingly, the results presented here are by no means comprehensive. Rather, these data represent “snapshots” of what can be observed for MDR Salmonella prevalence in diverse locations overtime, which provide baseline information for further investigations into these complex systems.
In all, 16,218 Salmonella cattle hide and carcass isolates were collected and screened for antimicrobial resistance. The most commonly observed MDR Salmonella serotypes were Newport, Typhimurium, and Uganda, which collectively made up 80.6% of all MDR Salmonella strains characterized (Fig. 1). MDR Salmonella Newport resistance phenotypes were dominated by the MDR-AmpC phenotype (Fig. 3), while MDR Salmonella Typhimurium exhibited four basic resistance phenotypes (Fig. 4). These included the ACSSuTe phenotype that is typically associated with DT104, an expanded version of this phenotype that included resistance to aminoglycosides, the AKSSuTe phenotype that is frequently associated with DT193 or 208 (40, 61), and an expanded version of this phenotype including cephalosporin resistance. The isolates in the last group demonstrated XbaI PFGE profiles similar to those of the recently described MDR Salmonella Typhimurium clade WA-TYP035/187, reported by Adhikari et al., who also found these Salmonella Typhimurium strains to exhibit cephalosporin resistance (1), a phenotype that is generally uncommon in MDR Salmonella Typhimurium (52). The third most frequently isolated MDR Salmonella serotype was Uganda, and these isolates were found to demonstrate resistance to 12 of the 15 antimicrobials screened. All MDR Salmonella Uganda strains characterized were resistant to the cephalosporins ceftiofur and cefoxitin and showed either decreased sensitivity or resistance to ceftriaxone (Fig. 1).
In the past decade, MDR Salmonella strains have received increased attention from the medical community, especially those resistant to the extended-spectrum cephalosporin ceftriaxone, the drug of choice for the treatment of invasive salmonellosis in children (42). The emergence of cephalosporin resistance in Salmonella has been attributed to the spread of a large resistance plasmid containing the blaCMY2 gene (18, 38, 52, 59, 76). It has been suggested that extensive therapeutic use of the veterinary cephalosporin ceftiofur has been a major driving force for the dissemination of this resistance (20, 32), and there is concern that ceftiofur-resistant Salmonella strains may develop cross-resistance to ceftriaxone, because of the structural similarity of these two drugs. In this study, we found that 83.7% of the MDR Salmonella strains characterized were resistant to ceftiofur. Ceftriaxone resistance was observed less frequently, with only 12.1% of the isolates examined showing resistance to this antimicrobial; however, decreased susceptibility to ceftriaxone was detected in 66.9% of the isolates. Overall, the percentage of the total number of Salmonella strains characterized in this study found to be resistant to ceftriaxone was low (∼5%, or 819 of 16,218 isolates). Nevertheless, veterinarians and cattle producers should take appropriate measures to minimize the spread of resistant pathogens when treating cattle with ceftiofur (28), as the selective pressure of antimicrobial exposure will cause subpopulations of any cephalosporin-resistant bacteria present to surge in number, possibly leading to an increase in the attempts to transfer these resistance determinants among bacterial populations.
Repetitive sampling at cattle slaughter establishments over the course of 10 months provided a unique opportunity to observe the diversity of MDR Salmonella strains present in these settings over time. Characterization of the serotypes, MDR phenotypes, and XbaI PFGE profiles of the Salmonella strains isolated revealed the existence of biotypes that appeared to be persistent and widely disseminated (observed in multiple seasons and from plants in multiple regions of the United States, as seen for Salmonella Newport and Typhimurium clusters I in Fig. 3 and 4, respectively) or endemic (observed in limited seasons or regions of the United States, as seen for Salmonella Newport cluster II [Fig. 3] and Salmonella Typhimurium clusters II to V [Fig. 4]). Hoelzer et al. recently reported a similar observation on the persistence and regional distribution of certain subtypes of Salmonella Newport and Typhimurium strains isolated from cattle and humans in two geographic regions of the United States (46). Salmonella Typhimurium and Newport are noted as host “generalists,” meaning they are able to infect and cause disease in a wide range of host species (although Salmonella Typhimurium is also the mouse host-adapted serovar, causing murine typhoid fever). Accordingly, these serovars may be better able to spread or disseminate from one environment to the next, either by wild-animal movements or contaminated feed sources or by manure or agricultural waste runoff. Numerous studies have documented the ability of Salmonella Newport and Typhimurium strains to survive in manure and terrestrial environments for extended periods (upwards of 9 months) (10, 13, 25, 77). Of course, genome-wide analyses would be needed to establish the relatedness of isolates in the clusters identified here, as a single PFGE profile is not a definitive indicator of lineage or relatedness (33, 46). However, the observation of potentially epidemic and endemic biotypes in this study further demonstrates the need, mentioned above, for research into the ability of pathogens to persist in epidemiologically complex, agricultural settings.
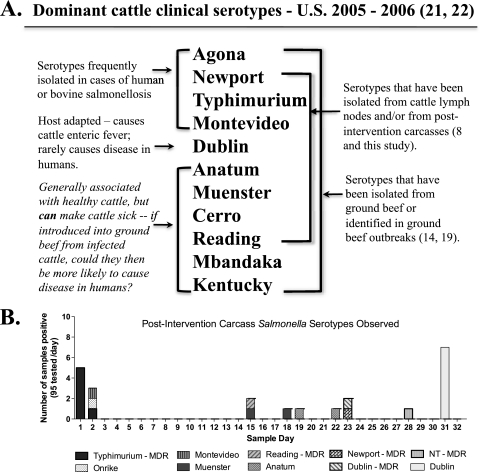
A final observation concerns comparisons of the Salmonella serotypes isolated from cattle hides versus those isolated from postintervention carcasses in this study. Preliminary typing data on all Salmonella strains collected in this study indicate that the dominant serotypes found entering processing environments on cattle hides were Montevideo, Anatum, and Muenster. These data are in keeping with the observations of Kunze et al., who found Salmonella Anatum and Montevideo to be the predominant serotypes on cattle hides at harvest (51). These serotypes are noted as being frequently isolated from healthy cattle (17, 74), and yet they were not the predominant serotypes found on postintervention carcasses here. As seen in Fig. 7B, postintervention carcass contamination was generally sporadic. However, on two separate occasions, clusters of postintervention carcasses contaminated with Salmonella were observed. The Salmonella strains found contaminating these carcasses were those of MDR Salmonella Typhimurium in one event and pansusceptible Salmonella Dublin in the other, both serotypes known for causing clinical illness in cattle.
FIG. 7.
(A) Comparison of the serotypes most frequently isolated from clinically infected cattle with those noted for frequently causing disease in humans and those isolated from cattle lymph nodes, ground beef, and postintervention carcasses. (B) Graph depicting the distribution of Salmonella serotypes isolated from postintervention carcasses by sample day.
This observation prompted a comparison of the Salmonella serotypes reported in a number of studies, including serotypes found in ground beef (14, 75, 79), those isolated from cattle lymph nodes (8), and those found on final carcasses in this study, along with serotypes noted for being isolated from clinically infected cattle (21, 22). The last group was identified from the Salmonella Surveillance Annual Summary reports for 2005 and 2006, which summarize the Salmonella strains isolated and serotyped from clinical cases of animal disease that were reported to the Centers for Disease Control and Prevention (CDC) and the National Veterinary Services Laboratory (NVSL) in those time periods. The CDC cautions that samples from nonhuman sources that are tested for Salmonella are obtained in a variety of ways and that sampling is neither complete nor random and undoubtedly has biases. Nevertheless, the same 11 serotypes were identified as causing more than 70% of reported bovine salmonellosis in 2005 (n = 2,674) and 2006 (n = 3,770) (82.6% and 72.9%, respectively). In comparing these 11 serotypes with those found in ground beef, in cattle lymph nodes, and on final carcasses, we found considerable overlap in the serotype distribution of these data sets (Fig. 7A).
Salmonella strains entering beef processing environments on the hides or in the feces of cattle, weather they are MDR or not, are undoubtedly present in a variety of metabolic states. The metabolic state of an organism, in combination with its genetic makeup (in terms of the stress response and virulence genes present), strongly influences the ability of that organism to respond to environmental stressors. One of the ways that Salmonella enters beef processing environments is possibly via cattle that are Salmonella carriers. It is well known that Salmonella infection can lead to the development of a carrier state (47, 69). After primary challenge, cattle may become passive carriers (immune animals demonstrating no active pathology that excrete Salmonella acquired from contaminated environmental sources), active carriers (animals excreting high levels of Salmonella, often in the absence of clinical signs), or latent carriers (asymptomatic animals with persistent Salmonella infection present in tissues) (69). Salmonella carriers that enter beef processing environments are likely shedding Salmonella strains that have survived the host environment. These Salmonella strains could, consequently, be adapted to acidic pH or exposure to thermal extremes (16) and as a result would be better equipped to survive carcass processing interventions. This could account for the preponderance of serotypes noted for infecting cattle on postintervention carcasses and in ground beef. The data presented here highlight the need for a better understanding of the biology of Salmonella carrier status in cattle and of the metabolic state(s) of the Salmonella being shed by carriers. Research in these areas will provide vital information for future efforts aimed at controlling Salmonella in cattle production and processing environments.
Acknowledgments
This project was funded in part by The Beef Checkoff.
We thank the plant personnel for their kind assistance and cooperation. We also thank Michael Guerini for his contribution to this project, Julie Dyer, Frank Reno, Bruce Jasch, Greg Smith, Kim Kucera, Jade Franklin, Casey Trambly, Sara Schumacher, and Emily Griess for their technical support, and Joan Rosch for administrative assistance.
The use of product names is necessary for factual reporting on available data; however, the USDA neither guarantees nor warrants the standard of the product, and the use of the name by the USDA does not imply approval of the product to the exclusion of others that may also be suitable.
Footnotes
Published ahead of print on 14 January 2011.
REFERENCES
- 1.Adhikari, B., et al. 2010. Multilocus variable-number tandem-repeat analysis and plasmid profiling to study the occurrence of blaCMY-2 within a pulsed-field gel electrophoresis-defined clade of Salmonella enterica serovar Typhimurium. Appl. Environ. Microbiol. 76:69-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alam, M. J., et al. 2009. Antimicrobial susceptibility profiles of Salmonella enterica serotypes recovered from pens of commercial feedlot cattle using different types of composite samples. Curr. Microbiol. 58:354-359. [DOI] [PubMed] [Google Scholar]
- 3.Alcaine, S. D., L. D. Warnick, and M. Wiedmann. 2007. Antimicrobial resistance in nontyphoidal Salmonella. J. Food Prot. 70:780-790. [DOI] [PubMed] [Google Scholar]
- 4.Alexander, K. A., L. D. Warnick, and M. Wiedmann. 2009. Antimicrobial resistant Salmonella in dairy cattle in the United States. Vet. Res. Commun. 33:191-209. [DOI] [PubMed] [Google Scholar]
- 5.Arthur, T. M., et al. 2008. Source tracking of Escherichia coli O157:H7 and Salmonella contamination in the lairage environment at commercial U.S. beef processing plants and identification of an effective intervention. J. Food Prot. 71:1752-1760. [DOI] [PubMed] [Google Scholar]
- 6.Arthur, T. M., et al. 2007. Effects of a minimal hide wash cabinet on the levels and prevalence of Escherichia coli O157:H7 and Salmonella on the hides of beef cattle at slaughter. J. Food Prot. 70:1076-1079. [DOI] [PubMed] [Google Scholar]
- 7.Arthur, T. M., et al. 2004. Escherichia coli O157 prevalence and enumeration of aerobic bacteria, Enterobacteriaceae, and Escherichia coli O157 at various steps in commercial beef processing plants. J. Food Prot. 67:658-665. [DOI] [PubMed] [Google Scholar]
- 8.Arthur, T. M., et al. 2008. Prevalence and characterization of Salmonella in bovine lymph nodes potentially destined for use in ground beef. J. Food Prot. 71:1685-1688. [DOI] [PubMed] [Google Scholar]
- 9.Bacon, R. T., J. N. Sofos, K. E. Belk, D. R. Hyatt, and G. C. Smith. 2002. Prevalence and antibiotic susceptibility of Salmonella isolated from beef animal hides and carcasses. J. Food Prot. 65:284-290. [DOI] [PubMed] [Google Scholar]
- 10.Baloda, S. B., L. Christensen, and S. Trajcevska. 2001. Persistence of a Salmonella enterica serovar Typhimurium DT12 clone in a piggery and in agricultural soil amended with Salmonella-contaminated slurry. Appl. Environ. Microbiol. 67:2859-2862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barkocy-Gallagher, G. A., et al. 2003. Seasonal prevalence of Shiga toxin-producing Escherichia coli, including O157:H7 and non-O157 serotypes, and Salmonella in commercial beef processing plants. J. Food Prot. 66:1978-1986. [DOI] [PubMed] [Google Scholar]
- 12.Barkocy-Gallagher, G. A., et al. 2005. Methods for recovering Escherichia coli O157:H7 from cattle fecal, hide, and carcass samples: sensitivity and improvements. J. Food Prot. 68:2264-2268. [DOI] [PubMed] [Google Scholar]
- 13.Berge, A. C., J. M. Adaska, and W. M. Sischo. 2004. Use of antibiotic susceptibility patterns and pulsed-field gel electrophoresis to compare historic and contemporary isolates of multi-drug-resistant Salmonella enterica subsp. enterica serovar Newport. Appl. Environ. Microbiol. 70:318-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bosilevac, J. M., M. N. Guerini, N. Kalchayanand, and M. Koohmaraie. 2009. Prevalence and characterization of salmonellae in commercial ground beef in the United States. Appl. Environ. Microbiol. 75:1892-1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brichta-Harhay, D., et al. 2008. Salmonella and Escherichia coli O157:H7 contamination on hides and carcasses of cull cattle presented for slaughter in the United States: an evaluation of prevalence and bacterial loads by immunomagnetic separation and direct plating methods. Appl. Environ. Microbiol. 74:6289-6297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Buchmeier, N. A., and F. Heffron. 1990. Induction of Salmonella stress proteins upon infection of macrophages. Science 248:730-732. [DOI] [PubMed] [Google Scholar]
- 17.Callaway, T. R., et al. 2005. Fecal prevalence and diversity of Salmonella species in lactating dairy cattle in four states. J. Dairy Sci. 88:3603-3608. [DOI] [PubMed] [Google Scholar]
- 18.Carattoli, A., et al. 2002. Characterization of plasmids carrying CMY-2 from expanded-spectrum cephalosporin-resistant Salmonella strains isolated in the United States between 1996 and 1998. Antimicrob. Agents Chemother. 46:1269-1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.CDC. 2009. National Antimicrobial Resistance Monitoring System for Enteric Bacteria (NARMS): human isolates final report, 2007. CDC, U.S. Department of Health and Human Services, Atlanta, GA. www.cdc.gov/narms/annual/2007/NARMSAnnualReport2007.pdf.
- 20.CDC. 2002. Outbreak of multidrug-resistant Salmonella Newport—United States, January-April 2002. MMWR Morb. Mortal. Wkly. Rep. 51:545-548. [PubMed] [Google Scholar]
- 21.CDC. 2007. Salmonella surveillance: annual summary, 2005. CDC, U.S. Department of Health and Human Services, Atlanta, GA.
- 22.CDC. 2008. Salmonella surveillance: annual summary, 2006. CDC, U.S. Department of Health and Human Services, Atlanta, GA.
- 23.Chen, S., et al. 2004. Characterization of multiple-antimicrobial-resistant Salmonella serovars isolated from retail meats. Appl. Environ. Microbiol. 70:1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Childs, K. D., et al. 2006. Molecular characterization of Escherichia coli O157:H7 hide contamination routes: feedlot to harvest. J. Food Prot. 69:1240-1247. [DOI] [PubMed] [Google Scholar]
- 25.Clegg, F. G., S. N. Chiejina, A. L. Duncan, R. N. Kay, and C. Wray. 1983. Outbreaks of Salmonella Newport infection in dairy herds and their relationship to management and contamination of the environment. Vet. Rec. 112:580-584. [DOI] [PubMed] [Google Scholar]
- 26.Cohen, M. L., and R. V. Tauxe. 1986. Drug-resistant Salmonella in the United-States—an epidemiologic perspective. Science 234:964-969. [DOI] [PubMed] [Google Scholar]
- 27.Cooke, F. J., et al. 2008. Characterization of the genomes of a diverse collection of Salmonella enterica serovar Typhimurium definitive phage type 104. J. Bacteriol. 190:8155-8162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cummings, K. J., et al. 2010. Temporal clusters of bovine Salmonella cases at a veterinary medical teaching hospital, 1996-2007. Vector Borne Zoonotic Dis. 10:471-479. [DOI] [PubMed] [Google Scholar]
- 29.Cummings, K. J., et al. 2009. The incidence of salmonellosis among dairy herds in the northeastern United States. J. Dairy Sci. 92:3766-3774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dargatz, D. A., et al. 2003. Prevalence and antimicrobial susceptibility of Salmonella spp. isolates from US cattle in feedlots in 1999 and 2000. J. Appl. Microbiol. 95:753-761. [DOI] [PubMed] [Google Scholar]
- 31.Davis, M. A., et al. 2007. Multidrug-resistant Salmonella Typhimurium, Pacific Northwest, United States. Emerg. Infect. Dis. 13:1583-1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Davis, M. A., et al. 2007. Antimicrobial resistance in Salmonella enterica serovar Dublin isolates from beef and dairy sources. Vet. Microbiol. 119:221-230. [DOI] [PubMed] [Google Scholar]
- 33.Davis, M. A., et al. 2003. Correlation between geographic distance and genetic similarity in an international collection of bovine faecal Escherichia coli O157:H7 isolates. Epidemiol. Infect. 131:923-930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dodd, C. C., et al. 2010. Genetic relatedness of Escherichia coli O157 isolates from cattle feces and preintervention beef carcasses. Foodborne Pathog. Dis. 7:357-365. [DOI] [PubMed] [Google Scholar]
- 35.Edrington, T. S., et al. 2008. Investigation into the seasonal salmonellosis in lactating dairy cattle. Epidemiol. Infect. 136:381-390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fedorka-Cray, P. J., D. A. Dargatz, L. A. Thomas, and J. T. Gray. 1998. Survey of Salmonella serotypes in feedlot cattle. J. Food Prot. 61:525-530. [DOI] [PubMed] [Google Scholar]
- 37.Fricke, W. F., et al. 2009. Antimicrobial resistance-conferring plasmids with similarity to virulence plasmids from avian pathogenic Escherichia coli strains in Salmonella enterica serovar Kentucky isolates from poultry. Appl. Environ. Microbiol. 75:5963-5971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Frye, J. G., P. J. Fedorka-Cray, C. R. Jackson, and M. Rose. 2008. Analysis of Salmonella enterica with reduced susceptibility to the third-generation cephalosporin ceftriaxone isolated from U.S. cattle during 2000-2004. Microb. Drug Resist. 14:251-258. [DOI] [PubMed] [Google Scholar]
- 39.Galland, J. C., et al. 2001. Diversity of Salmonella serotypes in cull (market) dairy cows at slaughter. J. Am. Vet. Med. Assoc. 219:1216-1220. [DOI] [PubMed] [Google Scholar]
- 40.Gebreyes, W. A., and C. Altier. 2002. Molecular characterization of multidrug-resistant Salmonella enterica subsp. enterica serovar Typhimurium isolates from swine. J. Clin. Microbiol. 40:2813-2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Greene, S. K., A. M. Stuart, F. M. Medalla, and J. M. Whichard. 2008. Distribution of multidrug-resistant human isolates of MDR-ACSSuT Salmonella Typhimurium and MDR-AmpC Salmonella Newport in the United States, 2003-2005. Foodborne Pathog. Dis. 5:669-680. [DOI] [PubMed] [Google Scholar]
- 42.Guerrant, R. L., et al. 2001. Practice guidelines for the management of infectious diarrhea. Clin. Infect. Dis. 32:331-351. [DOI] [PubMed] [Google Scholar]
- 43.Gupta, A., et al. 2003. Emergence of multidrug-resistant Salmonella enterica serotype Newport infections resistant to expanded-spectrum cephalosporins in the United States. J. Infect. Dis. 188:1707-1716. [DOI] [PubMed] [Google Scholar]
- 44.Harbottle, H., D. G. White, P. F. McDermott, R. D. Walker, and S. Zhao. 2006. Comparison of multilocus sequence typing, pulsed-field gel electrophoresis, and antimicrobial susceptibility typing for characterization of Salmonella enterica serotype Newport isolates. J. Clin. Microbiol. 44:2449-2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Helms, M., J. Simonsen, and K. Molbak. 2004. Quinolone resistance is associated with increased risk of invasive illness or death during infection with Salmonella serotype Typhimurium. J. Infect. Dis. 190:1652-1654. [DOI] [PubMed] [Google Scholar]
- 46.Hoelzer, K., et al. 2010. The prevalence of multidrug resistance is higher among bovine than human Salmonella enterica serotype Newport, Typhimurium, and 4,5,12:i:- isolates in the United States but differs by serotype and geographic. Appl. Environ. Microbiol. 76:5947-5959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.House, J. K., B. P. Smith, and D. Kamiya. 2001. Serological distinction of bovine Salmonella carriers from vaccinated and acutely infected cows. J. Vet. Diagn. Invest. 13:483-488. [DOI] [PubMed] [Google Scholar]
- 48.Hunter, S. B., et al. 2005. Establishment of a universal size standard strain for use with the PulseNet standardized pulsed-field gel electrophoresis protocols: converting the national databases to the new size standard. J. Clin. Microbiol. 43:1045-1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jacob, M. E., D. G. Renter, and T. G. Nagaraja. 2010. Animal- and truckload-level associations between Escherichia coli O157:H7 in feces and on hides at harvest and contamination of preevisceration beef carcasses. J. Food Prot. 73:1030-1037. [DOI] [PubMed] [Google Scholar]
- 50.Karon, A. E., J. R. Archert, M. J. Sotir, T. A. Monson, and J. J. Kazmierczakt. 2007. Human multidrug-resistant Salmonella Newport infections, Wisconsin, 2003-2005. Emerg. Infect. Dis. 13:1777-1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kunze, D. J., et al. 2008. Salmonella enterica burden in harvest-ready cattle populations from the southern high plains of the United States. Appl. Environ. Microbiol. 74:345-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lindsey, R. L., P. J. Fedorka-Cray, J. G. Frye, and R. J. Meinersmann. 2009. Inc A/C plasmids are prevalent in multidrug-resistant Salmonella enterica isolates. Appl. Environ. Microbiol. 75:1908-1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Matiasovicova, J., et al. 2007. Identification of putative ancestors of the multidrug-resistant Salmonella enterica serovar Typhimurium DT104 clone harboring the Salmonella genomic island 1. Arch. Microbiol. 187:415-424. [DOI] [PubMed] [Google Scholar]
- 54.McEvoy, J. M., et al. 2000. The relationship between hide cleanliness and bacterial numbers on beef carcasses at a commercial abattoir. Lett. Appl. Microbiol. 30:390-395. [DOI] [PubMed] [Google Scholar]
- 55.Mead, P. S., et al. 1999. Food-related illness and death in the United States. Emerg. Infect. Dis. 5:607-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nou, X., et al. 2006. Improvement of immunomagnetic separation for Escherichia coli O157:H7 detection by the PickPen magnetic particle separation device. J. Food Prot. 69:2870-2874. [DOI] [PubMed] [Google Scholar]
- 57.Nou, X. W., et al. 2003. Effect of chemical dehairing on the prevalence of Escherichia coli O157 : H7 and the levels of aerobic bacteria and Enterobacteriaceae on carcasses in a commercial beef processing plant. J. Food Prot. 66:2005-2009. [DOI] [PubMed] [Google Scholar]
- 58.Nucera, D. M., C. W. Maddox, P. Hoien-Dalen, and R. M. Weigel. 2006. Comparison of API 20E and invA PCR for identification of Salmonella enterica isolates from swine production units. J. Clin. Microbiol. 44:3388-3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Poole, T. L., et al. 2009. Conjugative transferability of the A/C plasmids from Salmonella enterica isolates that possess or lack bla(CMY) in the A/C plasmid backbone. Foodborne Pathog. Dis. 6:1185-1194. [DOI] [PubMed] [Google Scholar]
- 60.Poppe, C., et al. 2006. Characterization of antimicrobial resistance of Salmonella Newport isolated from animals, the environment, and animal food products in Canada. Can. J. Vet. Res. 70:105-114. [PMC free article] [PubMed] [Google Scholar]
- 61.Rabatsky-Ehr, T., et al. 2004. Multidrug-resistant strains of Salmonella enterica Typhimurium, United States, 1997-1998. Emerg. Infect. Dis. 10:795-801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rahn, K., et al. 1992. Amplification of an invA gene sequence of Salmonella Typhimurium by polymerase chain reaction as a specific method of detection of Salmonella. Mol. Cell. Probes 6:271-279. [DOI] [PubMed] [Google Scholar]
- 63.Reicks, A. L., et al. 2007. Impact of transportation of feedlot cattle to the harvest facility on the prevalence of Escherichia coli O157:H7, Salmonella, and total aerobic microorganisms on hides. J. Food Prot. 70:17-21. [DOI] [PubMed] [Google Scholar]
- 64.Rhoades, J. R., G. Duffy, and K. Koutsoumanis. 2009. Prevalence and concentration of verocytotoxigenic Escherichia coli, Salmonella enterica and Listeria monocytogenes in the beef production chain: a review. Food Microbiol. 26:357-376. [DOI] [PubMed] [Google Scholar]
- 65.Ribot, E. M., et al. 2006. Standardization of pulsed-field gel electrophoresis protocols for the subtyping of Escherichia coli O157:H7, Salmonella, and Shigella for PulseNet. Foodborne Pathog. Dis. 3:59-67. [DOI] [PubMed] [Google Scholar]
- 66.Scharff, R. L. 2010. Health-related costs from foodborne illness in the United States, p. 1-27. The Produce Safety Project at Georgetown University. Georgetown University, Washington, DC.
- 67.Simpson, E. H. 1949. Measurement of diversity. Nature 163:688. [Google Scholar]
- 68.Spika, J. S., et al. 1987. Chloramphenicol-resistant Salmonella Newport traced through hamburger to dairy farms. A major persisting source of human salmonellosis in California. N. Engl. J. Med. 316:565-570. [DOI] [PubMed] [Google Scholar]
- 69.Stevens, M. P., T. J. Humphrey, and D. J. Maskell. 2009. Molecular insights into farm animal and zoonotic Salmonella infections. Philos. Trans. R. Soc. Lond. B Biol. Sci. 364:2709-2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Troutt, H. F., and B. I. Osburn. 1997. Meat from dairy cows: possible microbiological hazards and risks. Rev. Sci. Tech. 16:405-414. [DOI] [PubMed] [Google Scholar]
- 71.Varma, J. K., et al. 2005. Hospitalization and antimicrobial resistance in Salmonella outbreaks, 1984-2002. Emerg. Infect. Dis. 11:943-946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Varma, J. K., et al. 2006. Highly resistant Salmonella Newport-MDRAmpC transmitted through the domestic US food supply: a FoodNet case-control study of sporadic Salmonella Newport infections, 2002-2003. J. Infect. Dis. 194:222-230. [DOI] [PubMed] [Google Scholar]
- 73.Venturini, C., S. A. Beatson, S. P. Djordjevic, and M. J. Walker. 2010. Multiple antibiotic resistance gene recruitment onto the enterohemorrhagic Escherichia coli virulence plasmid. FASEB J. 24:1160-1166. [DOI] [PubMed] [Google Scholar]
- 74.Wells, S. J., P. J. Fedorka-Cray, D. A. Dargatz, K. Ferris, and A. Green. 2001. Fecal shedding of Salmonella spp. by dairy cows on farm and at cull cow markets. J. Food Prot. 64:3-11. [DOI] [PubMed] [Google Scholar]
- 75.White, D. G., et al. 2001. The isolation of antibiotic-resistant Salmonella from retail ground meats. N. Engl. J. Med. 345:1147-1154. [DOI] [PubMed] [Google Scholar]
- 76.Winokur, P. L., D. L. Vonstein, L. J. Hoffman, E. K. Uhlenhopp, and G. V. Doern. 2001. Evidence for transfer of CMY-2 AmpC beta-lactamase plasmids between Escherichia coli and Salmonella isolates from food animals and humans. Antimicrob. Agents Chemother. 45:2716-2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.You, Y., et al. 2006. Survival of Salmonella enterica serovar Newport in manure and manure-amended soils. Appl. Environ. Microbiol. 72:5777-5783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhao, S., et al. 2005. Characterization of Salmonella Typhimurium of animal origin obtained from the National Antimicrobial Resistance Monitoring System. Foodborne Pathog. Dis. 2:169-181. [DOI] [PubMed] [Google Scholar]
- 79.Zhao, S., et al. 2006. Antimicrobial resistance and genetic relatedness among Salmonella from retail foods of animal origin: NARMS retail meat surveillance. Foodborne Pathog. Dis. 3:106-117. [DOI] [PubMed] [Google Scholar]
- 80.Zhao, S., et al. 2007. Characterization of multidrug resistant Salmonella recovered from diseased animals. Vet. Microbiol. 123:122-132. [DOI] [PubMed] [Google Scholar]