Abstract
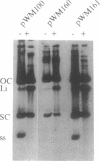
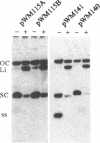
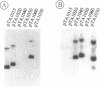
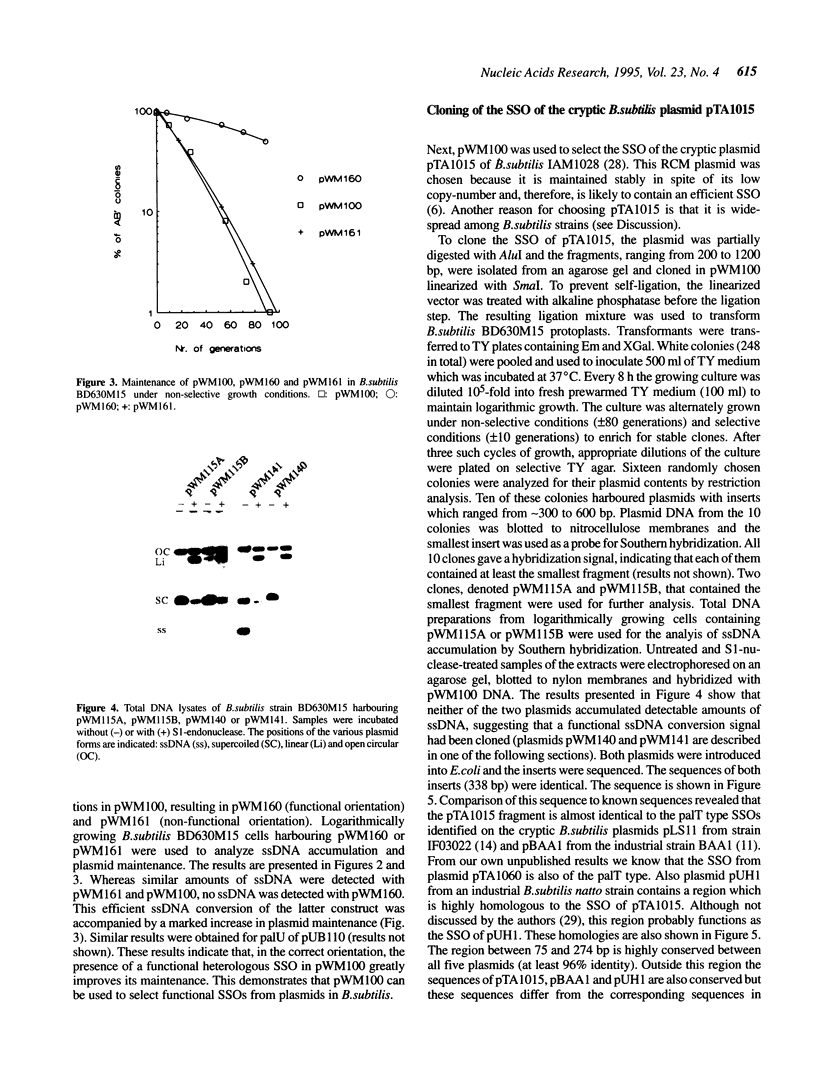
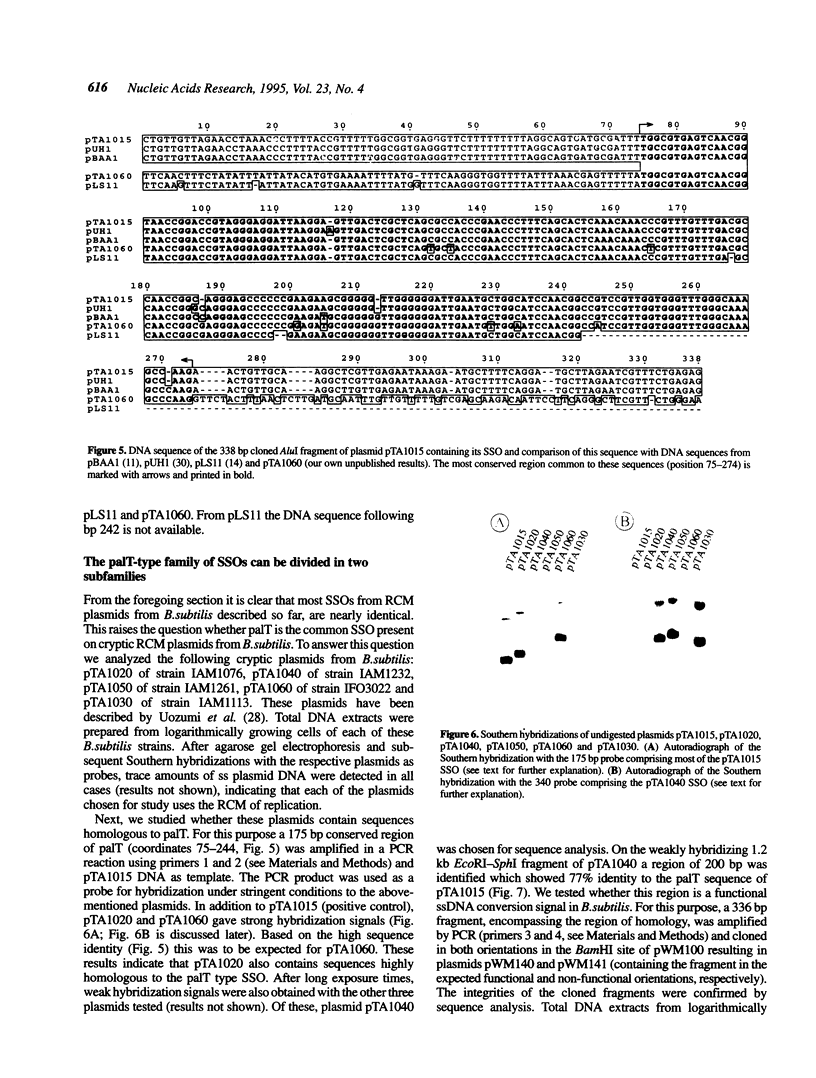
In this paper we describe the isolation and characterization of single strand origins (SSOs) of several cryptic Bacillus subtilis plasmids which use the rolling-circle mechanism of replication. The plasmids used in this study involved pTA1015, pTA1020, pTA1030, pTA1040, pTA1050 and pTA1060. The SSO of pTA1015 was isolated by shotgun cloning in a specially designed vector, pWM100, which has no SSO of its own. Sequence analysis revealed that the SSO of pTA1015 is almost identical to formerly described palT type SSOs. Also pTA1020 and pTA1060 were shown to contain SSOs highly homologous to palT. Using Southern hybridization with the palT of pTA1015 as a probe, the SSO of pTA1040 was cloned. Sequence analysis revealed a region of 200 bp which is 77% identical to the palT of pTA1015. The plasmids pTA1030 and pTA1050 contain an SSO which is highly homologous to the SSO of pTA1040. The majority of the SSOs of rolling-circle plasmids from B.subtilis seem to belong to two related families which we denote as palT1 (present on pTA1015, pTA1020 and pTA1060) and palT2 (present on pTA1030, pTA1040 and pTA1050). Both families of SSOs are highly efficient single-strand-conversion signals in B.subtilis.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Albano M., Breitling R., Dubnau D. A. Nucleotide sequence and genetic organization of the Bacillus subtilis comG operon. J Bacteriol. 1989 Oct;171(10):5386–5404. doi: 10.1128/jb.171.10.5386-5404.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boe L., Gros M. F., te Riele H., Ehrlich S. D., Gruss A. Replication origins of single-stranded-DNA plasmid pUB110. J Bacteriol. 1989 Jun;171(6):3366–3372. doi: 10.1128/jb.171.6.3366-3372.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bron S., Bosma P., van Belkum M., Luxen E. Stability function in the Bacillus subtilis plasmid pTA 1060. Plasmid. 1987 Jul;18(1):8–15. doi: 10.1016/0147-619x(87)90073-4. [DOI] [PubMed] [Google Scholar]
- Bron S., Holsappel S., Venema G., Peeters B. P. Plasmid deletion formation between short direct repeats in Bacillus subtilis is stimulated by single-stranded rolling-circle replication intermediates. Mol Gen Genet. 1991 Apr;226(1-2):88–96. doi: 10.1007/BF00273591. [DOI] [PubMed] [Google Scholar]
- Bron S., Luxen E., Swart P. Instability of recombinant pUB110 plasmids in Bacillus subtilis: plasmid-encoded stability function and effects of DNA inserts. Plasmid. 1988 May;19(3):231–241. doi: 10.1016/0147-619x(88)90041-8. [DOI] [PubMed] [Google Scholar]
- Burdett V. Identification of tetracycline-resistant R-plasmids in Streptococcus agalactiae (group B). Antimicrob Agents Chemother. 1980 Nov;18(5):753–760. doi: 10.1128/aac.18.5.753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang S., Chang S. Y., Gray O. Structural and genetic analyses of a par locus that regulates plasmid partition in Bacillus subtilis. J Bacteriol. 1987 Sep;169(9):3952–3962. doi: 10.1128/jb.169.9.3952-3962.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang S., Cohen S. N. High frequency transformation of Bacillus subtilis protoplasts by plasmid DNA. Mol Gen Genet. 1979 Jan 5;168(1):111–115. doi: 10.1007/BF00267940. [DOI] [PubMed] [Google Scholar]
- Devine K. M., Hogan S. T., Higgins D. G., McConnell D. J. Replication and segregational stability of Bacillus plasmid pBAA1. J Bacteriol. 1989 Feb;171(2):1166–1172. doi: 10.1128/jb.171.2.1166-1172.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruss A. D., Ross H. F., Novick R. P. Functional analysis of a palindromic sequence required for normal replication of several staphylococcal plasmids. Proc Natl Acad Sci U S A. 1987 Apr;84(8):2165–2169. doi: 10.1073/pnas.84.8.2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruss A., Ehrlich S. D. The family of highly interrelated single-stranded deoxyribonucleic acid plasmids. Microbiol Rev. 1989 Jun;53(2):231–241. doi: 10.1128/mr.53.2.231-241.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haima P., van Sinderen D., Bron S., Venema G. An improved beta-galactosidase alpha-complementation system for molecular cloning in Bacillus subtilis. Gene. 1990 Sep 1;93(1):41–47. doi: 10.1016/0378-1119(90)90133-c. [DOI] [PubMed] [Google Scholar]
- Hara T., Nagatomo S., Ogata S., Ueda S. The DNA sequence of gamma-glutamyltranspeptidase gene of Bacillus subtilis (natto) plasmid pUH1. Appl Microbiol Biotechnol. 1992 May;37(2):211–215. doi: 10.1007/BF00178173. [DOI] [PubMed] [Google Scholar]
- Ish-Horowicz D., Burke J. F. Rapid and efficient cosmid cloning. Nucleic Acids Res. 1981 Jul 10;9(13):2989–2998. doi: 10.1093/nar/9.13.2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leer R. J., van Luijk N., Posno M., Pouwels P. H. Structural and functional analysis of two cryptic plasmids from Lactobacillus pentosus MD353 and Lactobacillus plantarum ATCC 8014. Mol Gen Genet. 1992 Aug;234(2):265–274. doi: 10.1007/BF00283847. [DOI] [PubMed] [Google Scholar]
- Madsen S. M., Andrup L., Boe L. Fine mapping and DNA sequence of replication functions of Bacillus thuringiensis plasmid pTX14-3. Plasmid. 1993 Sep;30(2):119–130. doi: 10.1006/plas.1993.1039. [DOI] [PubMed] [Google Scholar]
- Novick R. P. Staphylococcal plasmids and their replication. Annu Rev Microbiol. 1989;43:537–565. doi: 10.1146/annurev.mi.43.100189.002541. [DOI] [PubMed] [Google Scholar]
- Seery L., Devine K. M. Analysis of features contributing to activity of the single-stranded origin of Bacillus plasmid pBAA1. J Bacteriol. 1993 Apr;175(7):1988–1994. doi: 10.1128/jb.175.7.1988-1994.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T., Koshikawa T. Isolation and characterization of four types of plasmids from Bacillus subtilis (natto). J Bacteriol. 1977 Aug;131(2):699–701. doi: 10.1128/jb.131.2.699-701.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T., Kuroda M., Sakaguchi K. Isolation and characterization of four plasmids from Bacillus subtilis. J Bacteriol. 1977 Mar;129(3):1487–1494. doi: 10.1128/jb.129.3.1487-1494.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uozumi T., Ozaki A., Beppu T., Arima K. New cryptic plasmid of Bacillus subtilis and restriction analysis of other plasmids found by general screening. J Bacteriol. 1980 Apr;142(1):315–318. doi: 10.1128/jb.142.1.315-318.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira J., Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982 Oct;19(3):259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- Viret J. F., Alonso J. C. A DNA sequence outside the pUB110 minimal replicon is required for normal replication in Bacillus subtilis. Nucleic Acids Res. 1988 May 25;16(10):4389–4406. doi: 10.1093/nar/16.10.4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Solar G. H., Puyet A., Espinosa M. Initiation signals for the conversion of single stranded to double stranded DNA forms in the streptococcal plasmid pLS1. Nucleic Acids Res. 1987 Jul 24;15(14):5561–5580. doi: 10.1093/nar/15.14.5561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Solar G., Kramer G., Ballester S., Espinosa M. Replication of the promiscuous plasmid pLS1: a region encompassing the minus origin of replication is associated with stable plasmid inheritance. Mol Gen Genet. 1993 Oct;241(1-2):97–105. doi: 10.1007/BF00280206. [DOI] [PubMed] [Google Scholar]
- van der Lelie D., Bron S., Venema G., Oskam L. Similarity of minus origins of replication and flanking open reading frames of plasmids pUB110, pTB913 and pMV158. Nucleic Acids Res. 1989 Sep 25;17(18):7283–7294. doi: 10.1093/nar/17.18.7283. [DOI] [PMC free article] [PubMed] [Google Scholar]