Abstract
Solvent production by Clostridium acetobutylicum collapses when cells are grown in pH-uncontrolled glucose medium, the so-called “acid crash” phenomenon. It is generally accepted that the fast accumulation of acetic acid and butyric acid triggers the acid crash. We found that addition of 1 mM formic acid into corn mash medium could trigger acid crash, suggesting that formic acid might be related to acid crash. When it was grown in pH-uncontrolled glucose medium or glucose-rich medium, C. acetobutylicum DSM 1731 containing the empty plasmid pIMP1 failed to produce solvents and was found to accumulate 0.5 to 1.24 mM formic acid intracellularly. In contrast, recombinant strain DSM 1731 with formate dehydrogenase activity did not accumulate formic acid intracellularly and could produce solvent as usual. We therefore conclude that the accumulation of formic acid, rather than acetic acid and butyric acid, is responsible for the acid crash of acetone-butanol-ethanol fermentation.
Clostridium acetobutylicum is a group of Gram-positive, spore-forming, obligate anaerobes that possess pathways capable of converting sugars into solvent, which is known as acetone-butanol-ethanol (ABE) fermentation (10). This process has recently been revived because of the potential application of butanol as an advanced biofuel. The metabolism of C. acetobutylicum is typically biphasic in batch culture, starting from an acidogenic phase and followed by a solventogenic phase. Acetic acid and butyric acid are produced during the acidogenic phase, while ethanol, butanol, and acetone are produced during the solventogenic phase, when acids are reutilized to produce solvent. Transition from acidogenesis to solventogenesis is a prerequisite for a successful ABE fermentation process (7, 8, 18, 25, 34). However, when C. acetobutylicum is growing at or close to its maximum growth rate or when the metabolic rate of C. acetobutylicum is very high, excess acid instead of solvent is produced and the switch from acidogenesis to solventogenesis fails to occur (22). This phenomenon is termed “acid crash” (13). It is generally accepted that acid crash is caused by the fast accumulation of acetic acid and butyric acid during the fermentation, as inferred from the fermentation results (13).
Formic acid is a weak acid produced by C. acetobutylicum at the pyruvate node. In clostridia, formic acid is usually reutilized as a one-carbon-unit donor (5, 26, 27). The pyruvate-formate lyase (PFL; encoded by CAC0980) involved in the conversion of pyruvic acid into formic acid was expressed, according to a previous DNA microarray study (1, 11), and was also detected in the proteome reference map of C. acetobutylicum that we published (14). Besides this, the physiological role of formic acid in C. acetobutylicum remains unclear. We found that addition of a very low concentration of sodium formate resulted in an acid crash phenomenon of ABE fermentation. We therefore proposed a hypothesis that formic acid might trigger the acid crash of ABE fermentation. This hypothesis was confirmed by constructing a recombinant strain with formate dehydrogenase (FDH) activity which was able to overcome the acid crash phenomenon.
MATERIALS AND METHODS
Strain and plasmid construction.
The bacterial strains and plasmids used in this study are listed in Table 1. The fdh gene, encoding a formate dehydrogenase, was amplified by PCR from the genomic DNA of Candida boidinii using primers fdh1 and fdh2 (fdh1, CGTGGATCCATGAAGATCGTTTTAGTC; fdh2, GCGGAATTCTTATTTCTTATCGTGTTTAC) containing BamHI and EcoRI restriction sites (underlined), respectively. The promoter region of the thiolase gene (CAC2873) was selected (24) and amplified from the genomic DNA of C. acetobutylicum DSM 1731 using primers Pthl1 and Pthl2 (Pthl1, AGTGTCGACTATATTGATAAAAATAATAATAGTGGG; Pthl2, CGTGGATCCTTCTTTCATTCTAACTAACCTCC) containing SalI and BamHI restriction sites (underlined), respectively. The mixture of pIMP1, the PCR products of fdh, and Pthl were digested by EcoRI, BamHI, and SalI in NEB buffer 3. The digested products were purified by use of an EZNA cycle-pure kit (Omega Biotek Inc., Guangzhou, China) and then ligated together by T4 DNA ligase to generate pITF. Prior to transformation into C. acetobutylicum, pIMP1 and pITF were methylated in Escherichia coli ER2275 containing pAN1 (as shown in Table 1). Electrotransformation of C. acetobutylicum was carried out according to the protocol developed previously (15). Transformants were confirmed by colony PCR and sequencing, and the confirmed clone was designated C. acetobutylicum DSM 1731(pITF). Similarly, the empty vector pIMP1 was transformed into C. acetobutylicum DSM 1731, resulting in a plasmid-control strain, DSM 1731(pIMP1).
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristicsa | Source or reference |
|---|---|---|
| Strains | ||
| Candida boidinii | Source of fdh gene | Lab collection |
| C. acetobutylicum | ||
| DSM 1731 | Wild-type strain | DSMZ |
| DSM 1731(pIMP1) | Plasmid control strain of DSM 1731 harboring control vector pIMP1 | This work |
| DSM 1731(pITF) | Recombinant strain of DSM 1731 harboring fdh expression vector pITF | This work |
| Escherichia coli | ||
| TOP10 | mcrA trio (mrr-hsdRMS-mcrBC) recA1 | Invitrogen |
| ER2275 | recA mcrBC hsdR | New England Biolabs |
| Plasmids | ||
| pAN1 | Cmr, Φ3T I gene, p15A origin | 15 |
| pIMP1 | MLSr Apr, shuttle vector of E. coli-C. acetobutylicum | 15 |
| pITF | fdh expression vector; the fdh gene from C. boidinii was ligated to pIMP1 | This work |
Cmr, chloramphenicol/thiamphenicol resistance; Apr, ampicillin resistance; MLSr, macrolide, lincosamide, and streptogramin B resistance; Φ3T I gene, Φ3T I methyltransferase gene of Bacillus subtilis phage Φ3T I.
Media and culture conditions.
E. coli strains were routinely grown aerobically at 37°C in Luria-Bertani medium supplemented, when necessary, with ampicillin (100 μg/ml) and/or chloramphenicol (25 μg/ml). All C. acetobutylicum strains were grown anaerobically at 37°C in different media supplemented, when necessary, with erythromycin (25 μg/ml on a solid plate or 100 μg/ml in liquid culture). C. acetobutylicum strains were grown in RCM medium (9) for routine growth, and in mRCM medium (RCM with 20 g/liter glucose as the sole carbohydrate) for preparation of competent cells. CGM medium (21) was used for fermentation in a 7.5-liter bioreactor with an initial work volume of 3.0 liters and an agitation speed of 150 rpm. The initial pH of all fermentations was adjusted to 6.5.
Acid inhibition experiments.
Corn mash (75 g/liter) was used as a test medium, to which different concentrations of formate, acetate, and butyrate in the form of the sodium salts were added at the beginning of fermentation. The resulting effects on solvent production were determined.
Analysis of cell growth and metabolites.
Cell density was measured at 600 nm using a UV-visible spectrophotometer (UV-2802PC; Unico, Shanghai, China). The dry cell weight (DCW) could be calculated from A600 using the equation DCW (g/liter) = 0.3A600 (2). The concentrations of glucose, acetone, acetate, butyrate, butanol, and ethanol were determined by high-pressure liquid chromatography (HPLC) analysis (1200; Agilent, Santa Clara, CA) (4, 30). A Bio-Rad (Hercules, CA) Aminex HPX-87H ion-exchange column (7.8 by 300 mm) was used with a mobile phase of 0.05 mM sulfuric acid and a flow rate of 0.50 ml/min at 15°C. A refractive index (RI) detector (Agilent) was used for signal detection at 30°C.
Formate dehydrogenase activity assay.
Cells were harvested from 20 ml culture broth by centrifugation at 5,000 × g and 4°C for 10 min. Cell pellets were washed and resuspended in 10 ml of precooled 10 mM sodium phosphate buffer containing 0.1 M β-mercaptoethanol (pH 7.5) and then sonicated for 10 min with 5-s pulses and at 200 W by a cell sonicator (JY92-II D; Xinchen, China). The sonicated cells were centrifuged at 20,000 × g and 4°C for 20 min to remove cell debris, resulting in a cell extract. The formate dehydrogenase activity of the cell extract was detected as previously described (33).
Assay for intracellular formic acid concentration.
CGM medium and CGM rich medium (10 g/liter Bacto Casamino Acids and 10 g/liter beef extract added to CGM) were used as the culture media. Three hundred milliliters of culture was sampled quickly and cooled thoroughly on ice. Cells were collected by centrifugation at 5,000 × g and 4°C for 10 min. The cell pellet was washed with precooled ultrapure water and resuspended in 2 ml ice-cooled ultrapure water. The cell suspension was sonicated for 1 h with 5-s pulses and at 400 W by a cell sonicator (JY92-II D; Xinchen). One milliliter of the upper aqueous phase was withdrawn and subsequently centrifuged at 12,000 × g and 4°C for 10 min. The supernatant was withdrawn and filtered for HPLC analysis (Agilent 1200). A Bio-Rad Aminex HPX-87H ion-exchange column (7.8 h 300 mm) was used with a mobile phase of 5.0% (vol/vol) acetonitrile in 0.0035 mol/liter H2SO4 at a flow rate of 0.6 ml/min and a temperature of 60°C (28). A UV detector (UVD; Agilent) adjusted to a wavelength of 210 nm and to a sensitivity of 0.4 absorbance units was used for signal detection. The intracellular formic acid concentration (C) was calculated as follows: C (μmol/g DCW cell) = 1,000C0/DCW, where C0 is the formic acid concentration in the supernatant detected by HPLC (represented in mmol/liter).
Determination of intracellular NAD+ and NADH concentrations.
The intracellular NAD+ and NADH were extracted by the phenol-chloroform-isoamyl alcohol method as described previously (17), with some modifications. Cells were harvested from 30 ml fermentation broth by centrifugation at 5,000 × g and 4°C for 10 min and resuspended in 3 ml ice-cooled pure water. Four milliliters of the ice-cooled mixture of phenol, chloroform, and isoamyl alcohol (34:24:1, vol/vol/vol) containing 0.6 ml of EDTA solution (66 mM, pH 7.4) was added to the suspension, and the mixture was thoroughly shaken for 90 s and centrifuged at 5,000 × g and 4°C for 10 min to achieve phase separation. Two milliliters of the upper aqueous phase was withdrawn and extracted twice with 4 ml of water-saturated diethyl ether by shaking the mixture for 30 s and removing the lower aqueous phase with a syringe. The aqueous phase was frozen in liquid nitrogen and stored at −80°C until HPLC analysis. Samples were analyzed by HPLC (Agilent 1200) with an Agilent Zorbax SB-Aq column (4.6 by 250 mm). The mobile phase was 50 mM sodium phosphate buffer containing 10 mM tetrabutyl ammonium bromide and acetonitrile (86:14) with a flow rate of 1 ml/min. A UVD (Agilent) was used for signal detection. Intracellular NADH and NAD+ concentrations were calculated as follows: C (μmol/g DCW cell) = C1/DCW, where C1 refers to the NADH or NAD+ titer in the fermentation broth (represented in μmol/liter).
Comparative proteomic analysis.
Cells of strains DSM 1731(pIMP1) and DSM 1731(pITF) grown in CGM for 16 h were collected. Sample preparation, two-dimensional polyacrylamide gel electrophoresis, gel image analysis, and protein expression profile analysis were all performed according to the published protocol (14).
ORP detection method.
Oxidoreduction potential (ORP) was measured with an ORP electrode (Pt4805-DPAS-SC-K85; Mettler-Toledo, Switzerland) combined with a pH/ORP controller (PC-350; Suntex, Taiwan), and the values were corrected according to the standard electrode value (ORP = ORPmeasure + ORPref, where ORPmeasure is the measured ORP and the reference ORP [ORPref] is equal to 210 mV at 37°C). Before measurement, the electrode was calibrated with redox standard solution (HI 7021L; Hanna Company, Italy).
RESULTS
Typical fermentation profiles of acid crash.
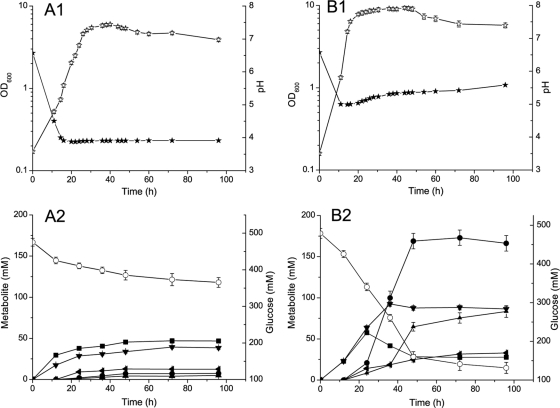
Acid crash of ABE fermentation often occurs when glucose is used as the sole carbon source. A typical profile of acid-crashed ABE fermentation can be observed when CGM was used as the medium and the pH was not controlled (Fig. 1 A1 and A2). In such a situation, butyrate could not be reassimilated and very little solvent was produced, resulting in a very high residual glucose concentration. When CGM was used and pH was controlled above 5.0, reassimilation of butyrate occurred and both glucose consumption and solvent production (including the ratio of ABE) exhibited regular profiles (Fig. 1B1 and B2).
FIG. 1.
Fermentation profile of DSM 1731(pIMP1) grown in CGM medium. (A1 and A2) pH uncontrolled; (B1 and B2) pH controlled above 5.0. Symbols: ⋆, optical density at 600 nm (OD600); ★, pH; ○, glucose; •, butanol; ▴, acetone; ◂, ethanol; ▪, butyrate; ▾, acetate. Values shown are means of triplicate determinations. Error bars represent standard deviations (n = 3).
Addition of low concentration of formate triggered acid crash of ABE fermentation.
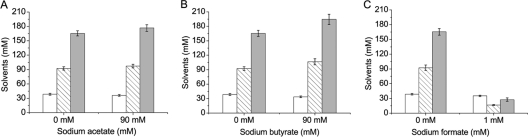
When corn mash was used as the medium for ABE fermentation, acid crash does not occur, no matter whether the pH is controlled above 5.0 or not controlled (data not shown). As it is generally accepted that acetic acid and butyric acid cause the acid crash of ABE fermentation, we wondered if acid crash could occur in corn mash medium when a certain concentration of acetic acid or butyric acid is added. Formic acid was included in this experiment, as the gene encoding pyruvate-formate lyase is present in the genome of C. acetobutylicum, but its role in acid crash was unclear. The effects of different concentrations of acetate, butyrate, and formate on ABE fermentation are shown in Fig. 2. Production of solvent was not inhibited by addition of acetate and butyrate up to a concentration of 90 mM (Fig. 2A and B), which is much higher than the titers of acetate and butyrate that can be produced during ABE fermentation. Moreover, addition of 90 mM butyrate slightly increased solvent production by 14% (Fig. 2B), suggesting that acetate and butyrate might not be major factors responsible for acid crash. Surprisingly, addition of 1 mM formate resulted in a dramatic decrease in solvent production (Fig. 2C), exhibiting a fermentation profile (data not shown) similar to that shown in Fig. 1A2. This suggests that addition of formate to corn mash medium might have triggered the acid crash.
FIG. 2.
Effects of acetate, butyrate, and formate on solvent production by C. acetobutylicum DSM 1731(pIMP1). Symbols: □, ethanol; ▧, acetone; ░⃞, butanol. The strain was cultivated anaerobically at 37°C in 7.5% corn mash with different concentrations of acetate, butyrate, and formate. Values shown are means of triplicate determinations. Error bars represent standard deviations (n = 3).
Expression of fdh gene relieved acid crash.
Addition of 1 mM formate to corn mash medium resulted in an acid-crashed ABE fermentation. We hypothesized that the formic acid might be taken up by the cells and that when the intracellular formic acid accumulates to a certain threshold concentration, it might inhibit the solvent biosynthetic pathways, resulting in decreased solvent production. If the intracellular formic acid could be utilized so that the formic acid concentration can be decreased, acid crash may not happen. To test this hypothesis, we cloned an NAD+-dependent FDH from Candida boidinii and introduced it into C. acetobutylicum DSM 1731 under the control of the thl promoter. The highest specific activity of FDH in strain DSM 1731(pITF) reached 5.73 U/mg during the acidogenic phase, while FDH activity could not be detected in strain DSM 1731(pIMP1).
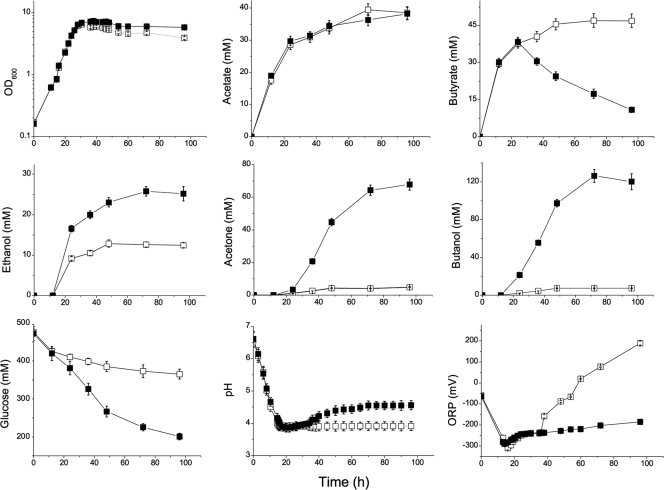
By expressing the fdh gene, we obtained a recombinant strain with formate dehydrogenase activity. We compared the fermentation profiles of strains DSM 1731(pITF) and DSM 1731(pIMP1) growing in CGM medium with the pH uncontrolled. As shown in Fig. 3, both strains grew at similar rates and produced similar amounts of acetate and butyrate in the first 24 h of fermentation. However, significant differences in the production of ethanol, acetone, and butanol as well as glucose consumption could be observed at 24 h. In the culture of strain DSM 1731(pITF), production of butanol is associated with the reassimilation of butyrate after 24 h, while DSM 1731(pIMP1) exhibited a typical profile of acid crash. Moreover, we observed a sharp increase of the ORP in the culture of strain DSM 1731(pIMP1) starting from 36 h, while the ORP of the culture of strain DSM 1731(pITF) could be stably maintained between −200 mV and −250 mV. The sharp increase in ORP might suggest that cell death of strain DSM 1731(pIMP1) occurred. Collectively, Fig. 3 shows that acid crash in glucose medium can be prevented when FDH activity is introduced into C. acetobutylicum.
FIG. 3.
Fermentation profiles of DSM 1731(pIMP1) and DSM 1731(pITF) in pH-uncontrolled CGM medium. Symbols: □, DSM 1731(pIMP1); ▪, DSM 1731(pITF). Values are means of triplicate determinations, and error bars represent standard deviations (n = 3). OD600, optical density at 600 nm.
To further demonstrate that the introduced FDH plays a role in reducing the formic acid concentration, strain DSM 1731(pITF) was grown in corn mash medium with 1 mM formate. Interestingly, it exhibited a fermentation profile similar to that shown in Fig. 1B2 (data not shown). This indicates that addition of 1 mM formate did not trigger acid crash of strain DSM 1731(pITF), suggesting that the introduced FDH is functional in vivo.
Expression of fdh gene decreased intracellular formic acid concentration.
The intracellular concentrations of formic acid in strains DSM 1731(pIMP1) and DSM 1731(pITF) grown in pH-uncontrolled CGM medium and CGM rich medium were determined. As shown in Table 2, intracellular formic acid was detected in strain DSM 1731(pIMP1) grown in both media during the acidogenic phase (16 h), transition phase (24 h), and solventogenic phase (36 h). Cells of DSM 1731(pIMP1) tend to produce approximately 50% more formic acid in CGM rich medium than in CGM medium during all three phases. Moreover, Table 2 shows that the production of formic acid in strain DSM 1731(pIMP1) occurred only during the acidogenic phase, as the intracellular concentration of formic acid did not significantly change from the transition phase (24 h) to the solventogenic phase (36 h). On the contrary, in strain DSM 1731(pITF) carrying the formate dehydrogenase, a very low intracellular concentration of formic acid was detected only during the acidogenic phase but was not detected during the transition and solventogenic phases. This suggests that the formic acid that should have been produced when the strain was grown in such medium was removed by the FDH in strain DSM 1731(pITF). We also tried to detect the extracellular concentration of formic acid under such conditions, but no formic acid could be detected in the fermentation broth.
TABLE 2.
Intracellular formic acid changes during fermentation
| Time (h) | Intracellular formic acid concn (μmol/g DCW)a |
|||
|---|---|---|---|---|
| DSM 1731(pIMP1) |
DSM 1731(pITF) |
|||
| CGM medium | CGM rich medium | CGM medium | CGM rich medium | |
| 16 | 2.15 ± 0.12 | 3.17 ± 0.15 | 0.03 ± 0.01 | 0.05 ± 0.02 |
| 24 | 3.54 ± 0.15 | 5.29 ± 0.18 | ND | ND |
| 36 | 3.39 ± 0.14 | 5.06 ± 0.14 | ND | ND |
Values shown are means of triplicates determinations (n = 3). ND, not detected.
Expression of fdh gene did not affect intracellular NADH/NAD+ level.
Usually, FDH was used to regenerate NADH by using formic acid as a substrate (29). Functional expression of fdh might alter the intracellular NADH/NAD+ ratio, thus affecting the switch from acidogenesis to solventogenesis. To rule out this possibility, we determined the intracellular concentrations of NADH and NAD+ in strains DSM 1731(pITF) and DSM 1731(pIMP1). As shown in Table 3, expression of fdh did not significantly affect the intracellular NADH and NAD+ pools, leading to an unaltered NADH/NAD+ ratio. This suggests that the intracellular redox level of strain DSM 1731(pITF) was not disturbed by the expression of the fdh gene.
TABLE 3.
Intracellular NADH and NAD+ pools and NADH/NAD+ ratios for DSM 1731(pIMP1) and DSM 1731(pITF)a
| Time (h) | Concn (μmol/g DCW) |
NADH/NAD+ (mol/mol) |
||||
|---|---|---|---|---|---|---|
| NADH |
NAD+ |
|||||
| DSM 1731(pIMP1) | DSM 1731(pITF) | DSM 1731(pIMP1) | DSM 1731(pITF) | DSM 1731(pIMP1) | DSM 1731(pITF) | |
| 16 | 4.71 ± 0.13 | 4.93 ± 0.14 | 4.74 ± 0.13 | 4.94 ± 0.16 | 1.01 ± 0.08 | 1.00 ± 0.08 |
| 36 | 0.42 ± 0.02 | 1.15 ± 0.04 | 0.53 ± 0.01 | 1.48 ± 0.06 | 0.79 ± 0.06 | 0.78 ± 0.08 |
Values shown are means of triplicate determinations (n = 3).
Expression of fdh gene did not affect expression level of pyruvate-formate lyase.
PFL (encoded by CAC0980), responsible for converting pyruvic acid into formic acid, was detected in the proteome reference map of C. acetobutylicum DSM 1731 that we established (14). We therefore wondered if the expression of the fdh gene in strain DSM 1731(pITF) would affect the expression of PFL. To test this, we performed a comparative proteomic analysis on strains DSM 1731 (pIMP1) and DSM 1731(pITF) using the established protocol (14). The proteomic data showed that the level of PFL expression did not alter upon introduction of FDH (data not shown), suggesting that the very low concentration or undetected intracellular formic acid in strain DSM 1731(pITF) is not due to the reduced level of PFL expression.
DISCUSSION
Maize and molasses are the main substrates for industrial ABE fermentation (10). A defined medium with glucose as the main carbon source for ABE fermentation has been developed (12, 16, 19), but the acid crash that occurred in such a medium, according to our experimental results and other reports (13), remains an interesting physiological phenomenon to explore.
Fast accumulation of acetic acid and butyric acid was believed to be responsible for acid crash (13). However, addition of 90 mM acetate or butyrate at the beginning of fermentation did not result in acid crash, suggesting that the dissociated acetate or butyrate would not cause acid crash. Addition of 1 mM formate to corn mash medium led to a failed ABE fermentation similar to acid crash. This inspires us to propose that the accumulation of formic acid, rather than the accumulation of acetic acid and butyric acid, is a key for acid crash. Functional expression of the fdh gene relieved acid crash, either in glucose medium or in corn mash supplemented with 1 mM formate, suggesting that formic acid plays an important role in acid crash. Confirmation of the accumulation of formic acid in the cells of DSM 1731(pIMP1) but not in those of DSM 1731(pITF) further led to the conception that formic acid is the key for acid crash.
Formate dehydrogenase is widely present in prokaryotes, yeasts, and plants (29) but not in clostridia. Heterologous expression of fdh can increase the intracellular NADH level in E. coli when glucose is used as a substrate (3) or can increase both the intracellular NADH and NAD+ levels in Klebsiella oxytoca when glycerol is used as a substrate (33). Functional expression of fdh did not lead to a significant change of the NADH/NAD+ ratio in C. acetobutylicum. The intracellular formate concentration in strain DSM 1731(pIMP1) ranged from 2.15 to 5.29 μmol/g DCW, which is equivalent to an intracellular concentration of 0.5 to 1.24 mM (assuming that 1 g DCW is equivalent to 4.27 ml wet cell volume, calculated on the basis of the fact that wet cells generally contain 75% water and water occupies 70% of wet cell volume [32]). The intracellular concentration of formic acid is therefore too low to significantly affect the NADH pool and the NADH turnover. This also proved that the relieved acid crash in strain DSM 1731(pITF) is not due to the redox change.
From proteomic data, the PFL in strain DSM 1731 is expressed during the acidogenic phase (14), and its expression was not affected by the introduction of FDH. Time series microarray data (1, 11) showed that the expression of PFL-encoding gene CAC0980 in C. acetobutylicum ATCC 824 was upregulated by 2-fold during the acidogenic phase (10 to 12 h), which is consistent with our observation that the production of formic acid increases during the acidogenic phase. This suggests that the PFL is indeed functional in C. acetobutylicum.
We were not the first to discover the inhibition effect of formic acid on ABE fermentation. About 80 years ago, the inhibitory effects of different acids, including formic acid, acetic acid, and butyric acid, on ABE fermentation were studied (31). On the basis of the molar concentration, the order of the inhibition effects of these three fatty acids is as follows: formic acid has the strongest inhibition effect, followed by butyric acid, while acetic acid showed the least inhibition effect. However, the acid crash phenomenon was not discovered at that time, so the authors were not able to show that formic acid actually plays a role in triggering acid crash. We suspect that accumulation of formic acid when a strain is grown in corn mash medium is slower than that when the strain is grown in CGM medium; therefore, acid crash would not occur in corn mash medium. This hypothesis was not tested because it is difficult to determine the formic acid concentration in cells grown in corn mash medium.
Interestingly, it has been shown that the concentrated cell-free extract (CCFE) of C. acetobutylicum is more inhibitory to ABE fermentation than the concentrated culture filtrate (CCF) (23). Fermentation with the CCFE of C. acetobutylicum showed a more pronounced effect than that with CCF. Compared with the control, butanol production was reduced by 15 to 20% when CCFE was added, while butanol production was reduced by only 5 to 10% when CCF was added. These results indicate that there are intracellular inhibitors in C. acetobutylicum, and our results suggested that formic acid might well be such an inhibitor.
Formic acid is a weak acid that can cause cytotoxic reactive oxygen species (ROS) in mammalian and eukaryotic cells, leading to cell death (6, 20). In anaerobic fermentation, ORP of the fermentation broth represents the oxidation-reduction state of the system. The ORP burst during the solventogenic phase of DSM 1731(pIMP1) suggested that ROS might be produced. As anaerobic bacteria usually cannot grow when ORP is higher than −100 mV, the sharp increase of the ORP indicates that cell death may have occurred in the fermentation broth, leading to the cessation of the fermentation. Elimination of formic acid by the expression of the fdh gene resulted in a relatively stable ORP below −200 mV, suggesting that the cells are still metabolically active.
Taken together, we conclude that formic acid plays a major role in triggering the acid crash of ABE fermentation, and the deleterious effect of formic acid on ABE fermentation might be mediated via oxidative stress. In addition, as small-molecule metabolites are produced and secreted by most prokaryotes and eukaryotes, this study illustrated how a small-molecule metabolite can affect cell physiology, providing a new perspective from which to study and eliminate such metabolite stress in the production of useful chemicals.
Acknowledgments
We thank Eleftherios Terry Papoutsakis (University of Delaware) for providing pIMP1 and E. coli ER2275(pAN1).
This work was supported by the National High Technology Research and Development Program of China (863 Project, no. 2006AA02Z237), the National Basic Research Program of China (973 Project, no. 2007CB707803), the Knowledge Innovation Program of the Chinese Academy of Sciences (no. KSCXZ-YW-G-007), and the Hundreds Talents Program of the Chinese Academy of Sciences.
Footnotes
Published ahead of print on 7 January 2011.
REFERENCES
- 1.Alsaker, K. V., and E. T. Papoutsakis. 2005. Transcriptional program of early sporulation and stationary-phase events in Clostridium acetobutylicum. J. Bacteriol. 187:7103-7118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Assobhei, O., A. E. Kanouni, M. Ismaili, M. Loutfi, and H. Petitdemange. 1998. Effect of acetic and butyric acids on the stability of solvent and spore formation by Clostridium acetobutylicum ATCC 824 during repeated subculturing. J. Ferment. Bioeng. 85:209-212. [Google Scholar]
- 3.Berrios-Rivera, S. J., G. N. Bennett, and K. Y. San. 2002. Metabolic engineering of Escherichia coli: increase of NADH availability by overexpressing an NAD+-dependent formate dehydrogenase. Metab. Eng. 4:217-229. [DOI] [PubMed] [Google Scholar]
- 4.Buday, Z., J. C. Linden, and M. N. Karima. 1990. Improved acetone-butanol fermentation analysis using subambient HPLC column temperature. Enzyme Microb. Technol. 12:24-27. [Google Scholar]
- 5.Daniel, A.-N., et al. 2010. Systems-level metabolic flux profiling elucidates a complete, bifurcated TCA cycle in Clostridium acetobutylicum. J. Bacteriol. 192:4452-4461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Du, L., et al. 2008. Formic acid induces Yca1p-independent apoptosis-like cell death in the yeast Saccharomyces cerevisiae. FEMS Yeast Res. 8:531-539. [DOI] [PubMed] [Google Scholar]
- 7.Grupe, H., and G. Gottschalk. 1992. Physiological events in Clostridium acetobutylicum during the shift from acidogenesis to solventogenesis in continuous culture and presentation of a model for shift induction. Appl. Environ. Microbiol. 58:3896-3902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harris, L. M., N. E. Welker, and E. T. Papoutsakis. 2002. Northern, morphological, and fermentation analysis of spo0A inactivation and overexpression in Clostridium acetobutylicum ATCC 824. J. Bacteriol. 184:3586-3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hirsch, A., and E. Grinsted. 1954. Methods for the growth and enumeration of anaerobic spore-formers from cheese, with observations on the effect of nisin. J. Dairy Res. 21:101-110. [Google Scholar]
- 10.Jones, D. T., and D. R. Woods. 1986. Acetone-butanol fermentation revisited. Microbiol. Rev. 50:484-524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jones, S. W., et al. 2008. The transcriptional program underlying the physiology of clostridial sporulation. Genome Biol. 9:R114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lampen, J. O., and W. H. Peterson. 1943. Growth factor requirements of clostridia. Arch. Biochem. 2:443-449. [Google Scholar]
- 13.Maddox, I. S., et al. 2000. The cause of “acid-crash” and “acidogenic fermentations” during the batch acetone-butanol-ethanol (ABE-) fermentation process. J. Mol. Microbiol. Biotechnol. 2:95-100. [PubMed] [Google Scholar]
- 14.Mao, S., et al. 2010. Proteome reference map and comparative proteomic analysis between a wild type Clostridium acetobutylicum DSM 1731 and its mutant with enhanced butanol tolerance and butanol yield. J. Proteome Res. 9:3046-3061. [DOI] [PubMed] [Google Scholar]
- 15.Mermelstein, L. D., N. E. Welker, G. N. Bennett, and E. T. Papoutsakis. 1992. Expression of cloned homologous fermentative genes in Clostridium acetobutylicum ATCC 824. Nat. Biotechnol. 10:190-195. [DOI] [PubMed] [Google Scholar]
- 16.Monot, F., J. R. Martin, H. Petitdemange, and R. Gay. 1982. Acetone and butanol production by Clostridium acetobutylicum in a synthetic medium. Appl. Environ. Microbiol. 44:1318-1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Noack, H., W. S. Kunz, and W. Augustin. 1992. Evaluation of a procedure for the simultaneous determination of oxidized and reduced pyridine nucleotides and adenylates in organic phenol extracts from mitochondria. Anal. Biochem. 202:162-165. [DOI] [PubMed] [Google Scholar]
- 18.Ravagnani, A., et al. 2000. Spo0A directly controls the switch from acid to solvent production in solvent-forming clostridia. Mol. Microbiol. 37:1172-1185. [DOI] [PubMed] [Google Scholar]
- 19.Reyes-Teodoro, R., and M. N. Mickelson. 1945. Growth factor requirements of saccharolytic butyl alcohol-acetone bacteria. Arch. Biochem. 6:471-477. [Google Scholar]
- 20.Richter, C., et al. 1995. Oxidants in mitochondria: from physiology to diseases. Biochim. Biophys. Acta 1271:67-74. [DOI] [PubMed] [Google Scholar]
- 21.Roos, J. W., J. K. McLaughlin, and E. T. Papoutsakis. 1985. The effect of pH on nitrogen supply, cell lysis, and solvent production in fermentations of Clostridium acetobutylicum. Biotechnol. Bioeng. 27:681-694. [DOI] [PubMed] [Google Scholar]
- 22.Schuster, K. C., R. Goodacre, J. R. Gapes, and M. Young. 2001. Degeneration of solventogenic Clostridium strains monitored by Fourier transform infrared spectroscopy of bacterial cells. J. Ind. Microbiol. Biotechnol. 27:314-321. [DOI] [PubMed] [Google Scholar]
- 23.Soni, B. K., K. Das, and T. K. Ghose. 1987. Inhibitory factors involved in acetone-butanol fermentation by Clostridium saccharoperbutylacetonicum. Curr. Microbiol. 16:61-67. [Google Scholar]
- 24.Stim-Herndon, K. P., D. J. Petersen, and G. N. Bennett. 1995. Characterization of an acetyl-CoA C-acetyltransferase (thiolase) gene from Clostridium acetobutylicum ATCC 824. Gene 154:81-85. [DOI] [PubMed] [Google Scholar]
- 25.Terracciano, J. S., and E. R. Kashket. 1986. Intracellular conditions required for initiation of solvent production by Clostridium acetobutylicum. Appl. Environ. Microbiol. 52:86-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thauer, R. K., F. H. Kirchniawy, and K. A. Jungermann. 1972. Properties and function of the pyruvate-formate-lyase reaction in clostridiae. Eur. J. Biochem. 27:282-290. [DOI] [PubMed] [Google Scholar]
- 27.Thauer, R. K., E. Rupprecht, and K. Jungermann. 1970. The synthesis of one-carbon units from CO2 via a new ferredoxin dependent monocarboxylic acid cycle. FEBS Lett. 8:304-307. [DOI] [PubMed] [Google Scholar]
- 28.Thomas, J. M., P. Nicholas, and K. C. Lucille. 1985. Influence of pH on organic acid production by Clostridium sporogenes in test tube and fermentor cultures. Appl. Environ. Microbiol. 49:733-736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tishkova, V. I., and V. O. Popovb. 2006. Protein engineering of formate dehydrogenase Biomol. Eng. 23:89-110. [DOI] [PubMed] [Google Scholar]
- 30.Tomas, C. A., N. E. Welker, and E. T. Papoutsakis. 2003. Overexpression of groESL in Clostridium acetobutylicum results in increased solvent production and tolerance, prolonged metabolism, and changes in the cell's transcriptional program. Appl. Environ. Microbiol. 69:4951-4965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wynne, A. M. 1931. Inhibition of the acetone-butyl alcohol fermentation by acids. J. Bacteriol. 22:209-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yerushalmi, L., B. Volesky, and J. Votruba. 1986. Modelling of culture kinetics and physiology for C. acetobutylicum. Can. J. Chem. Eng. 64:607-616. [Google Scholar]
- 33.Zhang, Y., Z. Huang, C. Du, Y. Li, and Z. Cao. 2009. Introduction of an NADH regeneration system into Klebsiella oxytoca leads to an enhanced oxidative and reductive metabolism of glycerol. Metab. Eng. 11:101-106. [DOI] [PubMed] [Google Scholar]
- 34.Zhao, Y., C. A. Tomas, F. B. Rudolph, E. T. Papoutsakis, and G. N. Bennett. 2005. Intracellular butyryl phosphate and acetyl phosphate concentrations in Clostridium acetobutylicum and their implications for solvent formation. Appl. Environ. Microbiol. 71:530-537. [DOI] [PMC free article] [PubMed] [Google Scholar]