Abstract
The aims of this study were, firstly, to compare five published methods for the isolation of Arcobacter spp. from animal feces in order to determine the most sensitive and specific method. Second, we analyzed the resulting isolates by multilocus sequence typing (MLST) in order to investigate the diversity of the isolates recovered. Third, we investigated the ability to recover Arcobacter spp. from frozen fecal samples. Seventy-seven fecal samples from cattle, sheep, and badgers were subjected to five isolation methods, based on published methods for the isolation of Arcobacter and Campylobacter spp. Thirty-nine Arcobacter butzleri isolates were analyzed using a multilocus sequence typing scheme. The survival of Arcobacter spp. in frozen samples was investigated by freezing the fecal samples at −80°C for 7 days and then applying the same five isolation methods. The most sensitive and specific method used an Arcobacter-specific broth in conjunction with modified charcoal cefoperazone deoxycholate agar (mCCDA) with added antibiotics. Freezing of fecal samples led to a reduction in the recovery of Arcobacter spp. by approximately 50%. The 39 allelic profiles obtained by MLST could be divided into 11 sequence types (STs). We have identified the most sensitive and specific method for the isolation of Arcobacter spp. from animal feces and demonstrated that the freezing of fecal samples prior to isolation reduces arcobacter recovery. MLST analysis of the isolates revealed a high level of diversity.
Arcobacter spp. are Gram-negative bacteria that differ from the closely related Campylobacter spp. in that they are able to grow at temperatures as low as 15°C and under aerobic conditions. The genus Arcobacter currently contains 10 species, of which seven may be considered emerging human food-borne pathogens. A. butzleri, A. skirrowii, A. cryaerophilus, A. cibarius, A. mytili, A. thereius, and A. trophiarum have all been isolated from foodstuffs, including meat, shellfish, and water, or from the feces of livestock (3, 4, 5, 7, 8, 9, 14, 17, 18, 20, 30, 32); A. butzleri, A. skirrowii, and A. cryaerophilus have been isolated from human fecal samples (15, 18, 24, 27, 29, 31, 33, 34, 35, 43).
It has been reported that Arcobacter spp., which were originally isolated from aborted bovine fetuses, can cause disease in cattle (7) although the true role of Arcobacter spp. as veterinary pathogens is yet to be definitively proven. Arcobacter spp. have also been isolated from healthy cattle in Belgium (37), Japan (20), Turkey (2, 23), the United States (8, 42), and New Zealand (25) and from beef and/or beef products from Thailand (39), Northern Ireland (32), Turkey (2, 28), Australia (30), Japan (21), Mexico (38), Czech Republic (40), the United States (8), and Netherlands (4). These strains show that the organism can be present in healthy animals and their products, illustrating the importance of cattle and beef as potential sources of Arcobacter infection of humans.
A variety of methods have been employed for the isolation of Arcobacter spp. from fecal samples, ranging from modified Campylobacter and Leptospira techniques to those involving Arcobacter-specific media. The first reported isolation of an Arcobacter used Leptospira Ellinghausen-McCullough-Johnson-Harris (EMJH) medium supplemented with 5-fluorouracil (7). Atabay and Corry (1) evaluated the use of Arcobacter broth (Oxoid, United Kingdom) with an added cefoperazone, amphotericin, and teicoplanin (CAT) supplement. Johnson and Murano (19) developed JM broth and plates, and more recently Houf et al. (13) developed an Arcobacter-specific isolation method involving the use of Arcobacter medium (Oxoid, United Kingdom) with a supplement consisting of five antibiotics (cefoperazone, trimethoprim, amphotericin, novobiocin, and 5-fluorouracil). This method has been used in a number of studies on the prevalence of Arcobacter spp. (12, 14, 15, 16, 17) and was modified for isolation of Arcobacter from animal feces by Van Driessche et al. (36). Other isolation methods used have included EMJH p-80 and brucella broth (28) and direct inoculation onto agar without antibiotics via filtration through a membrane (6).
Despite the range of isolation methods used previously, no single standard method for the isolation of Arcobacter spp. from fecal samples has been established. The lack of a standard method means that comparing data from different studies is difficult. It has been suggested that the lack of a standard Arcobacter isolation method may mean that many human cases go undetected and that such a method could lead to more accurate reporting of human infections (29) and thus improve efforts to control infection.
The main aims of this study were to compare five methods for the isolation of Arcobacter spp. from animal fecal samples and to investigate the diversity of a selection of the Arcobacter isolates obtained using multilocus sequence typing (MLST). Additionally, the effect of freezing on Arcobacter in fecal material was tested in order to determine the reliability of isolating Arcobacter from archived frozen samples.
MATERIALS AND METHODS
Comparison of Arcobacter isolation methods.
A total of 77 fecal samples were collected from cattle (n = 47), sheep (n = 18), and badgers (n = 12) on six farms in Cheshire and Lancashire and from a wildlife park in Gloucester, United Kingdom. Four cattle farms (comprising three dairy and one beef) and two sheep farms were sampled, along with the wildlife park (which contained farmland), which also had a large population of badgers. All of the locations were sampled once, with 6 to 12 fecal samples collected from each. Cattle samples were collected from unweaned calves, weaned calves, nonlactating adults, and lactating adults on each farm. Samples were collected using sterile plastic containers and were processed within 3 h of collection on all occasions, except for the badger samples, which were processed immediately after being received via post.
One gram of fecal material was transferred into 9 ml of enrichment broth, mixed by shaking, and incubated either aerobically or microaerobically, depending on the broth, for 24 h. The following broths were used. H broth was an Arcobacter-specific broth (Oxoid, Basingstoke, United Kingdom) with the addition of 5-fluorouracil (100 mg ml−1), amphotericin B (10 mg ml−1), cefoperazone (16 mg ml−1), novobiocin (32 mg ml−1), and trimethoprim (64 mg ml−1) (Sigma-Aldrich, Dorset, United Kingdom), as described by Houf et al. (12). AC broth was an Arcobacter-specific broth comprising Arcobacter broth (Oxoid, United Kingdom) with the addition of cefoperazone (8 mg liter−1), amphotericin B (10 mg liter−1), and teicoplanin (4 mg liter−1) (CAT supplement; Oxoid, United Kingdom), as described by Atabay and Corry (1). C broth was a Campylobacter-specific enrichment broth (Lab M, Bury, United Kingdom) containing 5% (vol/vol) defibrinated horse blood and cefoperazone (20 mg liter−1), vancomycin (20 mg liter−1), trimethoprim (20 mg liter−1), and cycloheximide (50 mg liter−1) (CVTC supplement; Lab M, Bury, United Kingdom) as described by Kemp et al. (23). H and AC broths were incubated aerobically at 30°C while C broth was incubated microaerobically at 37°C.
After incubation, 20 μl of enrichment broth was streaked onto solid medium and incubated again. The following solid media were used: H medium, which was the solid equivalent of H broth and contained the same five-antibiotic supplement (12); CC medium, which comprised modified charcoal agar (modified charcoal cefoperazone deoxycholate agar [mCCDA]; Lab M, Bury, United Kingdom) with added CAT supplement (Lab M, Bury, United Kingdom) (23); and C medium, which was a Campylobacter-specific isolation medium (Lab M, Bury, United Kingdom) containing mCCDA (Lab M, Bury, United Kingdom) with an added cefoperazone (32 mg liter−1) and amphotericin B (10 mg liter−1) supplement (Lab M, Bury, United Kingdom) as described by Kemp et al. (23). The five isolation methods used were named as follows: HH (Houf broth and Houf plates), HCC (Houf broth with mCCDA-CAT plates), ACH (Arcobacter broth-CAT broth with Houf plates), ACCC (Arcobacter broth-CAT broth with mCCDA-CAT plates) and CC (Campylobacter-specific broth and Campylobacter-specific plates). Enriched samples were plated onto solid media in duplicate, and each sample was subjected to each of the five isolation methods. Plates were incubated aerobically at 30°C (methods H and CC) or microaerobically at 37°C (method C) for 48 h. Even though the H method originally used an incubation temperature of 28°C, 30°C was used here after previous work found Arcobacter spp. to grow well at 30°C with little or no growth of contaminants (unpublished data). Up to 10 colonies per sample, per method, were then selected based on morphology (Gram-negative, small gray-white, round colonies) and streaked onto Columbia agar containing 5% (vol/vol) defibrinated horse blood and incubated as before, for 48 h. Table 1 shows the combinations of media used.
TABLE 1.
The five combinations of media and conditions used
| Method | Broth type | Solid medium (plate) | Incubation conditions |
|---|---|---|---|
| HH | H | H | 30°C, aerobic |
| HCC | H | CC | 30°C, aerobic |
| ACH | AC | H | 30°C, aerobic |
| ACCC | AC | CC | 30°C, aerobic |
| CC | C | C | 36°C, microaerobic |
The sensitivity of each method was calculated as the ability of a method to detect Arcobacter-positive animals. The maximum possible number of Arcobacter-positive animals was taken as the total number of animals testing positive during this study using all five of the methods. The number of Arcobacter-positive animals using a particular method was then calculated as a percentage of the maximum possible number. Specificity was calculated as the number of Arcobacter isolates obtained using each method as a percentage of the total number of isolates retrieved overall (which included Campylobacter spp. and other bacterial isolates). The difference in specificity of each isolation method was tested for significance using Fisher's exact test using the GraphPad QuickCalcs free online calculator (http://www.graphpad.com/quickcalcs/contingency1.cfm).
For each isolate, a cell lysate was prepared by creating a cell suspension in 150 μl of distilled water and heating the suspension at 100°C for 15 min before centrifuging at 13,000 rpm for 10 min; the supernatant was used as a template for PCRs.
Identification of isolates by PCR.
An Arcobacter genus-specific PCR assay (9) was applied to all isolates. Any isolates positive using this PCR assay were then further identified to species level using the Arcobacter multiplex PCR assay of Houf et al. (11). All isolates negative in the genus-specific PCR assay were discarded. Campylobacter isolates were identified using the multiplex PCR of Wang et al. (41). All PCR assays were carried out using ReddyMix PCR Master Mix (Abgene, Loughborough, United Kingdom), which contains 1.5 μM MgCl2, 1 U of ThermoPrime Taq DNA polymerase, and a 20 mM concentration of the deoxynucleoside triphosphates (dNTPs).
MLST analysis.
In order to assess diversity, MLST profiles were obtained for 39 randomly selected isolates. The isolates were subjected to the Arcobacter-specific MLST scheme as described by Miller et al. (26). Sequence data were aligned using the in-built ClustalW alignment tool, and dendrograms were constructed for each locus using the neighbor-joining bootstrap test of phylogeny.
Investigation into the effect of freezing.
All fecal samples were frozen at −80°C for 7 days and then defrosted and subjected to the five isolation methods described. Archived samples were available and stored at −80°C, so this investigation aimed to show whether these could be used for future work. The resulting isolates were tested using the Arcobacter genus-specific PCR assay (9).
RESULTS
Comparison of isolation methods.
In total, 1,260 isolates were recovered from 77 animal fecal samples using five isolation methods. Of these isolates, 483 (38.3%) were identified as Arcobacter and further assigned to species using PCR assays. Of the remainder, 231 were identified as Campylobacter and assigned to species using PCR; the rest (n = 546) did not belong to the Campylobacteraceae, and the majority of these did not comply with typical Arcobacter morphology when recultured. A small number of isolates (n = 24) gave a positive result in the genus-specific PCR assay but were then negative when the species-specific PCR was attempted. These isolates were subjected to groEL gene sequencing (22) and identified using BLAST analysis as some A. butzleri isolates as well as some fecal flora, including Pseudomonas sp., Acinetobacter sp., Psychrobacter sp., and Escherichia coli.
The five isolation methods (HH, HCC, ACH, ACCC, and CC) (Table 1) were compared for sensitivity and specificity using all of the samples from which Arcobacter spp. were isolated. Table 2 shows the sensitivity and specificity of each method. HCC had the greatest sensitivity (70.7%) and specificity (63.9%) of the five methods tested. The sensitivities of the other four methods were lower than the sensitivity of the HCC method (Table 2). The difference in the specificity of each method was tested for significance using Fisher's exact test. HCC was significantly more specific than all four other methods (P = 0.0139 compared with ACCC, ACH, and CC; P = 0.0249 compared with HH).
TABLE 2.
Sensitivity and specificity of each isolation method testeda
| Method | No. of Arcobacter- positive samples | No. of Arcobacter sp. isolates | No. of Campylobacter sp. and non-Campylobacteraceae isolates | Sensitivity (%) | Specificity (%) |
|---|---|---|---|---|---|
| HH | 17 | 92 | 62 | 41.5 | 59.7 |
| ACH | 18 | 63 | 201 | 43.9 | 23.9 |
| ACCC | 18 | 58 | 290 | 43.9 | 16.6 |
| HCC | 29 | 175 | 99 | 70.7 | 63.9 |
| CC | 18 | 95 | 125 | 43.9 | 43.2 |
The method with the greatest sensitivity and specificity is shown in boldface.
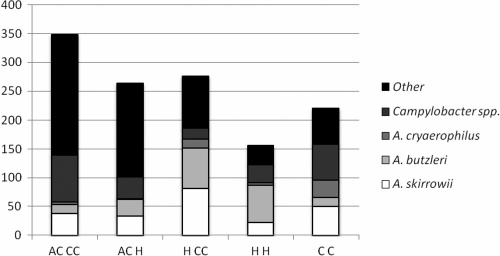
HCC isolated the largest proportion of Arcobacter (37.6%). Of these, almost equal proportions of A. skirrowii (29%) and A. butzleri (26%) were obtained, along with 6% A. cryaerophilus (Fig. 1). HCC was more selective for A. skirrowii than the HH method, which gave a much higher proportion of A. butzleri (41%) than A. skirrowii (15%). Of the isolates obtained using the CC method, the largest proportion of identifiable isolates (28%) were Campylobacter spp. (Fig. 1), as would be expected from a Campylobacter-specific method. This method isolated the largest proportion of A. cryaerophilus (14%). ACH, ACCC, and CC all appeared to isolate higher proportions of A. butzleri than A. skirrowii. Of these three, ACCC seemed to be more selective for A. skirrowii than A. butzleri, with 11% of the isolates being A. skirrowii and only 4% identified as A. butzleri (Fig. 1).
FIG. 1.
The proportion of species isolated using each of the five methods. Methods are as described in Table 1.
Overall, the most frequently isolated Arcobacter sp. was A. skirrowii, constituting 17.8% of the total isolates obtained (47% of all Arcobacter isolates). A. butzleri made up 15.5% of all isolates (41% of all Arcobacter isolates), and A. cryaerophilus comprised 4.5% (12% of all Arcobacter isolates). Campylobacter spp. accounted for 12.2% of all isolates.
Typing of the Arcobacter isolates by MLST.
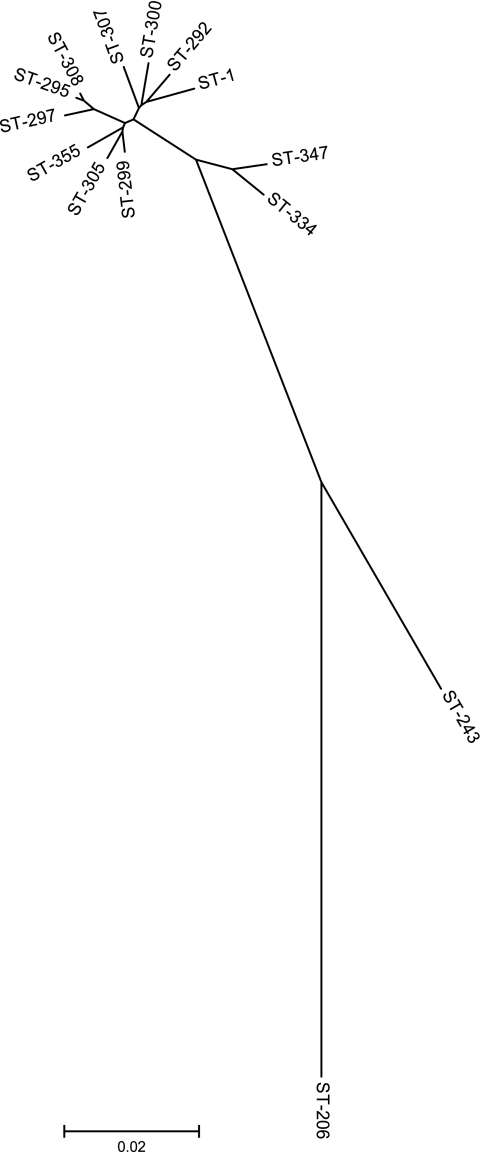
Thirty-nine allelic profiles were obtained by MLST, all of which belonged to the species A. butzleri and within which 11 sequence types (STs) were present (Table 3). Each ST was exclusive to one farm, meaning that no ST was found on multiple farms. Allelic density (number of alleles/number of strains) was as follows: aspA, 20.5%; atpA, 15.0%; glnA, 17.9%; gltA, 23.1%; glyA, 28.2%; pgm, 25.6%; and tkt, 20.5%. For the 11 A. butzleri sequence types identified, the allele sequences were concatenated in the order aspA, atpA, glnA, gltA, glyA, pgm, and tkt and aligned using ClustalW; a neighbor-joining tree was constructed from the aligned sequences (Fig. 2). ST-1 (A. butzleri), ST-206 (A. cryaerophilus), and ST-243 (A. skirrowii) from the MLST database were included in the tree for reference.
TABLE 3.
Distribution of the 11 A. butzleri sequence types present
| ST | No. of times present | Location |
|---|---|---|
| 292 | 11 | Mixed dairy/sheep farm |
| 295 | 2 | Mixed dairy/sheep farm |
| 297 | 1 | Dairy farm 1 |
| 299 | 2 | Dairy farm 1 |
| 300 | 2 | Beef farm |
| 305 | 1 | Mixed dairy/sheep farm |
| 307 | 3 | Mixed dairy/sheep farm |
| 308 | 11 | Dairy farm 2 |
| 334 | 1 | Dairy farm 2 |
| 347 | 2 | Beef farm |
| 355 | 3 | Dairy farm 2 |
FIG. 2.
Neighbor-joining tree constructed using alleles concatenated in the order aspA, atpA, glnA, gltA, glyA, pgm, and tkt from the 11 sequence types identified. ST-1 (A. butzleri), ST-206 (A. cryaerophilus), and ST-243 (A. skirrowii) were included from the MLST database for reference.
Use of frozen samples.
A total of 750 isolates were recovered from the same fecal samples after they had been frozen at −80°c for 1 week. Of these, 149 (19%) were identified as belonging to the Arcobacter genus using a PCR assay (9). These isolates were not assigned to species. Arcobacter recovery from frozen samples was approximately 49% compared with the recovery from fresh samples.
DISCUSSION
The five methods tested isolated different proportions of Arcobacter spp. These differences are most likely due to various sensitivities to the antibiotic supplements used in the media. A. skirrowii was the most frequently isolated species overall, followed by A. butzleri then A. cryaerophilus; HCC appeared to be more representative of these overall results than the next most specific method, HH. The fact that A. skirrowii was the most frequently isolated species of Arcobacter in this study is surprising as this species is reported as being the most susceptible to some antimicrobial agents used in selective media (13). A. skirrowii may have been present in large enough numbers to make it culturable despite its sensitivity, or possibly the isolates recovered in this study were less sensitive to agents in the media.
A. butzleri, A. skirrowii, and A. cryaerophilus were all isolated from cattle in this study, whereas A. butzleri was the only Arcobacter species isolated from sheep. Arcobacter spp. were not isolated from badgers. This is the first study to our knowledge to isolate all three species from the feces of cattle in the United Kingdom. A small number of isolates could not be assigned to species using the multiplex PCR assay (11) after testing positive in the Arcobacter genus-specific PCR assay (9). It was initially assumed that these isolates were likely to be either A. cibarius (14), A. thereius (17), or a novel Arcobacter sp. However, after groEL gene sequencing and BLAST analysis of the resulting sequences, it was determined that the isolates included further A. butzleri and A. skirrowii isolates, as well as Pseudomonas sp., Acinetobacter sp., E. coli, and a Saccharophagus sp., indicating some unreliability of both the Arcobacter genus-specific and species-specific PCR assays.
Previous studies outside the United Kingdom reported low prevalence of Arcobacter spp. in sheep (0% [2)]; 15% from lamb meat [29]; 16.1% from sheep feces [36]). The prevalence of Arcobacter spp. in the sheep samples in this study was 40% (n = 10), which is higher than found in a previous study in the United Kingdom, which isolated Arcobacter using the Campylobacter-specific method, CC (10). Season, climate, geographical location, and sampling and isolation methods may all contribute to the low prevalence of Arcobacter recovered from sheep.
MLST analysis revealed a high level of diversity among the isolates at all loci (data not shown). The glyA2 locus was not used as it was found to be of limited use by Miller et al. (26). The allelic density of A. butzleri isolates in this study is lower than that observed by Miller et al. (26), possibly as a result of the small sample size. The greatest allelic density was observed at the glyA locus (28.2%), followed by the pgm locus (25.6%), in agreement with the study of Miller et al. (26). The glyA and pgm loci show the greatest variation in both Arcobacter MLST studies to date, with the lowest allelic density at the atpA locus (15.0%) in this study. Further MLST studies on the diversity of Arcobacter spp. in cattle will elucidate whether allelic density is consistently lower in Arcobacter isolates from cattle.
Figure 2 shows that ST-334 and ST-347 form an outlying group from the main group of A. butzleri STs found in this study. ST-334 was identified on one occasion from the feces of a dairy cow on a farm in Lancashire. Other STs found on the same farm were STs 308 and 355, and one of three samples from this farm was coinfected with both ST-308 and ST-355. None of the STs from this farm was detected elsewhere. ST-347 was recovered from the feces of a beef bull on a farm in Cheshire; ST-300 was also found on the same farm but not in the same animal. Neither ST-347 nor ST-300 was found elsewhere. Overall, none of the STs identified in this study was found on more than one farm (Table 3). As might be expected, the A. butzleri isolates from this study form a cluster along with ST-1 (A. butzleri) from the MLST database while ST-206 (A. cryaerophilus) and ST-243 (A. skirrowii) form separate, distinct branches.
Freezing of the fecal samples resulted in a 50% reduction in recovery of Arcobacter spp. It is therefore recommended that for optimal isolation of Arcobacter spp. from fecal samples, the samples used must be fresh and not frozen. It is possible that one or more species of Arcobacter may be more or less susceptible to freezing; therefore, an investigation into the effect of freezing where species is taken into account would be of value.
In conclusion, this study has determined a sensitive and specific method for the isolation of Arcobacter spp. from animal feces, which is recommended for use as a standard Arcobacter isolation method, and determined that frozen fecal samples are not recommended for use in the isolation of Arcobacter. MLST showed that a large amount of diversity exists among Arcobacter isolates from cattle.
Acknowledgments
This work was funded by the Department for Environment, Food and Rural Affairs Veterinary Training and Research Initiative program.
We thank Dai Grove-White for help with sample collection and Thelma Roscoe, Trish Houghton, and Jo Sutherst for their valuable help with processing samples.
Footnotes
Published ahead of print on 30 December 2010.
REFERENCES
- 1.Atabay, H. I., and J. E. Corry. 1998. Evaluation of a new Arcobacter enrichment medium and comparison with two media developed for enrichment of Campylobacter spp. Int. J. Food Microbiol. 41:53-58. [DOI] [PubMed] [Google Scholar]
- 2.Aydin, F., K. S. Gumussoy, H. I. Atabay, T. Ica, and S. Abay. 2007. Prevalence and distribution of Arcobacter species in various sources in Turkey and molecular analysis of isolated strains by ERIC-PCR. J. Appl. Microbiol. 103:27-35. [DOI] [PubMed] [Google Scholar]
- 3.Collado, L., I. Cleenwerck, S. Van Trappen, P. De Vos, and M. J. Figueras. 2009. Arcobacter mytili sp. nov., an indoxyl acetate-hydrolysis-negative bacterium isolated from mussels. Int. J. Syst. Evol. Microbiol. 59:1391-1396. [DOI] [PubMed] [Google Scholar]
- 4.de Boer, E., J. J. Tilburg, D. L. Woodward, H. Lior, and W. M. Johnson. 1996. A selective medium for the isolation of Arcobacter from meats. Lett. Appl. Microbiol. 23:64-66. [DOI] [PubMed] [Google Scholar]
- 5.De Smet, S., et al. 2010. Arcobacter trophiarum sp. nov. isolated from fattening pigs. Int. J. Syst. Evol. Microbiol., in press. [DOI] [PubMed]
- 6.Diergaardt, S. M., S. N. Venter, A. Spreeth, J. Theron, and V. S. Brozel. 2000. The occurrence of campylobacters in water sources in South Africa. Water Res. 39:2589-2595. [DOI] [PubMed] [Google Scholar]
- 7.Ellis, W. A., S. D. Neill, J. J. O'Brien, H. W. Ferguson, and J. Hanna. 1977. Isolation of Spirillum/Vibrio-like organisms from bovine fetuses. Vet. Rec. 100:451-452. [DOI] [PubMed] [Google Scholar]
- 8.Golla, S. C., et al. 2002. Determination of the occurrence of Arcobacter butzleri in beef and dairy cattle from Texas by various isolation methods. J. Food Prot. 65:1849-1853. [DOI] [PubMed] [Google Scholar]
- 9.Gonzalez, I., T. Garcia, A. Antolin, P. E. Hernandez, and R. Martin. 2000. Development of a combined PCR-culture technique for the rapid detection of Arcobacter spp. in chicken meat. Lett. Appl. Microbiol. 30:207-212. [DOI] [PubMed] [Google Scholar]
- 10.Grove-White, D. H., A. J. Leatherbarrow, P. J. Cripps, P. J. Diggle, and N. P. French. 2010. Temporal and farm management-associated varation in the faecal-pat prevalence of Campylobacter jejuni in ruminants. Epidemiol. Infect. 138:549-558. [DOI] [PubMed] [Google Scholar]
- 11.Houf, K., A. Tutenel, L. De Zutter, J. Van Hoof, and P. Vandamme. 2000. Development of a multiplex PCR assay for the simultaneous detection and identification of Arcobacter butzleri, Arcobacter cryaerophilus and Arcobacter skirrowii. FEMS Microbiol. Lett. 193:89-94. [DOI] [PubMed] [Google Scholar]
- 12.Houf, K., L. A. Devriese, L. De Zutter, J. Van Hoof, and P. Vandamme. 2001. Development of a new protocol for the isolation and quantification of Arcobacter species from poultry products. Int. J. Food Microbiol. 71:189-196. [DOI] [PubMed] [Google Scholar]
- 13.Houf, K., L. A. Devriese, L. De Zutter, J. Van Hoof, and P. Vandamme. 2001. Susceptibility of Arcobacter butzleri, Arcobacter cryaerophilus and Arcobacter skirrowii to antimicrobial agents used in selective media. J. Clin. Microbiol. 39:1654-1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Houf, K., et al. 2005. Arcobacter cibarius sp. nov., isolated from broiler carcasses. Int. J. Syst. Evol. Microbiol. 55:713-717. [DOI] [PubMed] [Google Scholar]
- 15.Houf, K., and R. Stephan. 2007. Isolation and characterization of the emerging foodborne pathogen arcobacter from human stool. J. Microbiol. Methods 68:408-413. [DOI] [PubMed] [Google Scholar]
- 16.Houf, K., S. De Smet, J. Bare, and S. Daminet. 2008. Dogs as carriers of the emerging pathogen Arcobacter. Vet. Microbiol. 130:208-213. [DOI] [PubMed] [Google Scholar]
- 17.Houf, K., et al. 2009. Arcobacter thereius sp. nov., isolated from pigs and ducks. Int. J. Syst. Evol. Microbiol. 59:2599-2604. [DOI] [PubMed] [Google Scholar]
- 18.Hsueh, P. R., et al. 1997. Bacteremia caused by Arcobacter cryaerophilus 1B. J. Clin. Microbiol. 35:489-491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnson, L. G., and E. A. Murano. 1999. Development of a new medium for the isolation of Arcobacter spp. J. Food Prot. 62:456-462. [DOI] [PubMed] [Google Scholar]
- 20.Kabeya, H., et al. 2003. Distribution of Arcobacter species among livestock in Japan. Vet. Microbiol. 93:153-158. [DOI] [PubMed] [Google Scholar]
- 21.Kabeya, H., et al. 2004. Prevalence of Arcobacter species in retail meats and antimicrobial susceptibility of the isolates in Japan. Int. J. Food Microbiol. 90:303-308. [DOI] [PubMed] [Google Scholar]
- 22.Karenlampi, R. I., T. P. Tolvanen, and M. L. Hanninen. 2004. Phylogenetic analysis and PCR-restriction fragment length polymorphism identification of Campylobacter species based on partial groEL gene sequences. J. Clin. Microbiol. 42:5731-5738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kemp, R., et al. 2005. Prevalence and genetic diversity of Campylobacter spp. in environmental water samples from a 100-square-kilometer predominantly dairy farming area. Appl. Environ. Microbiol. 71:1876-1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kiehlbauch, J. A., et al. 1991. Campylobacter butzleri sp. nov. isolated from humans and animals with diarrheal illness. J. Clin. Microbiol. 29:376-385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McFadden, A. M., C. Heuer, R. Jackson, D. M. West, and T. J. Parkinson. 2005. Investigation of bovine venereal campylobacteriosis in beef cow herds in New Zealand. N Z Vet. J. 53:45-52. [DOI] [PubMed] [Google Scholar]
- 26.Miller, W. G., et al. 2009. First multi-locus sequence typing scheme for Arcobacter spp. BMC Microbiol. 9:196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.On, S. L., A. Stacey, and J. Smyth. 1995. Isolation of Arcobacter butzleri from a neonate with bacteraemia. J. Infect. 31:225-227. [DOI] [PubMed] [Google Scholar]
- 28.Ongor, H., B. Cetinkaya, M. N. Acik, and H. I. Atabay. 2004. Investigation of arcobacters in meat and faecal samples of clinically healthy cattle in Turkey. Lett. Appl. Microbiol. 38:339-344. [DOI] [PubMed] [Google Scholar]
- 29.Prouzet-Mauleon, V., L. Labadi, N. Bouges, A. Menard, and F. Megraud. 2006. Arcobacter butzleri: underestimated enteropathogen. Emerg. Infect. Dis. 122:307-309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rivas, L., N. Fegan, and P. Vanderlinde. 2004. Isolation and characterisation of Arcobacter butzleri from meat. Int. J. Food Microbiol. 91:31-41. [DOI] [PubMed] [Google Scholar]
- 31.Samie, A., C. L. Obi, L. J. Barrett, S. M. Powell, and R. L. Guerrant. 2007. Prevalence of Campylobacter species, Helicobacter pylori and Arcobacter species in stool samples from the Venda region, Limpopo, South Africa: studies using molecular diagnostic methods. J. Infect. 54:558-566. [DOI] [PubMed] [Google Scholar]
- 32.Scullion, R., C. S. Harrington, and R. H. Madden. 2006. Prevalence of Arcobacter spp. in raw milk and retail raw meats in Northern Ireland. J. Food Prot. 69:1986-1990. [DOI] [PubMed] [Google Scholar]
- 33.Tee, W., R. Baird, M. Dyall-Smith, and B. Dwyer. 1988. Campylobacter cryaerophila isolated from a human. J. Clin. Microbiol. 26:2469-2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vandamme, P., et al. 1992. Outbreak of recurrent abdominal cramps associated with Arcobacter butzleri in an Italian school. J. Clin. Microbiol. 30:2335-2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vandenberg, O., et al. 2004. Arcobacter species in humans. Emerg. Infect. Dis. 10:1863-1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Van Driessche, E., K. Houf, K. Van Hoof, L. De Zutter, and P. Vandamme. 2003. Isolation of Arcobacter species from animal faeces. FEMS Microbiol. Lett. 229:243-248. [DOI] [PubMed] [Google Scholar]
- 37.Van Driessche, E., K. Houf, F. Vangroenweghe, L. De Zutter, and J. Van Hoof. 2005. Prevalence, enumeration and strain variation of Arcobacter species in the faeces of healthy cattle in Belgium. Vet. Microbiol. 105:149-154. [DOI] [PubMed] [Google Scholar]
- 38.Villarruel-Lopez, A., et al. 2003. Isolation of Arcobacter spp. from retail meats and cytotoxic effects of isolates against Vero cells. J. Food Prot. 66:1374-1378. [DOI] [PubMed] [Google Scholar]
- 39.Vindigni, S. M., et al. 2007. Prevalence of foodborne microorganisms in retail foods in Thailand. Foodborne Pathog. Dis. 4:208-215. [DOI] [PubMed] [Google Scholar]
- 40.Vytrasova, J., M. Pejchalova, K. Harsova, and S. Binova. 2003. Isolation of Arcobacter butzleri and A. cryaerophilus in samples of meats and from meat-processing plants by a culture technique and detection by PCR. Folia Microbiol. (Praha) 48:227-232. [DOI] [PubMed] [Google Scholar]
- 41.Wang, G., et al. 2002. Colony multiplex PCR assay for identification and differentiation of Campylobacter jejuni, C. coli, C. lari, C. upsaliensis and C. fetus subsp. fetus. J. Clin. Microbiol. 40:4744-4747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wesley, I. V., et al. 2000. Fecal shedding of Campylobacter and Arcobacter spp. in dairy cattle. Appl. Environ. Microbiol. 66:1994-2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Woo, P. C., K. T. Chong, K. Leung, T. Que, and K. Yuen. 2001. Identification of Arcobacter cryaerophilus isolated from a traffic accident victim with bacteremia by 16S ribosomal RNA gene sequencing. Diagn. Microbiol. Infect. Dis. 40:125-127. [DOI] [PubMed] [Google Scholar]