Abstract
We studied the interaction between capsule production and hydrodynamic growth conditions on the internal and macroscopic structure of biofilms and spontaneously formed aggregates of Klebsiella pneumoniae. Wild-type and capsule-deficient strains were studied as biofilms and under strong and mild hydrodynamic conditions. Internal organization of multicellular structures was determined with a novel image-processing algorithm for feature extraction from high-resolution confocal microscopy. Measures included interbacterial spacing and local angular alignment of individual bacteria. Macroscopic organization was measured via the size distribution of aggregate populations forming under various conditions. Compared with wild-type organisms, unencapsulated mutant organisms formed more organized aggregates with less variability in interbacterial spacing and greater interbacterial angular alignment. Internal aggregate structure was not detectably affected by the severity of hydrodynamic growth conditions. However, hydrodynamic conditions affected both wild-type and mutant aggregate size distributions. Bacteria grown under high-speed shaking conditions (i.e., at Reynolds' numbers beyond the laminar-turbulent transition) formed few multicellular aggregates while clumpy growth was common in bacteria grown under milder conditions. Our results indicate that both capsule and environment contribute to the structure of communities of K. pneumoniae, with capsule exerting influence at an interbacterial length scale and fluid dynamic forces affecting overall particle size.
The morphology of biofilms or multicellular bacterial flocs is a function, in part, of bacterial species, the extracellular materials they produce, and local hydrodynamics. For biofilms, morphological quantification is an important means of phenotyping these materials and typically proceeds from images obtained by usual light, fluorescence, or confocal microscopy on a length scale many times longer than a single bacterium (10). For flocs, structural determinations are less common and usually consist of population-based assays such as distribution of particle diameter and sedimentation behavior (9). The internal structures of both forms of growth are of interest. While greater attention has been directed at structurally evaluating biofilms, suspended flocs are an important type of growth in a number of microbiological applications. The tendency for organisms to spontaneously form, or to assume upon treatment, multicellular aggregates is fundamental to industrial processes from beer brewing to wastewater treatment (reviewed in references 3 and 18). Clumpy growth is a critical survival strategy used by waterborne pathogens such as cholera (7). Additionally, biofilm fragmentation under hydrodynamic stress produces flocs which are a key means of disseminating biofilm-based organisms in nature and likely in human illness (17). For both biofilm and suspended aggregate growth, little has been reported about community structure at the length scale of individual constituent cells.
Presumably, for both suspended aggregates and biofilms, large-scale architectural phenotypes emerge from finer features of these communities including interbacterial spacing and, in the case of bacillary organisms, correlation of angular alignment. Measurement of aggregate microstructural properties such as bacterial location, orientation, and spacing would find immediate use in providing a better understanding fundamental bacteriological questions such as the contribution of extracellular matrix to overall structure and the diffusion of nutrients, antibiotics, and quorum-sensing molecules into, out of, and within biofilms and flocs.
Here, we report an analysis of the contribution of the capsule of Klebsiella pneumoniae, a common environmental and human-pathogenic organism, to the structure of biofilms and multicellular flocs formed by that species. The capsular exopolysaccharide is a nearly universal feature of environmental strains of Klebsiella and is essential to virulence in human hosts (14). Although in this report we consider fine structural features of both biofilms and flocs, we direct our attention primarily to aggregates growing in suspension These structures have received relatively less attention in the literature and are generally too small (∼5 to 50 um) to be evaluated meaningfully by the standard confocal microscopic techniques used for biofilms. Using a wild-type (wt) encapsulated strain and an isogenic capsule-deficient mutant, we asked several questions. First, what is the effect of capsule on structure within communities, including interbacterial spacing and alignment, and is this phenotype affected by the severity of hydrodynamic conditions under which the organisms are grown? Additionally, when grown under pro-flocculating conditions, what is the relative contribution of capsular polysaccharide and hydrodynamic environment on the size distribution of aggregates formed? As part of this work, we introduce a new technique for confocal laser scanning microscopic (CLSM) evaluation of bacterial communities at scales less than 1 μm. The strategy draws significantly from methods recently developed in colloid science to resolve individual submicrometer particles, including nonspherical particles analogous to the bacilli encountered in Klebsiella microcommunities, and has significant potential in advancing the structural characterization of both suspended aggregates and anchored biofilms (5, 13).
MATERIALS AND METHODS
Bacteria preparation and growth conditions.
K. pneumoniae LM21, an encapsulated K35 serotype wild-type strain, and LM21 Δcps, a capsular mutant created with a conditionally replicative plasmid, were kindly provided by Christiane Forestier (8; also M. Thornton et al., unpublished data). Both strains were cryopreserved at −80°C prior to next-day loop inoculation in Hanks balanced salt solution (HBSS; NaCl 137 mM, Na2HPO4 0.3 mM, KCl 5.4 mM, KH2PO4 0.4 mM, MgCl2 0.5 mM, MgSO4 0.4 mM, CaCl2 1.3 mM, glucose 5.5 mM) supplemented with 0.4% glutamine as a nitrogen source. HBSS was chosen for its ionic similarity to human plasma, a medium in which our group has interest. In flask experiments, oxygen transport into the medium was enhanced by using small-volume (10-ml) batch cultures in 250-ml Erlenmeyer flasks. All flask experiments were performed at 37°C in room air using a shaker/incubator with a 1-cm radius orbit (model 435; Thermo-Forma, Marlette, OH).
For biofilm studies, organisms were seeded onto 1-mm by 4-mm by 40-mm rectangular flow cells (Stovall, Greensboro, NC) for 1 h. Flow cells were then perfused for 24 h at a volumetric flow rate of 0.5 ml/min. With medium density (978 kg/m3) and viscosity (7.55 × 10−4 Pa-s at 37°C) experimentally determined using standard methods, generating an estimated wall shear of 9 × 10−3 Pa and Reynolds' number (Re) of 4.3. For flask experiments, Re was estimated as previously described for unbaffled flasks on orbital shakers (4). The estimated Re of our system at 50 rpm was 7,626, and at 200 rpm it was 30,504. For unbaffled flasks the Reynolds' number at which flow evolves from laminar to turbulent is approximately 10,000; our experimental conditions therefore bracketed this transition, and here will be referred to as subcritical (50 rpm) and supercritical (200 rpm) Re shaking flow.
To quantify the growth kinetics of the test organisms under these conditions, batch cultures were grown at 200 rpm (the most turbulent, least aggregating) for 6 h and assayed for bacterial content hourly using turbidimetry and total protein content of pelleted bacteria (Bio-Rad Protein Low-Concentration Microplate Assay; Bio-Rad, Hercules, CA). Both methods indicated comparable proliferation rates of the two strains through the first 3 h, at which point the organisms had reached early exponential growth; this time point was used to harvest aggregates in subsequent experiments.
A two-by-three factorial study was undertaken, examining bacterial strain (wt or capsular mutant) and hydrodynamic growth conditions (biofilm, subcritical Re shaking, and supercritical Re shaking) to examine the relative contributions of the extracellular polysaccharide (EPS) and the local hydrodynamic and mixing conditions.
Uronic acid analysis.
Capsular polysaccharides were extracted and quantified using a colorimetric assay for uronic acid, the most readily assayed component of the K. pneumoniae K35 capsule repeat unit, as described by Favre-Bonte et al. for the specific strains under consideration (8). Differing from the cited method, total uronic acid (in mg/ml) was standardized to sample total protein content (as mg/ml).
Confocal microscopy.
SYTO 9 (5 mM; 1 μl/10 ml of medium; Invitrogen, Carlsbad, CA) was added to biofilms and aggregate suspensions 10 min prior to imaging. This stain was chosen as it rendered organisms homogenously internally fluorescent, a prerequisite of the image processing algorithms to follow. Aggregates were added to chambers the bases of which were number 1 (thickness, 0.14 to 0.17 mm) coverslips. Aggregates were allowed to settle onto the coverslip. To enhance adhesion and minimize Brownian and convective motion during imaging, a layer (∼20 um) of poly(dimethyl siloxane) (PDMS; Dow Sylgard 184 Silicone Elastomer Kit) was spin coated at 2,400 rpm for 30 s onto each coverslip prior to its use. CLSM for biofilms was performed directly through the coverslip at the base of the flow cell.
Images were acquired using a 100× oil immersion objective (numerical aperture, 1.4) with a 488-nm argon confocal laser scanning microscope (Leica TCS SP-2 with a DMIRE-2 inverted microscope) detecting emitted wavelengths between 500 to 650 nm. Two-dimensional (2D) images were collected with voxel dimensions between 40 and 80 nm on a side. The total number of axial slices collected was dependent upon the size of the aggregate imaged but typically was in the range 150 to 400 slices.
Identification of bacterial position and orientation using image processing.
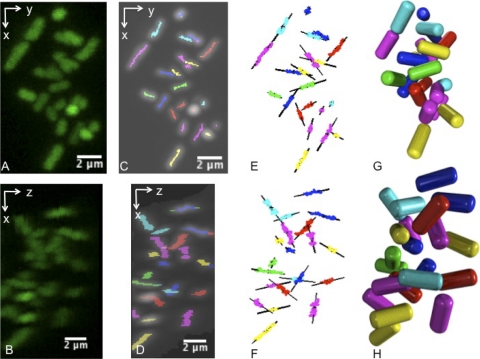
The lengths, geometric centers, and angles of orientation of individual bacilli were determined using a four-step process derived from a method used to analyze CLSM image volumes of nonbiological rod-shaped particles of a similar scale (13). These included preliminary removal of image noise using 3D Gaussian filtering of the raw CLSM image volumes, identifying all voxels contributing to the central axes (backbones) of any bacteria, resolving this collection of voxels into individual bacteria, and lastly determining the centroidal coordinates {x,y,z} and the angles of azimuthal and polar orientation {φ,θ} for each cell (Fig. 1). Renderings were produced using the open source ray-tracing software POV-Ray (www.povray.org). Computational details are provided in the supplemental material.
FIG. 1.
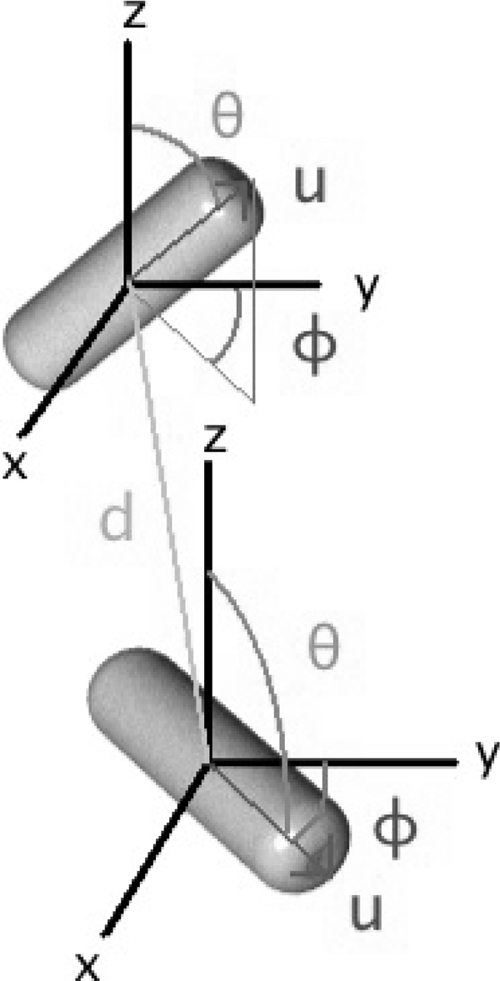
Spatial coordinates used to characterize biofilm and aggregate internal structure. For each bacterium, five parameters were determined: the {x,y,z} spatial location of their centers and their angles of polar (θ) and azimuthal (φ) orientation. The relationship between two organisms was measured by their nearest-neighbor distance (d) as well as a measure of spatial ordering.
Use of positions and orientation angles to quantify spatial organization.
Two spatial statistics were determined for communities grown under each experimental condition. The first was the nearest-neighbor distance, the distance in micrometers between the geometric center of each bacterium and the center of that bacterium's closest neighbor. The second spatial statistic was the nematic order parameter, S. This metric is taken from the physics literature, is calculated over all cells in an aggregate or biofilm sample, and quantifies the extent of angular alignment between organisms. For each aggregate, it may take a value from 0 (no local correlation) to 1 (complete “liquid crystalline” alignment).
Bacterial aggregate size distribution.
Size distribution of populations of particles in aggregated liquid cultures were measured using a Z2 particle counter (Beckman-Coulter, Fullerton, CA) equipped with a 100-μm aperture (allowing accurate sizing between 2 and 60 um). A 2- to 5-μm-diameter window was studied, with 240 size bins included. The device internally adjusts bin width, gain, and other electronic parameters during measurement, and these default settings were used in each case. Measurements for sizing were taken from five replicate flasks for each experimental condition.
Computational methods and statistical analysis.
All image analysis was performed in software written by our group in C++ and is available from the authors. All statistical analysis was performed in R, version 2.6.2 (15). Where appropriate, t tests were used for simple between-group comparisons. Distributions of nearest neighbor distances were compared using the Cramér von Mises goodness of fit test. Angular alignment (i.e., order parameter) estimates as well as population size distributions between strains and under different shaking conditions were analyzed using analysis of variance (ANOVA).
RESULTS
Both wild-type and capsular mutant strains of K. pneumoniae LM21 grew as aggregates under the shaking conditions tested. The two strains had similar batch culture growth kinetics by turbidimetry and total cellular protein content (Fig. 2A). As noted previously with these strains (8), chemical analysis of aggregates at mid-log growth showed an approximately 7-fold difference in capsular uronic acid content between wild-type and cps mutant strains as a function of total protein (0.92 ± 0.24 mg of uronic acid/mg of protein for the wild type and 0.13 ± 0.07 mg of uronic acid/mg of protein for the mutant; P < 0.01). The capsular mutant was observed to grow more slowly as a biofilm than the wild-type strain. However, as our interest was primarily on internal structure and as both strains grew to the extent necessary to perform CLSM, we did not formally evaluate bulk biofilm growth.
FIG. 2.
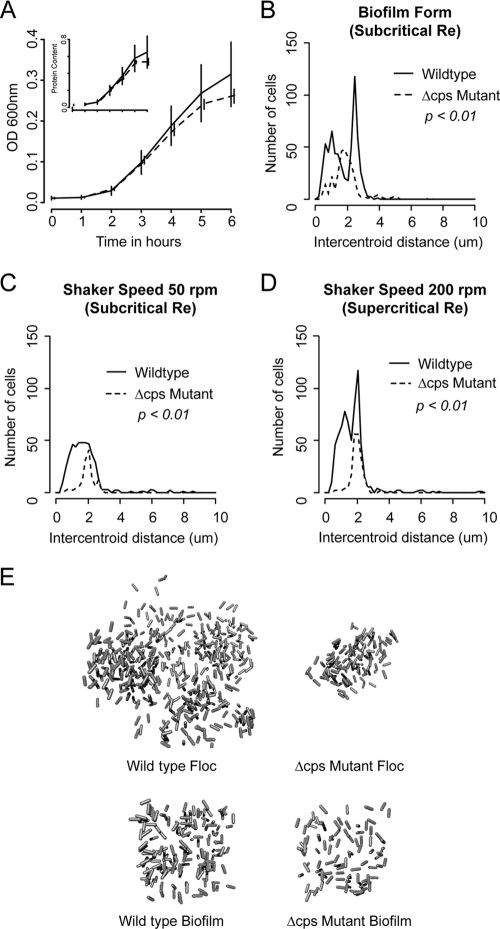
Various features of single-cellular and clumped growth of the two strains. (A) Growth of nonaggregated wild-type and cps mutant K. pneumoniae strains grown under 250 rpm to prevent any aggregation, as measured by turbidimetry. (Inset) Total cellular protein. No significant differences were seen, and up until the 3-h time point at which subsequent experiments were performed, the growth curves were essentially identical. (B, C, and D) Distribution of bacterial nearest-neighbor distances. For each cell within each biofilm or aggregate studied, the distance to its closest neighboring cell (in micrometers) was determined. Results were pooled across all samples of a given experimental condition (e.g., wild-type aggregates grown at 50 rpm). Comparisons between distributions were made using the Cramér von Mises test for distribution equivalence. Regardless of the hydrodynamic conditions under which they formed, wild-type aggregates demonstrated broader distributions of internal interbacterial distances than mutant flocs. (E) Reconstructions of representative floc and biofilm volumes. Margins of flocs are irregular as the entire volume of these structures was captured whereas only cubic samples were taken of larger biofilms. In each case, individual rods are one micrometer in length.
High-resolution measurements of individual bacteria within biofilms and aggregates were made using confocal microscopy followed by computational image analysis. An example result is shown in Fig. 3, wherein the process by which a community was evaluated is summarized.
FIG. 3.
Extraction of aggregate internal structure using confocal microscopy and image processing. (A and B) Small aggregate of K. pneumoniae projected from two view angles. (C and D) Identification of backbone voxels lying within individual cells. (E and F) Assignment of estimated position and angle of alignment. (G and H) Ray-tracing reconstruction of internal architecture of the aggregate from the projection angles used in panels A and B.
To quantify internal organization of these structures, we first examined the distribution of nearest-neighbor distances, the length in micrometers between the center of each bacterium and that of its closest adjacent cell. These measurements were taken from analysis of wild-type and cps mutant biofilms and aggregates and included over 5,000 interbacterial distance measurements. Results are shown in Fig. 2C and D. Whereas the interbacterial spacing between individual mutant organisms was narrowly distributed around 2 μm, wild-type organisms grew in a broader, and under some conditions, clearly biomodal distribution (P < 0.01 for a difference in distribution by Cramér von Mises). Within suspended aggregates, organization was not affected by the Reynolds' number under which the structures were grown.
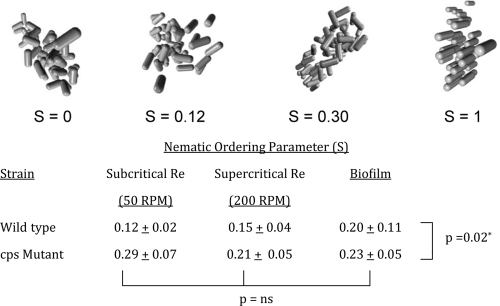
The image processing algorithm we used also allowed examination of angular alignment within particles. We chose the nematic ordering parameter, a common metric in colloidal and liquid crystal physics, to quantify relative alignment between bacteria within the aggregate (1). This variable may take values from 0 (uniformly random alignment between cells) to 1 (complete orientation of all bacteria) and is a measure of internal organization within the aggregate. Results are shown in Fig. 4, where we have also included images of simulated aggregates of various angular alignments to assist in interpreting the results. As with the nearest-neighbor measurements, the primary determinant of structural organization was the presence of absence of capsule. Whether as biofilms or aggregates, wild-type organisms assembled in a less organized fashion (P < 0.012 by two-way ANOVA). The fluid dynamic conditions under which the communities formed did not appear to contribute to internal organization.
FIG. 4.
Effect of capsule and growth conditions on internal angular ordering. The extent of interbacterial orientational alignment was quantified using the nematic ordering parameter, which spans from 0 (no angular organization) to 1 (complete liquid crystalline alignment). Compared to wild-type biofilms and aggregates, structures formed by capsule-deficient mutants demonstrated greater internal ordering (P = 0.02 by two-way ANOVA). No differences were seen as a function of hydrodynamic growth conditions. Images depict simulated aggregates with no ordering (S = 0), partial ordering of magnitudes comparable to what was seen in experimental samples, and a theoretical fully ordered aggregate (S = 1).
Figure 2E, provides three-dimensional reconstructions of representative aggregates and biofilm samples from our experiments to provide broader context to the structural metrics described above.
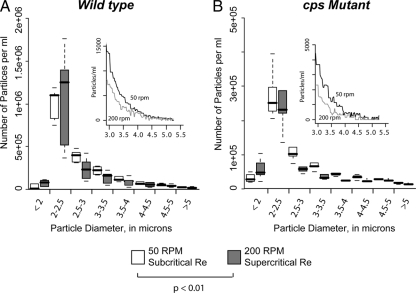
To relate the effect of capsule on internal aggregate structure with macroscopic population aggregative growth, we measured the distribution of aggregate sizes for each of the experimental conditions using a Coulter particle analyzer. This device provided details of aggregate diameter from 2 to 5 μm and outputs a distribution of particle sizes in this range. In distinction to what was seen with the intraparticle structural analysis, the primary determinant of particle population diameter within the 2- to 5-μm range was the rate at which flasks were shaking during growth (Fig. 5). The distribution of particle sizes was significantly skewed toward smaller diameters under 200-rpm conditions (i.e., Re greater than the turbulent transition) compared to results at 50 rpm (Re less than the transition), suggesting that smaller structures are favored under harsh hydrodynamic conditions although we note that some other feature correlated with shaking conditions (e.g., media oxygenation) was not involved.
FIG. 5.
Distribution of particle sizes assumed by wild-type and capsule-deficient K. pneumoniae aggregates. (A) Coulter-method sizing results for wild-type aggregates grown below (50 rpm) and above (200 rpm) the Reynolds number at which shaking flask flow becomes turbulent. Values in each size bracket are shown as box plots representing the 0th, 25th, 50th, 75th, and 100th percentile measures. The inset panel shows the mean value of the distributions in the larger diameter “tail” of the size distribution from 3 to 5 μm. Differences in the distributions of particle sizes were seen (P < 0.01 by ANOVA), with aggregates grown under turbulent conditions forming fewer large aggregates. (B) Same analysis as shown in panel A but for cps mutant organisms. The difference in aggregate size distribution is more pronounced (P < 0.01). Taken together, these data indicate that, unlike measures of internal order, particle size distributions for these strains are affected both by the presence of bacterial capsule and the local hydrodynamic conditions under which they form.
DISCUSSION
We describe a novel micrometer-scale structural analysis to evaluate the role of capsule in the architecture of biofilms or spontaneously formed bacterial aggregates in liquid culture. We found that the structural implication of capsular operon interruption was to render mutant communities quantitatively more uniform in both interbacterial spacing and angular orientation. The capsule's contribution thus was not merely to serve as a spacer between cells; on the contrary, the primary means by which capsule broadened the distribution of nearest-neighbor distances was by allowing tighter packing than was seen in capsule-deficient aggregates.
The Klebsiella cps mutant strain studied was generated by deletion of a conserved promoter region of a 13-open reading frame (ORF) polycistronic operon, resulting in an experimentally confirmed 7-fold reduction in uronic acid-to-total protein ratio during mid-log growth (8). The presence of wild-type quantities of extracellular material was not associated with what might be expected in this setting, namely, greater intercellular spacing within aggregates. The shorter distances between adjacent bacteria in the presence of capsule may be a result of daughter cells remaining embedded in a common EPS envelope after division or of differences in EPS density and water content from one region to another. Alternatively, capsule may interfere with the effects of other extracellular structures (e.g., pili or curli) that might contribute to space-filling between cells. This already is known to occur in Klebsiella, in which capsular EPS shields fimbrial adhesins (8, 16).
While the K35 structure is known, synthetic and export details are few (6). In the report from the Favre-Bonte laboratory originally describing the creation of the cps mutant, capsular synthesis by the parent strain was shown to be dependent on growth phase; the amount of recoverable uronic acid per bacterium fell sharply across mid-exponential growth. Thus, rather than being an all-or-none phenomenon, capsular expression is in part a function of growth phase. It remains to be determined whether EPS production by organisms deep within aggregates is more akin to fully developed, diffusion-limited biofilms or to fully dispersed liquid cultures. Given the uncertainties of the metabolic conditions within both biofilms and aggregates, an important next step in this work will be to develop techniques for simultaneously determining bacterial and EPS spatial location.
We foresee several near-term applications for the technique described. Detailed measures of interbacterial spacing have immediate use in providing a better understanding the nutrient and quorum-sensing environment within aggregates. Concentrations of both small and large molecules within communities are functions of production, consumption, and transport into and out of the community by convective and diffusive forces. Combined with measures of production and consumption by individual cells, our technique provides the spatial information needed to predict measurable intra-aggregate features such as oxygen tension and redox potential and to make forecasts regarding local environmental features (e.g., concentration of quorum-sensing molecules deep within a community) that are not easily obtained experimentally.
Divining the process of genesis of individual aggregates and of polydisperse bacterial microcommunities will also be assisted by this technique. In a previous theoretical consideration of this issue, our group described bacterial particulate populations as a dynamic equilibrium between aggregates enlarging—by bacterial cell division or by adhesive collisions between cells and aggregates—and aggregates fragmenting through erosion or fragmentation (2). Work in nonbiological particulate suspensions suggests that structural analysis of the kind described in this report might be used to distinguish growth by cell proliferation and growth by collision. Being able to identify the mechanism of particle growth by bacterial communities in hydrodynamically active environments is important to understanding what type of aggregates might be expected to form in settings where interaggregate collisions would be expected to be rare (e.g., very diffuse “open” water environments or blood streams of bacteremic hosts). Such measurements might also allow one to estimate an intra-aggregate equivalent to a rate constant for growth.
In addition to passive observations of community architecture, modifications of the currently described image analysis strategy might be used for more sophisticated structural analysis. We have recently described a microfluidic channel-based rheometer for evaluating biofilm viscoelasticity on structures of comparable sizes to those measured here (12). Our cell-locating algorithm, if applied repeatedly to a community of bacteria undergoing mechanical deformation over time, might be used to extract local viscoelasticity determinations (e.g., elastic modulus and Poisson ratio) within aggregates or biofilms. Such information is necessary for understanding the fate of these materials in flowing systems.
We would point out that the technique described herein is readily applied to other bacterial morphology. In fact, analyzing communities comprised of cocci is simpler than for bacilli in that only position and radius, and not angle of alignment, need be determined to fully describe a cell (11).
An important challenge going forward is identifying the most useful metrics for describing this collection of phenotypes. Fortunately, a number of strategies have been validated in the study of nonbiological colloids and two, nearest-neighbor distance and the nematic order parameter, were successfully used in the current work. What remains is the gathering of more experience with these metrics across a broader set of bacterial strains and experimental conditions such that the effect sizes observed in the current work—2-fold differences observed in both nearest-neighbor distance and the nematic order parameter by capsular depletion—can be placed in appropriate context.
Supplementary Material
Footnotes
Published ahead of print on 14 January 2011.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Allen, M., G. Evans, D. Frenkel, and B. Mulder. 1993. Hard convex body fluids. Adv. Chem. Physics 86:1-166. [Google Scholar]
- 2.Bortz, D., T. Jackson, K. Taylor, A. Thompson, and J. Younger. 2008. Flocculation dynamics of Klebsiella pneumoniae. Bull. Math. Biol. 70:745-768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bratby, J. 2006. Coagulation and flocculation in water and wastewater treatment, 2nd ed. IWA Publishing, London, United Kingdom.
- 4.Buchs, J., U. Maier, C. Milbradt, and B. Zoels. 2000. Power consumption in shaking flasks on rotary shaking machines: II. Nondimensional description of specific power consumption and flow regimes in unbaffled flasks at elevated liquid viscosity. Biotechnol. Bioeng. 68:594-601. [PubMed] [Google Scholar]
- 5.Crocker, J., and D. Grier. 1996. Methods of digital microscopy for colloidal studies. J. Colloid Interface Sci. 179:298-310. [Google Scholar]
- 6.Dutton, G., and A. Lim. 1985. Structural investigation of the capsular polysaccharide of Klebsiella serotype K35. Carbohydr. Res. 145:67-80. [DOI] [PubMed] [Google Scholar]
- 7.Faruque, S., et al. 2006. Transmissibility of cholera: in vivo-formed biofilms and their relationship to infectivity and persistence in the environment. Proc. Nat. Acad. Sci. U. S. A. 103:6350-6355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Favre-Bonte, S., B. Joly, and C. Forestier. 1999. Consequences of reduction of Klebsiella pneumoniae capsule expression on interactions of this bacterium with epithelial cells. Infect. Immun. 67:554-561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gregory, J. 2006. Particles in water: properties and processes. IWA Publishing, London, United Kingdom.
- 10.Heydorn, A., et al. 2000. Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology 146:2395-2407. [DOI] [PubMed] [Google Scholar]
- 11.Hohne, D. 2008. Characterization of industrial and biological complex fluids using confocal microscopy. Ph.D. dissertation. University of Michigan, Ann Arbor, MI.
- 12.Hohne, D., J. Younger, and M. Solomon. 2009. Flexible microfluidic device for mechanical property characterization of soft viscoelastic solids such as bacterial biofilms. Langmuir 25:7743-7751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mohraz, A., and M. Solomon. 2005. Direct visualization of colloidal rod assembly by confocal microscopy. Langmuir 21:5298-5306. [DOI] [PubMed] [Google Scholar]
- 14.Podschun, R., and U. Ullman. 1998. Klebsiella spp. as nosocomial pathogens: epidemiology, taxonomy, typing methods, and pathogenicity factors. Clin. Microbiol. Rev. 11:589-603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.R Development Core Team. 2008. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria.
- 16.Schembri, M., J. Blom, K. Krogfelt, and P. Klemm. 2005. Capsule and fimbria interaction in Klebsiella pneumoniae. Infect. Immun. 73:4626-4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stoodley, P., et al. 2001. Growth and detachment of cell clusters from mature mixed species biofilms. Appl. Environ. Microbiol. 67:5608-5613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Verstrepen, K., and F. Klis. 2006. Flocculation, adhesion, and biofiolm formation in yeast. Mol. Microbiol. 60:5-15. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.