Abstract
“Metallosphaera yellowstonensis” is a thermoacidophilic archaeon isolated from Yellowstone National Park that is capable of autotrophic growth using Fe(II), elemental S, or pyrite as electron donors. Analysis of the draft genome sequence from M. yellowstonensis strain MK1 revealed seven different copies of heme copper oxidases (subunit I) in a total of five different terminal oxidase complexes, including doxBCEF, foxABCDEFGHIJ, soxABC, and the soxM supercomplex, as well as a novel hypothetical two-protein doxB-like polyferredoxin complex. Other genes found in M. yellowstonensis with possible roles in S and or Fe cycling include a thiosulfate oxidase (tqoAB), a sulfite oxidase (som), a cbsA cytochrome b558/566, several small blue copper proteins, and a novel gene sequence coding for a putative multicopper oxidase (Mco). Results from gene expression studies, including reverse transcriptase (RT) quantitative PCR (qPCR) of cultures grown autotrophically on either Fe(II), pyrite, or elemental S showed that the fox gene cluster and mco are highly expressed under conditions where Fe(II) is an electron donor. Metagenome sequence and gene expression studies of Fe-oxide mats confirmed the importance of fox genes (e.g., foxA and foxC) and mco under Fe(II)-oxidizing conditions. Protein modeling of FoxC suggests a novel lysine-lysine or lysine-arginine heme B binding domain, indicating that it is likely the cytochrome component of a heterodimer complex with foxG as a ferredoxin subunit. Analysis of mco shows that it encodes a novel multicopper blue protein with two plastocyanin type I copper domains that may play a role in the transfer of electrons within the Fox protein complex. An understanding of metabolic pathways involved in aerobic iron and sulfur oxidation in Sulfolobales has broad implications for understanding the evolution and niche diversification of these thermophiles as well as practical applications in fields such as bioleaching of trace metals from pyritic ores.
Terminal oxidases, required for all aerobic organisms, perform the final step in electron transport, coupling the reduction of oxygen to proton translocation. There are two known types of terminal oxidases: the universal heme copper oxidases (HCOs) and the less ubiquitous, bd-type quinol oxidases found only in prokaryotes (17, 44). Terminal oxidase complexes often contain 3 to 4 subunits in prokaryotes, compared to 13 in mitochondria. However, the HCO (CuB-Fe center) required for the reduction of O2 is highly conserved across the three domains of life (consistently referred to as subunit I). Most prokaryotic terminal oxidase complexes also contain a conserved heme copper oxidase subunit II, responsible for binding and oxidation of either cytochrome c (with a CuA center) or ubiquinol (no CuA center), followed by electron transfer to subunit I (17, 45). Alternative mechanisms exist in some terminal oxidase complexes, such as the SoxM supercomplex, where a blue copper sulfocyanin protein is thought to transfer electrons directly to the HCO subunit II (33, 34). Redox potential and oxygen affinity vary among different terminal oxidase complexes, consistent with the numerous electron donors used by aerobic microorganisms (17, 45).
Reduced Fe and S species (e.g., Fe2+, HS−) are important electron donors in all natural environments but are especially important in acid mine drainage and hydrothermal systems (6, 19, 28). Metabolic pathways involved in the oxidation of ferrous iron [Fe(II)] have been studied extensively in the bacteria Acidithiobacillus ferrooxidans and Leptospirillum ferrooxidans (2, 10, 20, 49, 52, 53, 61, 62). The proposed pathway in A. ferrooxidans involves the blue copper protein rusticyanin and various c-type cytochromes (61, 62). A novel c-type cytochrome, cytochrome c579, has been implicated in Fe(II) oxidation in a Leptospirillum-dominated community at Iron Mountain, CA (53). In contrast, Ferroplasma acidiphilum, an archaeon present in the same community, is believed to oxidize Fe(II) directly via a small blue copper protein (15).
Several of the diverse terminal oxidase complexes from members of the order Sulfolobales (phylum Crenarchaeota) have been characterized and include the aa3-type quinol oxidase DoxBCEF complex from Acidianus ambivalens (47), the aa3 quinol oxidase SoxABC complex, and the SoxM supercomplex of Sulfolobus spp. (35, 40, 41). More recently, a novel terminal oxidase complex (the foxABCDEFGHIJ gene cluster) has been reported in Sulfolobus metallicus, Sulfolobus tokodaii, and Metallosphaera sedula, and this complex has been associated with the oxidation of Fe(II) (5, 8). Several genes in the fox cluster code for hypothetical proteins whose role in electron transport via Fe(II) is unknown at the current time. In fact, only foxA and foxB are clearly defined as similar to the subunits I and II found in other terminal oxidase complexes, respectively. Previous gene expression studies of M. sedula have shown increased expression of genes within the fox cluster as well as the cbsAB-soxLN cytochrome ba complex in cultures grown on Fe(II) or pyrite relative to S0 (5, 7, 8, 29). However, no studies have been conducted in Metallosphaera-like organisms found in acidic geothermal habitats of Yellowstone National Park (YNP), where the oxidation of Fe(II) results in the deposition of numerous different types of Fe(III)-oxyhydroxide and/or jarositic microbial mats, depending on other geochemical conditions (25, 36).
Prior 16S rRNA gene surveys (25, 36, 43) and recent metagenome sequence analyses (27) indicate that organisms highly similar (>99% identity to the 16S rRNA gene) to “Metallosphaera yellowstonensis” (strain MK1) are consistently one of the dominant community members in acidic thermophilic Fe(II)-oxidizing microbial mats of YNP. Metallosphaera yellowstonensis is ideal for studying relationships among different terminal oxidases and electron donors, because the organism grows autotrophically on Fe(II), elemental S, or complex organic C (e.g., yeast extract [YE]) as a sole C and energy source. Moreover, the role of these Fe(II)-oxidizing chemoautotrophs in contemporary geothermal habitats provides clues regarding the importance of oxygen in the metabolism and evolutionary history of the Sulfolobales. The specific objectives of the current study were to (i) characterize the terminal oxidase genes found in the draft genome sequence of Metallosphaera yellowstonensis strain MK1 and in acidic, thermophilic Fe(III)-oxyhydroxide microbial mats of YNP, (ii) quantify the expression of terminal oxidase genes in field samples as well as cultures grown autotrophically on Fe(II), pyrite, or elemental S and heterotrophically on YE, and (iii) evaluate the potential function of genes within the fox cluster by using bioinformatic approaches (e.g., protein modeling), with special focus on the putative blue copper and cytochrome b proteins that appear to play an important role in the oxidation of Fe(II).
MATERIALS AND METHODS
DNA extraction for genomic sequencing.
Metallosphaera yellowstonensis strain MK1 was cultivated at 70°C in 2 liters of growth medium as described by Kozubal et al. (36), amended with 2% pyrite, 0.005% yeast extract, and 15 mg/liter vancomycin (added to prevent contamination by thermophilic bacilli). Total DNA was extracted using the FastDNA spin kit for soil (Q-Biogene, Irvine, CA). Purity of the DNA sent for genomic sequencing was verified by denaturing gradient gel electrophoresis with universal 931F and 1392GC primers and by sequencing 36 clones of the PCR product amplified with near-full-length archaea-specific primers as described by Kozubal et al. (36). DNA concentration and integrity were determined using gel electrophoresis, prior to construction of small insert libraries (2 kb), subsequent cloning, and capillary sequencing (SymBio Corporation, Menlo Park, CA). Approximately 19 Mb of total sequence (∼500 bp per sequence read) was assembled into 174 contigs larger than 5 kb, with an average coverage of 4.8-fold.
Genome searches and primer design.
Previously identified Fe(II) and S oxidation genes within the Sulfolobales were used as queries to search translated open reading frames (ORFs) from the draft sequence of Metallosphaera yellowstonensis and included the following: the terminal oxidase complexes (soxABCD, soxM, doxBCEF, and foxABCDEFGHIJ), the cbsA-soxLN operons, thiosulfate oxidase (tqoAB), sulfite oxidase molybdopterin (som; Msed_0362), sulfide quinone oxidoreductase (sqr; SSO2261), sulfur oxidoreductase (sor), and cytochromes thought to be important in the oxidation of Fe(II) in A. ferrooxidans and Leptospirillum group II. Metallosphaera sedula blue copper proteins (Msed_0323 and Msed_1206) and a Rieske cytochrome b fusion protein (rcbf; Msed_1191) were also used as ORF search queries. Deduced amino acid sequences of the subunit I HCOs (and additional selected proteins) were aligned and used to develop qPCR primers (see Table S1 in the supplemental material), including those used for qPCR and for electrophoresis-based qualitative expression analysis. All qPCR primer pairs were designed to amplify approximately 200-bp fragments with melting temperatures of 58°C. The primers were analyzed using the IDT SciTools OligoAnalyzer 3.0 (Integrated DNA Technologies, Coralville, IA) for hairpins, dimers, and melting temperatures and were tested on DNA from a Metallosphaera sp. strain MK1 culture and a hydrous ferric oxide (HFO) mat sample from an acid-sulfate-chloride (ASC) geothermal spring, as well as negative controls.
Collection of environmental samples from YNP.
Four acidic Fe mat samples ranging from 54 to 72°C (ASC springs in the Norris Geyser Basin region of YNP) previously found to contain significant populations of M. yellowstonensis strain MK1 (25, 36) were chosen for analysis of mRNA: OSPA (One Hundred Spring Plain Spring, 72°C), OSPB (62°C), BED (Beowulf Spring, 62°C), and GAPB (Gap Spring, 84°C). Approximately 10 g of Fe-oxide microbial mat was removed from the primary flow channel with heat-sterilized spatulas and stored immediately in nuclease-free Falcon tubes on dry ice. RNA extraction took place within 8 h of sampling. A negative-control sample from a 75°C sulfur deposition zone at the source of Beowulf Spring (previously shown not to contain 16S rRNA sequences related to M. yellowstonensis strain MK1) did not result in amplification with primers used in this study.
Growth conditions for Metallosphaera yellowstonensis strain MK1.
Pure cultures of M. yellowstonensis strain MK1 were maintained at 65°C in synthetic growth medium as described by Kozubal et al. (36) at a pH of 3.0, adjusted with H2SO4. The cultures were grown in 20-ml serum bottles containing 15 ml of synthetic medium with 0.5 g (<0.15-mm size fraction) of one of the following substrates, sterilized prior to use: elemental S0, research-grade pyrite (FeS2; Wards Scientific), or Fe(II) adsorbed to ferrihydrite (the organism does not grow on 20 mM FeSO4 in the absence of a solid surface [36]). Cultures of MK1 were also grown with 0.2% YE with no mineral substrates. Cultures with no YE were grown autotrophically using a headspace composition of 25% O2, 50% CO2, and 25% air. Cultures were mixed twice daily to maintain microoxic conditions.
RNA extraction.
Samples for RNA extraction were prepared from the >0.2-μm fraction of cultures harvested at mid-log phase or from 0.5 g of a Fe-oxide microbial mat sampled from YNP. All samples were washed with pH 3.0 RNase/DNase-free water prior to RNA extraction. Total RNA was extracted with the FastRNA Pro Blue kit (MP Biomedicals, Irvine, CA) and treated with RQ1 RNase-free DNase (Promega, Madison, WI) for 1 h after extraction. Extracts were determined to be DNA-free by gel electrophoresis of PCR products with and without cDNA extension by reverse transcription for each primer to obtain quantitative estimates of gene expression (see Table S1 in the supplemental material). Concentrations and purity of RNA were verified with a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE) and by gel electrophoresis.
RT-PCR and qPCR.
RT-PCR was performed with the Access RT-PCR system (Promega, Madison, WI). RT-PCR was carried out with three controls for cultured cells: (i) RNA without reverse transcriptase to verify only RNA as the template, (ii) nuclease-free water as a negative-control template, and (iii) genomic DNA from M. yellowstonensis strain MK1 as a positive control for amplification. For environmental samples, a negative control from the source of Beowulf Spring was used; this source does not contain sequences related to strain MK1. RNasin RNase inhibitor (Promega, Madison, WI) was added (320 U) to 400 μl Promega GoTaq master mix and 40 U reverse transcriptase enzyme. Template RNA from specific culture treatments or from environmental samples was added to each master mix to a final concentration of approximately 0.1 ng/μl RNA. RT-qPCR was performed on eight sets of samples (one set for each culture treatment and spring site) along with negative controls without reverse transcriptase. The thermal cycler protocol was 45°C for 45 min, 94°C for 2 min; 35 cycles of 94°C (30 s) melting, 56°C (45 s) annealing, and 72°C (25 s) extension; and a final 5-min extension period at 72°C. qPCR analysis was performed with the Rotor-Gene RG-3000 apparatus (Corbett Life Science, Sydney, Australia), and expression results were normalized relative to the 16S rRNA gene according to the method described by Kappler et al. (29). qPCR of total 16S rRNA was performed according to Kozubal et al. (36) to verify that ribosome copies did not vary significantly across different culture treatments. Melt curve analysis was used to verify the integrity of the qPCR products and to compare melting temperatures among pure cultures and spring sites.
Genome sequence analysis.
Assembled contigs of M. yellowstonensis were compared to the M. sedula genome for similarity of sequence content and synteny. Sequence gaps were filled by primer walking where possible. Additional protein sequences for tree construction were obtained from the GenBank database by conserved domain sequence searches across all taxonomical lineages. Protein sequence alignments were performed using ClustalX (version 1.81); misalignments or gaps were edited with Se-Al v2.0a11. Distance analysis was performed using the Jukes and Cantor correction, followed by phylogenetic tree construction using the maximum likelihood method in PAUP*4.0 (Sinauer Associates, Sunderland, MA) with 1,000 bootstrap replicates. Several bioinformatic programs were used to verify putative functions of genes found in this study. Protein BLAST (blastp and PHI-BLAST) and conserved domain searches were used to identify obvious homologies with characterized sequences in the NCBI databases. Multiple protein sequence analysis tools were used to determine structural relatives for less tractable sequences. These tools included the Pfam database, SMART, HHpred, and the BioInfoBank Metaserver. Structurally related sequences found with HHpred or the BioInfoBank metaserver were aligned with ClustalX and the DeepView program (v. 3.7), and models were created with the SWISS-MODEL server (3). Model quality was tested with the ProQ (Stockholm Bioinformatics Center) (58), ProSA (59), MolProbity (14), and Verify3d (41) programs.
Accession number.
The Metallosphaera yellowstonensis protein and nucleotide sequences used in this study are available at the JGI IMG/M website under IMG submission ID 268.
RESULTS
Terminal oxidase genes of Metallosphaera yellowstonensis strain MK1.
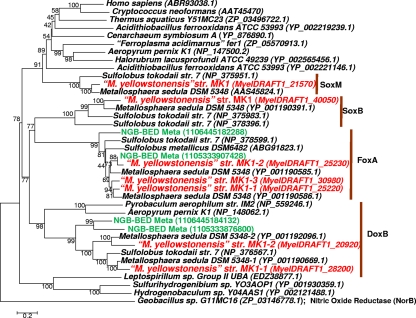
The genome sequence from M. yellowstonensis strain MK1 contains coding regions for five putative terminal oxidase complexes (Fig. 1): soxABC, doxBCE, the soxM supercomplex, the foxABCDEFGHIJ gene cluster, and a separate doxB-like subunit I transcribed as a bicistronic mRNA with a polyferredoxin-like subunit (referred to as doxB-2/polyferredoxin in this study). The draft genome of M. yellowstonensis strain MK1 contains three foxA heme Cu oxidases (subunit I); two copies were found within the foxABCDEFGHIJ gene cluster (foxA-1 and foxA-2), while a third (foxA-3) was associated with an insertion sequence elsewhere in the genome.
FIG. 1.
Protein tree of heme copper oxidases (subunit I) from all three domains of life. The extensive diversity of heme copper oxidases within the archaeal order Sulfolobales and Metallosphaera yellowstonensis strain MK1 includes DoxB, DoxB-2, FoxA, SoxB, and SoxM (M. yellowstonensis entries are labeled in red). Entries from the metagenome sequence collected at Beowulf Spring (NGB-BED) are labeled in green. GenBank protein accession numbers are shown in parentheses (IMG locus tag IDs are given for M. yellowstonensis, and JGI/IMG IDs are given for the metagenome samples). (The neighbor-joining tree was constructed with 1,000 bootstraps; nitric oxide reductase [NorB] from Geobacillus sp. G11MC16 was used as an outgroup.)
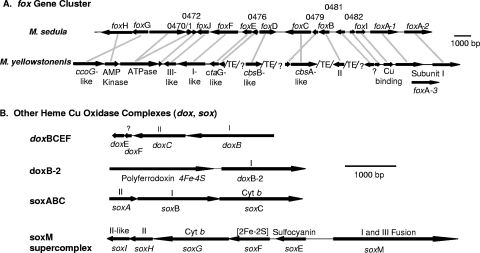
The gene arrangement of the soxABCD, soxM, and dox terminal oxidase complexes is conserved between M. yellowstonensis strain MK1 and M. sedula DSM5348 (Fig. 2). However, the foxABCDEFGHIJ gene cluster exhibits more variation in both gene order and content between the two organisms. Both M. sedula and M. yellowstonensis have additional hypothetical proteins within the foxABCDEFGHIJ gene cluster (M. sedula genes msed0471, -0472, -0476, -0479, and -0482) not found in S. metallicus or S. tokodaii (8). The ATPase-like sequence (msed0470) is fused to the homologue of msed0471 in M. yellowstonensis, and the foxGH and foxC/msed 0479 genes are encoded on opposite strands. In addition to a lack of synteny across the fox gene clusters in the two Metallosphaera organisms, the draft genome of M. yellowstonensis contains a high frequency of transposable elements (perhaps up to 10%), similar to the frequency of transposases observed in Sulfolobus solfataricus (55).
FIG. 2.
Gene arrangement of terminal oxidase complexes in Metallosphaera yellowstonensis (strain MK1). (A) Comparison of the fox terminal oxidase complex between M. yellowstonensis and M. sedula, showing annotation in M. sedula (top) and corresponding genes found in M. yellowstonensis (an additional description is provided in Table 1). foxC is the subject of further analysis and discussion. (B) Gene arrangement of additional terminal oxidase complexes (doxBCEF, doxB-2, soxABC, and the soxM supercomplex) present in M. yellowstonensis. Note: gene arrangements are identical to those in M. sedula. ?, the putative protein function cannot be inferred; TE, transposable element.
Other genes found in M. yellowstonensis that may play a role in the oxidation of Fe(II) include genes that appear to encode a blue multicopper protein (Mco), a Rieske-cytochrome b fusion protein (Rcbf), a cytochrome b558/566 (CbsA), and two sulfocyanin subunits (SoxE-1 and SoxE-2). Putative sulfur-oxidizing genes are also present in M. yellowstonensis strain MK1 and include sequences similar to a thiosulfate oxidase complex, tqoAB (also referred to as doxAD in A. ambivalens [30]) and a sulfite-oxidase molybdopterin (som) located directly upstream of tqoAB.
Metagenome sequence.
Four highly related heme Cu oxidases (subunit I) were found in the metagenome sequence obtained from an acidic Fe-oxide microbial mat in Norris Geyser Basin (YNP), where M. yellowstonensis-like organisms are known to represent approximately 25 to 30% of the archaeal community (Fig. 1) (27, 36). Two of the environmental sequences are closely related to the foxA gene from strain MK1, while the other two cluster with the M. yellowstonensis doxB subunit I. These results have now been corroborated from metagenome sequences collected from two additional thermophilic, acidic Fe microbial mats in Norris Geyser Basin (data not shown). Both sites contain M. yellowstonensis-like foxA gene sequences highly similar to those entries shown for Beowulf Spring (Fig. 1), indicating that the metabolic potential for encoding Fox cluster proteins is common in these habitat types. Importantly, fox genes were not found in numerous other geothermal sites where M. yellowstonensis-like populations are absent and where Fe(II) oxidation is not a dominant process (27).
Gene expression studies in pure culture and environmental samples.
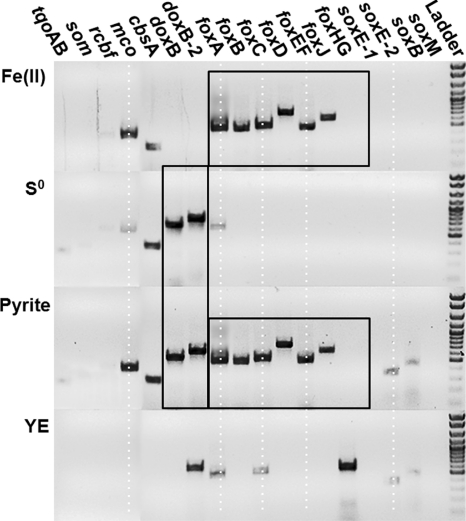
Expression assays (mRNA) of numerous genes found within the different types of terminal oxidase complexes (including five subunit I heme Cu oxidases as well as additional candidates involved in S and/or Fe oxidation) were performed using RT-PCR on the same amount of total RNA extracted from M. yellowstonensis cultures grown autotrophically on Fe(II)-ferrihydrite, pyrite, and elemental S and heterotrophically on YE (Fig. 3). All gene transcripts (mRNA) were confirmed by sequencing. Results indicated that expression levels of foxA, foxB, foxC, foxD, foxEF, foxJ, and mco genes were all highly increased in cultures grown on Fe(II)-ferrihydrite or pyrite (Fig. 3). Conversely, doxB- and doxB-2-like transcripts were the dominant mRNA products observed in cells grown on elemental S. The doxB and doxB-2 subunit I were also present in treatments with pyrite (as expected in the presence of reduced S) but were clearly absent in cultures grown on Fe(II). Transcripts of the cbsA gene were detected in all treatments except cultures grown on YE; consequently, the activity of this gene was independent of whether Fe(II) or reduced S served as the electron donor. Cells grown on YE exhibited increased expression of foxHG, which was not observed in cultures grown on Fe(II) or elemental S. Cells grown on YE also showed expression of doxB-2 and weaker products of foxA and foxC.
FIG. 3.
Agarose gels showing mRNA transcripts of target genes (listed above the gel) after cultivation of Metallosphaera yellowstonensis on either Fe(II) adsorbed to ferrihydrite, pyrite, S0, or YE, all at 70°C and pH 3.0. The black frames highlight doxB and doxB-2 expression with S0 and pyrite as electron donors, versus foxABCDEFGHIJ with pyrite and Fe(II) as donors. Target genes were heme Cu oxidases (subunit I), hypothetical fox genes, a multicopper oxidase (mco), thiosulfate oxidase (tqo), a Rieske cytochrome b fusion protein (rcbf), and a sulfite oxidase molybterin (som).
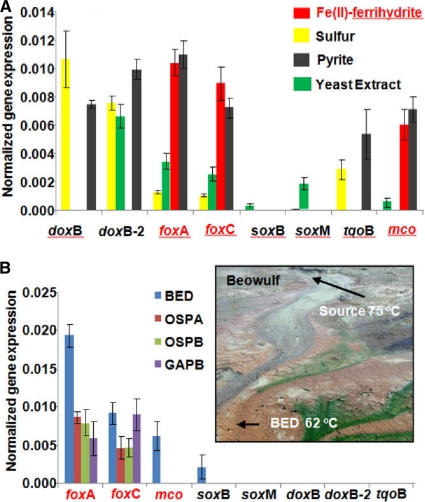
Priority gene transcripts identified in expression assays (described above) were then quantified in additional experiments using RT-qPCR, including the heme Cu oxidases (subunit I; e.g., doxB, doxB-2, foxA, soxB, and soxM), a putative Fe(II)-oxidizing cytochrome b558/566 (foxC), a multicopper oxidase (mco), and a thiosulfate oxidase (tqoB). Results from RT-qPCR were consistent with expression assays and showed that M. yellowstonensis strain MK1 produces levels nearly an order of magnitude higher of foxA, foxC, and mco gene transcripts when grown on Fe(II) or pyrite compared to cells grown on elemental S or YE (Fig. 4 A). Conversely, doxB transcripts were only observed in cells grown on elemental S or pyrite, whereas expression of doxB-2 increased in cells grown on elemental S (or pyrite) or YE (Fig. 4A). Cultures grown on YE also showed weak to modest increased expression of foxA, foxC, and soxM subunit I. Expression of the putative thiosulfate oxidase (tqoB) was increased in cultures grown on elemental S (or pyrite) but not on Fe(II).
FIG. 4.
Normalized quantitative gene expression (relative to 16S rRNA) for heme copper oxidases (subunit Is include doxB, doxB-2, foxA, soxB, soxM), foxC, mco and tqoB genes. (A) Metallosphaera yellowstonensis grown on Fe(II), pyrite, S0, or YE. (B) Four high-temperature, acidic Fe(III) mats in Norris Geyser Basin (YNP). The inset shows a site photograph (Beowulf Spring) of one of three acidic geothermal systems sampled, indicating the location of geothermal discharge and one of the sampling locations (BED) discussed in this report. Gene expression bars without values were below the detection limit.
Quantitative RT-PCR was also performed on samples from several acidic Fe-oxide mats in YNP, where organisms highly similar to M. yellowstonensis strain MK1 (>98% 16S rRNA gene nucleotide identity) are known to be important members of the microbial community (27, 36). The only HCO (subunit I) expressed in four different Fe-oxide mat samples (Fig. 4B) was foxA-1, with the exception of a considerably lower level of soxB in Beowulf Spring (BED sample). At this location, mRNA transcripts of foxA-1 were found at concentrations nearly 10 times greater than soxB. mRNA transcript expression levles of foxC were also increased in all Fe mat samples (Fig. 4B), which would be expected for a functional Fox complex. Moreover, with the exception of higher foxA-1 levels in site BED, the other three sites yielded ratios of foxA-1 mRNA similar to that of Metallosphaera yellowstonensis-like 16S rRNA. Additional copies of foxA found in strain MK1 (foxA-2 and foxA-3) were not significantly expressed under any conditions (data not shown).
Protein sequence analysis.
Bioinformatic analysis was conducted on coding genes within the five HCO complexes to predict putative functions based on motif searches, structural analysis, and protein modeling. Of the genes in the fox gene cluster, foxC is an excellent candidate for protein sequence analysis (Table 1). Exprssion of this gene is greatly increased in all cultures and environmental samples where Fe(II) is used as an electron donor. The only close relatives (∼25% amino acid identity) of this gene are cytochrome b558/566 (cbsA) sequences found in several members of the Sulfolobales and Caldivirga maquilingensis (within the order Thermoproteales). Three sequences related to foxC (>60% amino acid [aa] identity) were found in GenBank and belong to M. sedula (Msed_0478), S. tokodaii (ST2592/2593), and S. metallicus (accession number ABG91824). The foxC gene in S. tokodaii is composed of two proteins (ST2592 and ST2593), which are fused in the other organisms. The foxC sequence from strain MK1 represents a fourth sequence and is related to Msed_0478, with 86% aa identity.
TABLE 1.
Domain and structural predictions of putative proteins encoded by genes in the fox heme copper oxidase gene cluster
| Gene | Modeled? | Closest protein domain | Additional comment(s) |
|---|---|---|---|
| foxA | Yes | Heme Cu oxidase, subunit I | Three copies in genome of M. yellowstonensis |
| foxB | Yes | Cytochrome oxidase, subunit II | Contains conserved Cu-binding ligands for CuA center similar to SoxH |
| foxC | Yes (also see Results) | Cytochrome b558/566, subunit A (pairs with FoxD) | Candidate for Fe(II) oxidation (also see Results) |
| foxD | No | Cytochrome b558/566, subunit B (pairs with FoxC) | 10 transmembrane helices |
| foxE | No | Cytochrome c oxidase assembly factor (CtaG) | 6 transmembrane helices |
| foxF | No | Faint structural similarity to a cbb3-type cytochrome oxidase, subunit I (also similar to FoxA) | 12 transmembrane helices |
| foxG | Yes | Structurally similar to the CcoG protein required for biogenesis of cbb3-type cytochrome c oxidase | 9 transmembrane helices; NapH ferredoxin fold |
| foxH | Partial | Two cystathionine-beta-synthase domains, bind adenosyl group ligands (e.g., AMP, ATP S-AdoMet) | Senses changes in AMP/ATP ratios |
| foxI | Yes | Cu resistance protein, CopC | Binds Cu |
| foxJ | No | Cytochrome oxidase subunit III | 7 transmembrane helices |
Protein modeling of FoxC.
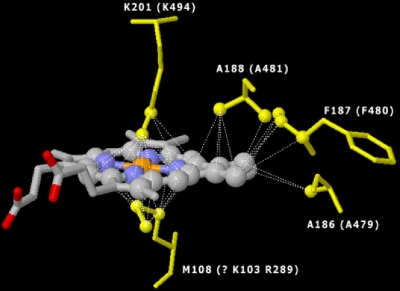
Sequence alignment of foxC and cbsA showed highly conserved regions (Fig. 5) at alignment positions 466 to 500, matching a heme B binding region of the gamma subunit of the ethylbenzene dehydrogenase (EBDH) crystal described for Azoarcus sp. EbN1 (31, 32). The EBDH heme B binding motif involves an unusual Lys-Met axial coordination. The conserved lysine in this coordination (K484 in our alignments) is shared in both cbsA and foxC (see Fig. S1 in the supplemental material). However, a conserved methionine is not found in either foxC or cbsA. Two candidate conserved amino acids for axial heme coordination with K484 are lysine and arginine (K103 and R289). Further evidence for this novel heme coordination was obtained by modeling the heme binding region with the EBDH crystal (Fig. 6), which suggested the spatial location of heme B in relation to axial coordinating residues, arginines and phenylalanine. Conserved residues also include three tyrosines with possible roles in heme coordination.
FIG. 5.
Protein alignment of FoxC and CbsA from Sulfolobales species and Caldivirga maquilingensis (CbsA only). Boxes marked by solid lines indicate conserved histidine (H) and methionine (M) residues required for binding heme B and also illustrate a conserved cysteine (C) found only in the FoxC sequences, which may be used to bind Cu, Fe, or heme B in a novel configuration. Conserved FoxC sequences are at alignment positions C152, H97, M119, H311, H312, M313, H360, and H464. Conserved histidines, methionines, and cysteines are labeled with boxes and may indicate additional metal binding sites. The box marked by a dashed line is a region high in threonine (T) that is used in glycosylation and is not found in FoxC.
FIG. 6.
Model of the FoxC putative heme B binding domain (orange, Fe; blue, N), indicating the spatial location of heme B relative to axial coordinating residues. The model shows lysine (K201) and methionine (M108) axial coordination for the EBDH crystal and corresponding lysine (K494) and arginine (R289) or lysine (K103) for the FoxC model. The model also shows conserved alanine (A188 and A186 for the EDBH crystal; A481 and A479 for FoxC) and phenylalanine (F187 for the EDBH crystal; F480 for FoxC).
Differences between cbsA and foxC include two conserved methionine residues 13 amino acids apart that are found in cbsA at alignment positions 293 and 307 and which may be involved in coordination of an additional heme. All four foxC sequences contain multiple conserved residues (amino acids) necessary for heme, Fe, or copper binding that are not found in cbsA. These include one cysteine (C132), four histidines (H96, H312, H360, and H465), and three methionines (M119, M312, and M313). All cbsA genes contain a region high in threonine (T) at alignment positions 507 to 535, indicating the possibility of a region of high glycosylation or a site for phosphorylation. This region is not found in foxC sequences. The threonine mol% composition is about 7 mol% for foxC, compared to about 11 mol% for cbsA from strain MK1.
Sequence analysis of Mco.
The novel multi-blue Cu oxidase gene (mco) found in M. yellowstonensis is also an excellent candidate for protein modeling. Expression of this gene was greatly increased (along with foxC and other fox cluster genes) under Fe(II)-oxidizing conditions. Bioinformatic analysis suggested that the hypothetical structure of Mco is related to eukaryal laccases and Fet3p proteins, which have been directly linked with the oxidation of Fe(II) (48, 54). The predicted protein (Mco) contains two putative plastocyanin copper binding sites at sequence positions His35, Cys93, His96, and Met100 and His314, Cys373, His376, and Met381 (18). However, the predicted Mco in M. yellowstonensis strain MK1 does not have type II or III copper domains that are typical of eukaryal blue multicopper proteins and are responsible for binding and reducing oxygen. The protein was modeled accurately around the plastocyanin domains, but a crystal structure is not yet available to allow accurate modeling of the remainder of the protein.
DISCUSSION
Metallosphaera yellowstonensis strain MK1 contains up to seven heme copper oxidase (subunit I) genes, which to our knowledge is more than any other described microorganism. The putative FoxA (subunit I) appears to be involved in the final step of electron transfer from Fe(II) to O2(aq). The fact that three copies of this gene are present in the draft genome sequence of strain MK1 further support its importance in the metabolism of this thermo-acidophile. The DoxBCEF complex was expressed only in treatments containing reduced S and therefore appears to be important in sulfur oxidation, as does the doxB-2/polyferredoxin bicistronic transcript, a novel putative terminal oxidase. This operon, known only in strain MK1 and M. sedula, does not contain an HCO subunit II, but rather has a novel gene that encodes a putative polyferredoxin. Polyferredoxins have negative redox potentials that range from −100 to −300 mV (21), and this range overlaps with values of half-reactions for the oxidation of reduced sulfur species. The potential utilization of a polyferrodoxin protein rather than a conventional heme copper cytochrome c or quinol oxidase subunit II (e.g., DoxC in the DoxBCEF complex) is a possibility, and this two-protein complex would be one of the simplest electron transport systems in known aerobic organisms. doxB-2 is also expressed in cultures grown on YE and may, therefore, be involved in heterotrophic metabolism, along with the soxM supercomplex. The foxA, doxB, doxB-2, and tqoB genes are all expressed in pyrite-grown cultures, consistent with the mechanism of pyrite dissolution by Fe(II)-oxidizing microorganisms involving the release of thiosulfate and Fe(II) (52).
Two foxA- and two doxB-like sequences were found in the metagenome sequence obtained from Beowulf Spring (position BED), which corresponds to the location of the original inoculum used to isolate strain MK1 (27, 36). These sequences confirmed that M. yellowstonensis-like populations present in situ exhibit metabolic potential for the oxidation of both Fe(II) and elemental S. Although dominated by high-As amorphous Fe(III)-hydroxide (25), elemental S formed upstream in sulfidic zones can be transported into the Fe mats and thus could be used as an electron donor in these environments (25, 26, 43). However, quantitative expression results show clearly that fox genes (including foxA and foxC) are the dominant terminal oxidase genes expressed in all the Fe(III) microbial mats sampled. Although elemental S is abundant near the source of Beowulf Spring, M. yellowstonensis-like organisms do not appear until the Fe(III)-oxide depositional zone, where oxygenation of the aqueous phase is sufficient to support Fe(II) oxidation (26).
The significantly increased expression of fox cluster genes under Fe(II)-oxidizing conditions in our current study, as well as in previous studies (4, 8, 29), suggests that the putative proteins encoded by these genes are excellent candidates for mediating electron transport from Fe(II) (29). Prior work on the cytochrome spectra of thermo-acidophilic Sulfolobales cultivated on Fe(II) or pyrite suggested that CbsA may be responsible for a unique highly expressed absorption peak at 573 nm (9, 29). However, other findings on protein extracts indicate that this absorption peak is not due to CbsA (22). Instead, FoxC may be responsible for this spectral signature resulting from an additional heme or metal binding site(s) not shared with CbsA (the predicted structure of FoxC contains four conserved histidines, three methionines, and a cysteine not found in CbsA). cbsA-soxNL2 homologues are found in species that are not capable of oxidizing iron, and this study shows that the M. yellowstonensis cbsA was equally expressed in the presence or absence of Fe(II) (Fig. 3). Interestingly, expression of cbsA-soxNL2 was shown to be increased in M. sedula when grown on FeSO4 compared to growth on elemental sulfur or tetrathionate, suggesting a role in Fe(II) oxidation. Therefore, cbsA-soxNL2 cannot be ruled out as important to Fe(II) oxidation in Metallosphaera yellowstonensis and may show increased expression under growth conditions not tested in this study.
Although His-His, His-Met, or Met-Met axial coordination of heme B has been well-documented (13, 46), Lys-Met coordination has been described for the EBDH gamma subunit, nitrate reductases, selenate reductase, chlorate reductases, dimethyl sulfoxide reductase heme B subunit from halophilic Euryarchaeota, and hypothetical cytochromes from Sulfurihydrogenibium spp. (31, 32). FoxC and CbsA align most closely to the EBDH gamma subunit (see Fig. S2 in the supplemental material), but alignment and protein modeling indicate a novel Lys-Lys or Lys-Arg coordination of heme B (Fig. 6) that represents a new mechanism for axial coordination of heme with no obvious domain relatives found in GenBank. Lys-Met coordination imparts a more positive redox potential than other heme B proteins, because lysine (and arginine) are less nucleophilic than methionine or histidine, which stabilizes the Fe(II) state. The Lys-Lys or Lys-Arg coordination proposed here would be expected to have an even higher Fe(II)-stabilizing potential. In fact, previous studies have measured a very high redox potential for CbsA protein fractions at around +400 mV (22). A higher redox potential would also be expected for FoxC to be effective in oxidizing Fe(II) at lower pH values (1). These properties appear to correspond quite well with the potentials necessary for an energetic electron transport from Fe(II) as an electron donor. Calculated redox potentials for the Fe(II)/Fe(III) half-reaction in these environments range between ∼470 and 590 mV, depending on oxygen levels and pH (26).
CbsA has recently been implicated as the cytochrome b component of an archaeal bc1 subunit III electron transport complex analogue (7). Given the structural relatedness of components of FoxC and CbsA, it is expected that these proteins indeed play similar roles in electron transport. However, FoxC appears to be the heme B subunit of a larger protein complex with a function similar to the gamma subunit of the EBDH heterodimer from Azoarcus sp. EbN1 (31). The EBDH complex contains a molybdopterin protein (alpha subunit) that contains a catalytic domain involved directly in the oxidation of ethylbenzene and an Fe-S cluster (beta subunit) that is similar to ferredoxins. FoxG contains a ferredoxin fold (Table 1) and is therefore a candidate beta subunit for a hypothetical complex with FoxC. Finally, the predicted FoxH is similar to ATP/AMP sensor proteins, and the putative FoxH may regulate expression of Fox components, such as FoxC, based on energy requirements.
The predicted Mco of M. yellowstonensis represents another novel protein and an interesting candidate for future study. Transcripts of mco were not observed in all HFO spring sites (found in only one of four). Only two sequences similar to the M. yellowstonensis strain MK1 mco were found in public databases, including M. sedula and Sulfolobus islandicus. Interestingly, mco-like sequences were found in two of seven Sulfolobus islandicus strains, corresponding to those isolated from YNP (51). However, it is not known whether these S. islandicus strains are capable of oxidizing Fe(II). Of the described Sulfolobales, only Metallosphaera spp., Acidianus brierleyi, Sulfolobus metallicus, and Sulfolobus tokodaii are capable of chemolithoautotrophic growth on Fe(II), with limited growth reported for S. tokodaii (8). Although not required for iron oxidation in the Sulfolobales, Mco may impart an advantage for chemolithotrophic growth on Fe(II).
The translated 550-aa sequence contains two plastocyanin domains and models well with laccase proteins. Many type I domains in multicopper oxidases are known to be nonspecific in one-electron transfers from a variety of chemical species, including organic compounds (e.g., lignin phenolic groups), Fe(II), and Mn(II). However, a specific Fe(II) binding site has been found in Fet3P proteins, which show stronger Fe(II) oxidation activities than other blue multicopper proteins (48, 54). Binding is believed to take place via a type I domain with two acidic residues (glutamic and aspartic acids). Whether Mco has a similar Fe(II)-specific binding region will require further protein analysis by either purification and specific activity assays or modeling with structures not yet in the PDB. The Mco in M. yellowstonensis may represent an additional example of a fusion between two small blue copper proteins, believed to be an evolutionary step toward enzymes such as laccase in fungi and ceruloplasmin in higher mammals (44). However, the putative Mco does not contain type II or III copper binding domains, which are typical of blue multicopper proteins found in eukaryal species and are responsible for binding and reducing oxygen.
The predicted Mco may be involved in a variety of one-electron transfers besides Fe(II) oxidation. Alternatively, Mco and/or other blue copper proteins (SoxE-1 and SoxE-2) may function (like rusticyanin in A. ferrooxidans) as temporary storage for electrons (52) or as an electron carrier between redox components of the Fox complex (i.e., FoxC and FoxA) (35). However, with the exception of the plastocyanin domains, Mco has little similarity to any known protein sequence and undoubtedly represents a novel family of multicopper proteins that are quite different from small blue copper proteins, such as rusticyanin, which is only 187 aa long in A. ferrooxidans.
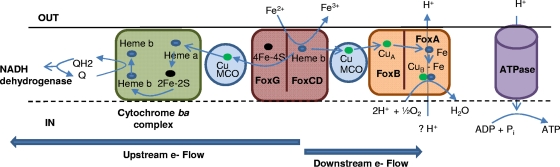
Members of the Sulfolobales do not have c-type cytochromes, which are important for Fe(II) oxidation in A. ferrooxidans and Leptospirillum group II, and this further supports the hypothesis that a different evolutionary path is responsible for Fe(II) oxidation in M. yellowstonensis. Cytochrome b (i.e., FoxC and/or CbsA) and blue multicopper (Mco) proteins likely have similar functions to cytochrome c and rusticyanin for direct Fe(II) oxidation and/or upstream/downstream electron transport. Expression and modeling results along with comparative genome analysis provided evidence to support a hypothetical model for Fe(II) oxidation in M. yellowstonensis strain MK1 (Fig. 7). This model assumes that FoxC is the direct oxidant of Fe(II) and that Mco acts much like a rusticyanin analogue in transporting electrons either upstream for anabolic processes such as carbon fixation or downstream to the Fox HCO for energy gain and ATP synthesis. M. yellowstonensis and the other Sulfolobales do not have a periplasmic space and Mco does not have transmembrane domains, and therefore the cellular location is unknown. Mco is not part of the Fox operon and, therefore, may not be bound to the HCO complex, similar to sulfocyanin. Instead, Mco may be a soluble electron shuttle within the cytoplasm.
FIG. 7.
Hypothetical model of Fe(II) oxidation in M. yellowstonensis strain MK1, based on annotation of the draft genome sequence, expression results, and functional modeling of putative proteins encoded by the fox supercomplex. Blue arrows indicate paths of electrons. Q, quinone; QH2, hydroquinone.
The work presented in this study strongly suggests that the acidic Fe(II)-oxidizing niche realized by M. yellowstonensis-like organisms in geothermal springs of YNP is linked directly with the metabolic capabilities imparted by putative proteins of the Fox complex, as well as possible roles for a unique Cu oxidase containing two plastocyanin domains. These proteins have only been observed (to date) in high-temperature Fe(II)-oxidizing habitats and are not present in other Sulfolobales habitats (27). Consequently, these putative proteins are strong candidates for further study, including verification of protein transcription in situ, as well as in vitro studies aimed at the isolation and characterization of these proteins.
Supplementary Material
Acknowledgments
This work was supported by the National Science Foundation Microbial Observatory Program (MCB-0132022), the National Aeronautic and Space Administration (NASA) via funds provided to the Thermal Biology Institute at Montana State University (project numbers NAG5-8807 and NNG04GR46G), and the Montana Agricultural Experiment Station (projects 911398 and 911300).
We appreciate assistance from C. Hendrix and T. Olliff for permitting this work in Yellowstone National Park (permit YELL-2006-2008-SCI-1976).
Footnotes
Published ahead of print on 14 January 2011.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Amend, J. P., and E. L. Shock. 2001. Energetics of overall metabolic reactions of thermophilic and hyperthermophilic Archaea and Bacteria. FEMS Microbiol. Rev. 25:175-243. [DOI] [PubMed] [Google Scholar]
- 2.Appia-Ayme, C., N. Guiliani, J. Ratouchniak, and V. Bonnefoy. 1999. Characterization of an operon encoding two c-type cytochromes, an aa3-type cytochrome oxidase, and rusticyanin in Thiobacillus ferrooxidans ATCC 33020. Appl. Environ. Microbiol. 65:4781-4787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arnold, K., L. Bordoli, J. Kopp, and T. Schwede. 2006. The SWISS-MODEL workspace: a web-based environment for protein structure homology modeling. Bioinformatics 22:195-201. [DOI] [PubMed] [Google Scholar]
- 4.Auernik, K. S., Y. Maezato, P. H. Blum, and R. M. Kelly. 2008. The genome sequence of the metal-mobilizing, extremely thermoacidophilic archaeon Metallosphaera sedula provides insights into bioleaching-associated metabolism. Appl. Environ. Microbiol. 74:682-692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Auernik, K. S., and R. M. Kelly. 2008. Identification of components of electron transport chains in the extremely thermoacidophilic crenarchaeon Metallosphaera sedula through iron and sulfur compound oxidation transcriptomes. Appl. Environ. Microbiol. 74:7723-7732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baker, B. J., and J. F. Banfield. 2003. Microbial communities in acid mine drainage. FEMS Microbiol. Ecol. 44:139-152. [DOI] [PubMed] [Google Scholar]
- 7.Bandeiras, T. M., et al. 2009. The cytochrome ba complex from the thermoacidophilic crenarchaeote Acidianus ambivalens is an analog of bc(1) complexes. Biochim. Biophys. Acta 1787:37-45. [DOI] [PubMed] [Google Scholar]
- 8.Bathe, S., and P. R. Norris. 2007. Ferrous iron- and sulfur-induced genes in Sulfolobus metallicus. Appl. Environ. Microbiol. 73:2491-2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blake, R. C., E. A. Shute, J. Waskovsky, and A. P. Harrison, Jr. 1993. Respiratory components in acidophilic bacteria that respire on iron. Geomicrobiol. J. 10:173-192. [Google Scholar]
- 10.Blake, R., and D. B. Johnson. 2000. Phylogenetic and biochemical diversity among acidophilic bacteria that respire on iron, p. 53-78. In L. Derek (ed.), Environmental microbe-metal interactions. ASM Press, Washington, DC.
- 11.Brierley, C. L., and J. A. Brierley. 1973. A chemoautotrophic and thermophilic microorganism isolated from an acid hot spring. Can. J. Microbiol. 19:183-188. [DOI] [PubMed] [Google Scholar]
- 12.Brock, T. D., K. M. Brock, R. T. Belly, and R. L. Weiss. 1972. Sulfolobus: a new genus of sulfur-oxidizing bacteria living at low pH and high temperature. Arch. Mikrobiol. 84:54-68. [DOI] [PubMed] [Google Scholar]
- 13.Cowan, J. A. 1993. Inorganic biochemistry: an introduction. VCH Publishers, New York, NY.
- 14.Davis, A., et al. 2007. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 35:W375-W383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dopson, M., C. Baker-Austin, and P. L. Bond. 2005. Analysis of differential protein expression during growth states of Ferroplasma strains and insights into electron transport for iron oxidation. Microbiology 151:4127-4137. [DOI] [PubMed] [Google Scholar]
- 16.Fuchs, T., H. Huber, K. Teiner, S. Burggraf, and K. O. Stetter. 1995. Metallosphaera prunae, sp. nov., a novel metal-mobilizing, thermoacidophilic archaeum, isolated from a uranium mine in Germany. Syst. Appl. Microbiol. 18:560-566. [Google Scholar]
- 17.García-Horsman, J. A., B. Barquera, J. Rumbley, J. Ma, and R. B. Gennis. 1994. The superfamily of heme-copper respiratory oxidases. J. Bacteriol. 176:5587-5600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Giri, A. V., S. Anishetty, and P. Gautam. 2004. Functionally specified protein signatures distinctive for each of the different blue copper proteins. BMC Bioinformatics 5:127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hallberg, K. B., and D. B. Johnson. 2001. Biodiversity of acidophilic prokaryotes. Adv. Appl. Microbiol. 49:37-84. [DOI] [PubMed] [Google Scholar]
- 20.Hallmann, R., et al. 1993. Physiological characteristics of Thiobacillus ferrooxidans and Leptospirillum ferrooxidans and physiochemical factors influence microbial metal leaching. Geomicrobiol. J. 10:193-206. [Google Scholar]
- 21.Hedderich, R. 2004. Energy-converting [NiFe] hydrogenases from archaea and extremophiles: ancestors of complex I. J. Bioenerg. Biomembr. 36:65-75. [DOI] [PubMed] [Google Scholar]
- 22.Hettmann, T., et al. 1998. Cytochrome b558/566 from the archaeon Sulfolobus acidocaldarius. A novel highly glycosylated, membrane-bound b-type hemoprotein. J. Biol. Chem. 273:12032-12040. [DOI] [PubMed] [Google Scholar]
- 23.Huber, G., C. Spinnler, A. Gambacorta, and K. O. Stetter. 1989. Metallosphaera sedula gen. nov. and sp. nov. represents a new genus of aerobic, metal-mobilizing, thermoacidophilic archaebacteria. Syst. Appl. Microbiol. 12:38-47. [Google Scholar]
- 24.Huber, G., and K. O. Stetter. 1991. Sulfolobus metallicus, sp. nov., a novel strictly chemolithoautotrophic thermophilic archaeal species of metal-mobilizers. Syst. Appl. Microbiol. 14:372-378. [Google Scholar]
- 25.Inskeep, W. P., R. E. Macur, G. Harrison, B. C. Bostick, and S. Fendorf. 2004. Biomineralization of As(V)-hydrous ferric oxyhydroxide in microbial mats of an acid-sulfate-chloride geothermal spring, Yellowstone National Park. Geochim. Cosmochim. Acta 68:3141-3155. [Google Scholar]
- 26.Inskeep, W. P., et al. 2005. On the energetics of chemolithotrophy in nonequilibrium systems: case studies of geothermal springs in Yellowstone National Park. Geobiology 3:297-320. [Google Scholar]
- 27.Inskeep, W. P., et al. 2010. Metagenomes from high-temperature chemotrophic systems reveal geochemical controls on microbial community structure and function. PLoS One 5:e9773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johnson, D. B., and K. B. Hallberg. 2003. The microbiology of acidic mine waters. Res. Microbiol. 154:466-473. [DOI] [PubMed] [Google Scholar]
- 29.Kappler, U., L. I. Sly, and A. G. McEwan. 2005. Respiratory gene clusters of Metallosphaera sedula: differential expression and transcriptional organization. Microbiology 151:35-43. [DOI] [PubMed] [Google Scholar]
- 30.Kletzin, A., T. Urich, F. Müller, T. M. Bandeiras, and C. M. Gomes. 2004. Dissimilatory oxidation and reduction of elemental sulfur in thermophilic archaea. J. Bioenerg. Biomembr. 36:77-91. [DOI] [PubMed] [Google Scholar]
- 31.Kloer, D. P., C. Hagel, J. Heider, and G. E. Schulz. 2006. Crystal structure of ethylbenzene dehydrogenase from Aromatoleum aromaticum. Structure 14:1377-1388. [DOI] [PubMed] [Google Scholar]
- 32.Kniemeyer, O., and J. Heider. 2001. (S)-1-Phenylethanol dehydrogenase of Azoarcus sp. strain EbN1, an enzyme of anaerobic ethylbenzene catabolism. Arch. Microbiol. 176:129-135. [DOI] [PubMed] [Google Scholar]
- 33.Komorowski, L., S. Anemüller, and G. Schäfer. 2001. First expression and characterization of a recombinant CuA-containing subunit II from an archaeal terminal oxidase complex. J. Bioenerg. Biomembr. 33:27-34. [DOI] [PubMed] [Google Scholar]
- 34.Komorowski, L., and G. Schäfer. 2001. Sulfocyanin and subunit II, two copper proteins with novel features, provide new insight into the archaeal SoxM oxidase supercomplex. FEBS Lett. 487:351-355. [DOI] [PubMed] [Google Scholar]
- 35.Komorowski, L., W. Verheyen, and G. Schäfer. 2002. The archaeal respiratory supercomplex SoxM from S. acidocaldarius combines features of quinole and cytochrome c oxidases. Biol. Chem. 383:1791-1799. [DOI] [PubMed] [Google Scholar]
- 36.Kozubal, M., et al. 2008. Isolation and distribution of a novel iron-oxidizing crenarchaeon from acidic geothermal springs in Yellowstone National Park. Appl. Environ. Microbiol. 74:942-949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kurosawa, N., Y. H. Itoh, and T. Itoh. 2003. Reclassification of Sulfolobus hakonensis Takayanagi et al. 1996 as Metallosphaera hakonensis comb. nov. based on phylogenetic evidence and DNA G+C content. Int. J. Syst. Evol. Microbiol. 53:1607-1608. [DOI] [PubMed] [Google Scholar]
- 38.Li, X., et al. 2007. Crystal structures of E. coli laccase CueO at different copper concentrations. Biochem. Biophys. Res. Commun. 354:21-26. [DOI] [PubMed] [Google Scholar]
- 39.Lübben, M., B. Kolmerer, and M. Saraste. 1992. An archaebacterial terminal oxidase combines core structures of two mitochondrial respiratory complexes. EMBO J. 11:805-812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lübben, M., S. Warne, S. Albracht, and M. Saraste. 1994. The purified SoxABCD quinol oxidase complex of Sulfolobus acidocaldarius. Mol. Microbiol. 13:327-335. [DOI] [PubMed] [Google Scholar]
- 41.Lübben, M., et al. 1994. A second terminal oxidase in Sulfolobus acidocaldarius. Eur. J. Biochem. 224:151-159. [DOI] [PubMed] [Google Scholar]
- 42.Lüthy, R., J. U. Bowie, and D. Eisenberg. 1992. Assessment of protein models with three-dimensional profiles. Nature 356:83-85. [DOI] [PubMed] [Google Scholar]
- 43.Macur, R. E., H. W. Langner, B. D. Kocar, and W. P. Inskeep. 2004. Linking geochemical processes with microbial community analysis: successional dynamics in an arsenic-rich, acid-sulfate-chloride geothermal spring. Geobiology 2:163-177. [Google Scholar]
- 44.Nakamura, K., and N. Go. 2005. Function and molecular evolution of multicopper blue proteins. Cell. Mol. Life Sci. 62:2050-2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pereira, M. M., et al. 2004. Respiratory chains from aerobic thermophilic prokaryotes. J. Bioenerg. Biomembr. 36:93-105. [DOI] [PubMed] [Google Scholar]
- 46.Poulos, T. L. 1996. The role of the proximal ligand in heme enzymes. J. Biol. Inorg. Chem. 1:356-359. [Google Scholar]
- 47.Purschke, W. G., C. L. Schmidt, A. Petersen, and G. Schäfer. 1997. The terminal quinol oxidase of the hyperthermophilic archaeon Acidianus ambivalens exhibits a novel subunit structure and gene organization. J. Bacteriol. 179:1344-1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Quintanar, L., M. Gebhard, T. P. Wang, D. J. Kosman, and E. I. Solomon. 2004. Ferrous binding to the multicopper oxidases Saccharomyces cerevisiae Fet3p and human ceruloplasmin: contributions to ferroxidase activity. J. Am. Chem. Soc. 126:6579-6589. [DOI] [PubMed] [Google Scholar]
- 49.Rawlings, D. E. 2002. Heavy metal mining using microbes. Annu. Rev. Microbiol. 56:65-91. [DOI] [PubMed] [Google Scholar]
- 50.Rawlings, D. E. 2005. Characteristics and adaptability of iron- and sulfur-oxidizing microorganisms used for the recovery of metals from minerals and their concentrates. Microb. Cell Fact. 4:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Reno, M. L., N. L. Held, C. J. Fields, P. V. Burke, and R. J. Whitaker. 2009. Biogeography of the Sulfolobus islandicus pan-genome. Proc. Natl. Acad. Sci. U. S. A. 106:8605-8610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rohwerder, T., T. Gehrke, K. Kinzler, and W. Sand. 2003. Bioleaching review part A: progress in bioleaching: fundamentals and mechanisms of bacterial metal sulfide oxidation. Appl. Microbiol. Biotechnol. 63:239-248. [DOI] [PubMed] [Google Scholar]
- 53.Singer, S. W., et al. 2008. Characterization of cytochrome 579, an unusual cytochrome isolated from an iron-oxidizing microbial community. Appl. Environ. Microbiol. 74:4454-4462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stoj, C. S., A. J. Augustine, L. Zeigler, E. I. Solomon, and D. J. Kosman. 2006. Structural basis of the ferrous iron specificity of the yeast ferroxidase, Fet3p. Biochemistry 45:12741-12749. [DOI] [PubMed] [Google Scholar]
- 55.Suzuki, T., et al. 2002. Sulfolobus tokodaii sp. nov. (f. Sulfolobus sp. strain 7), a new member of the genus Sulfolobus isolated from Beppu Hot Springs, Japan. Extremophiles 6:39-44. [DOI] [PubMed] [Google Scholar]
- 56.Takayanagi, S., et al. 1996. Sulfolobus hakonensis sp. nov., a novel species of acidothermophilic archaeon. Int. J. Syst. Bacteriol. 46:377-382. [DOI] [PubMed] [Google Scholar]
- 57.To, T. B., D. K. Nordstrom, K. M. Cunningham, J. W. Ball, and R. B. McClesky. 1999. New method for the direct determination of dissolved Fe(III) concentration in acid mine waters. Environ. Sci. Technol. 33:807-813. [Google Scholar]
- 58.van der Oost, J., et al. 1994. The heme-copper oxidase family consists of three distinct types of terminal oxidases and is related to nitric oxide reductase. FEMS Microbiol. Lett. 121:1-9. [DOI] [PubMed] [Google Scholar]
- 59.Wallner, B., and A. Elofsson. 2003. Can correct protein models be identified? Protein Sci. 12:1073-1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wiederstein, M., and M. J. Sippl. 2007. ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 35:W407-W410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yarzábal, A., G. Brasseur, and V. Bonnefoy. 2002. Cytochromes c of Acidithiobacillus ferrooxidans. FEMS Microbiol. Lett. 209:189-195. [DOI] [PubMed] [Google Scholar]
- 62.Yarzábal, A., et al. 2002. The high-molecular-weight cytochrome c Cyc2 of Acidithiobacillus ferrooxidans is an outer membrane protein. J. Bacteriol. 184:313-317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zähringer, U., H. Moll, T. Hettmann, Y. A. Knirel, and G. Schäfer. 2000. Cytochrome b558/566 from the archaeon Sulfolobus acidocaldarius has a unique Asn-linked highly branched hexasaccharide chain containing 6-sulfoquinovose. Eur. J. Biochem. 267:4144-4149. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.