Abstract
The introduction and survival of zoonotic bacterial pathogens in poultry farming have been linked to bacterial association with free-living protozoa. To date, however, no information is available on the persistence of protozoan communities in these environments across consecutive rearing cycles and how it is affected by farm- and habitat-specific characteristics and management strategies. We therefore investigated the spatial and temporal dynamics of free-living protozoa in three habitats (pipeline, water, and miscellaneous samples) in three commercial poultry houses across three rearing cycles by using the molecular fingerprinting technique denaturing gradient gel electrophoresis (DGGE). Our study provides strong evidence for the long-term (ca. 6-month) persistence of protozoa in broiler houses across consecutive rearing cycles. Various free-living protozoa (flagellates, ciliates, and amoebae), including known vectors of bacterial pathogens, were observed during the down periods in between rearing cycles. In addition, multivariate analysis and variation partitioning showed that the protozoan community structure in the broiler houses showed almost no change across rearing cycles and remained highly habitat and farm specific. Unlike in natural environments, protozoan communities inside broiler houses are therefore not seasonal. Our results imply that currently used biosecurity measures (cleaning and disinfection) applied during the down periods are not effective against many protozoans and therefore cannot prevent potential cross-contamination of bacterial pathogens via free-living protozoa between rearing cycles.
Free-living bacterivorous protozoa are increasingly implicated in the survival and transmission of bacterial pathogens (31). Food-borne pathogens like Campylobacter and Salmonella, important agents of (gastro)enteritis often related to the consumption of contaminated chicken meat, may survive, multiply, and be transported in the environment through association with various protozoan organisms (5, 6, 8, 10, 15, 20, 23, 30, 33, 40, 41). Some bacteria resist digestion by protozoans through adaptation of the intraprotozoan environment by alternation of the maturation pathway of food vacuoles (11, 22, 25), while others may survive or grow saprophytically in the extraprotozoan environment upon materials released from protozoan cells (2). To date, knowledge of poultry colonization by zoonotic pathogens and their survival in poultry environments is still limited (26, 44), and the presence and persistence of potential vectors such as free-living protozoa in poultry environments have been largely unexplored. In a recent study, we described free-living protozoan diversity and spatial distribution in commercial poultry houses during a single rearing cycle (7). In the present study, the dynamics of protozoan communities across and within three consecutive rearing cycles (down and rearing periods), in relation to habitat- and farm-specific management strategies, were examined. Information on the persistence of protozoan communities across rearing cycles is crucial to evaluate their potential as a transmission route for human bacterial pathogens, like Campylobacter and Salmonella, between consecutive poultry flocks.
MATERIALS AND METHODS
Sampling strategy and collection.
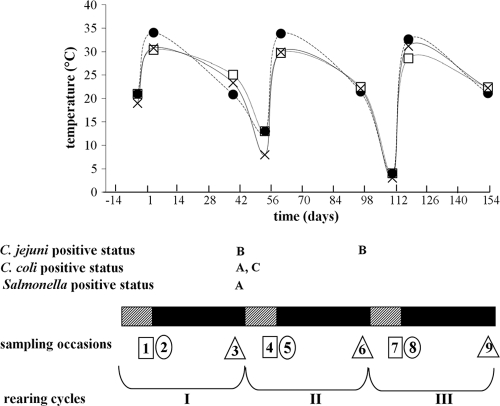
Three commercial poultry farms in Belgium (farms A, B, and C) were sampled on nine occasions between August 2006 and January 2007 (Fig. 1), encompassing the following three consecutive rearing cycles: I (sampling occasions 1 to 3), II (sampling occasions 4 to 6), and III (sampling occasions 7 to 9). Per sampling occasion, two broiler houses per farm were sampled. During each rearing cycle, samples were taken 1 to 4 days before the chickens arrived but after decontamination and disinfection (i.e., the down period; sampling occasions 1-4-7), 2 to 7 days after introduction of the chickens (sampling occasions 2-5-8), and between days 38 and 41 of the rearing period before slaughter of the chickens (sampling occasions 3-6-9). These will be referred to as the three different phases of each rearing cycle (Fig. 1). Air temperature inside the broiler houses varied between and within rearing cycles, being equal to the outside temperature during the down periods (from 21°C in the summer to 3°C in the winter) and maintained between 28.6 and 34.4°C and between 20.6 and 25.5°C when broilers were present during the second and third phases, respectively (Fig. 1).
FIG. 1.
Temperature and bacterial status of the broiler farms. Striped and black boxes represent the down periods (approximately 2 weeks) and rearing periods (6 weeks), respectively. Squares, circles, and triangles represent phases 1 (down period), 2 (beginning of rearing period), and 3 (end of rearing period), respectively; the numbers inside the symbols represent the sampling occasions (1 to 9). Letters represent the farms positive for C. jejuni, C. coli, or Salmonella species at the end of each rearing period. Temperature is expressed as the mean from two broiler houses per farm (•, farm A; □, farm B; ×, farm C).
All samples were taken with sterile equipment and collected in sterile flasks. Three different habitats were sampled inside each broiler house, as follows: pipeline, water, and miscellaneous habitats. The “pipeline” habitat consisted of 2- to 4-liter water samples from pipelines in the anterooms of the broiler houses before the water entered the rearing area of the chickens. The water at farms A and B came directly from a local well (60-m depth), while farm C used (chlorinated) main supply water. The “water” habitat represented 50-ml water samples (delivered via the pipelines) taken from drinking nipples (farm A) or cups (farms B and C) inside the rearing areas of the broiler houses. As the drinker systems were not yet connected to the water supply system during the down periods, this habitat could not be sampled during this phase. The “miscellaneous” habitat consisted of the following samples obtained from aerial habitats from the rearing areas of the broiler houses: dust, condensation water, films on the exterior surface of the water lines, and damp areas on walls and in litter. Farms A and B used commercial wood shavings, whereas farm C applied untreated straw as bedding material. Farm A stored the shavings in a shed, while farm B applied new packages when required. Upon entry in the laboratory (within 1 h after sampling), miscellaneous samples were diluted in 10 ml sterile demineralized water. All samples (pipeline, water, and miscellaneous) were then collected on 0.22-μm white GSWP filters (Millipore) using sterile equipment and stored at −20°C. Prior to molecular analyses, filters were pooled per habitat and per broiler house for each farm and sampling occasion, which resulted in a total of 140 pooled samples. Cecal droppings collected during the last phase of each rearing cycle were investigated for the presence of Campylobacter and Salmonella, according to the method of Rasschaert et al. (28). More detailed information on farm characteristics and management strategies can be found in the work of Baré et al. (7).
Molecular community profiling using denaturing gradient gel electrophoresis (DGGE).
DNA of the pooled samples was extracted using the bead-beating method, with phenol extraction and ethanol precipitation (36). Extracted DNA was purified on a Wizard column (Promega Corporation, Madison, WI), according to the manufacturer's recommendations. Given the polyphyletic nature of protozoa, the design of a universal set of protozoan-specific primers is impossible (35). Therefore, a universal set of eukaryote-specific 18S rRNA gene primers was used in order to capture as much eukaryotic (including protozoan) diversity as possible.
PCR amplification was carried out according to the study by van Hannen et al. (36), with the exception of performing 30 amplification cycles instead of 25. PCR amplification procedures were performed with a Bioman thermocycler (Westburg, Leusden, Netherlands). Each PCR mixture contained 1 to 4 μl of template DNA, 2 μl primers (12.5 pmol), 5 μl deoxynucleoside triphosphate (dNTP) (200 μM each; GeneAmp, Applied Biosystems Inc., CA), 2 μl bovine serum albumin (BSA) (400 ng; Roche Diagnostics GmbH, Germany), 5 μl of buffer I (Applied Biosystems), and 2.5 μl Taq DNA polymerase (2.5 U AmpliTaq DNA polymerase; Applied Biosystems) and was adjusted to a final volume of 50 μl with sterile water (Merck & Co., NJ). The presence of PCR products was determined by analyzing 5 μl of product on 1.66% agarose gels, staining it with ethidium bromide (Merck & Co.), and comparison with a molecular weight marker (SmartLadder; Eurogentec, Seraing, Belgium).
DGGE was performed with the D-Code system from Bio-Rad Laboratories (Hercules, CA), mainly as described by van Hannen et al. (36). Equal amounts of DNA (650 ng μl−1) were applied to a 28 to 57% gradient polyacrylamide gel (acrylamide/bisacrylamide ratio, 37.5:1; 100% denaturant corresponded to 7 M urea and 40% deionized formamide). Electrophoresis was performed for 990 min at 70 V; the temperature was set at 60°C. DGGE gels were stained with SYBR gold dye (Invitrogen, Paisley, United Kingdom), photographed, and processed by Quantity One 1-D analysis software (Bio-Rad Laboratories). Eukaryotic DNA obtained from previous studies performed in the laboratory was utilized to generate DGGE standards, for which three lanes were included per gel. The positions of the standards were applied to align the digitized DGGE images using BioNumerics 5.1 (Applied Maths, Sint-Martens-Latem, Belgium). Sequence information on the bands (see below) was used to check the grouping of bands into band classes or phylotypes. The number of different phylotypes within a (group of) sample(s) was defined as phylotype diversity. All data were combined in a matrix based on relative band intensities (i.e., the relative contribution of each band to the total band signal in the lane), which was then used for the data analyses.
Sequence information was obtained by sequencing DNA amplicons from purified excised DGGE bands. Sequencing was performed with the ABI Prism kit (PE Biosystems) using the 1427F primer (no GC clamp) (36) and an Applied Biosystems ABI 3130XL genetic analyzer. Bands were identified by screening the partial 18S rRNA gene sequence against GenBank sequences using BLAST (July 2009) (3). Protozoan phylotypes were classified by functional group (i.e., ciliate, flagellate, or amoeba) (13) and taxonomic group according the recent eukaryotic classification of Adl et al. (1).
Data analysis.
Principal components analysis (PCA; implemented using the program CANOCO 4.5 for Windows) (32), based on the covariance-variance matrix and with scaling focused on phylotype correlations, was used on log(x + 1)-transformed relative band intensities to assess spatial and temporal variations in phylotype composition of the samples. Four samples did not yield any protozoan DGGE bands and were removed from the analyses. The final data set thus consisted of 136 samples and 17 identified protozoan phylotypes. Variation in the relative abundances of the phylotypes was partitioned into spatially (farm and habitat) and temporally (rearing cycle and sampling phase within rearing cycles) structured components by performing partial regression analyses using redundancy analyses (RDA) in the CANOCO 4.5 for Windows program (9, 18, 39). This variation partitioning (VP) approach allowed separation of the pure effects of each component and their joint effects (39). The environmental matrix used in these analyses consisted of dummy variables for all four components (21). The forward selection procedure was used to select only those variables (per component) that contributed significantly to explaining the variation in the phylotype data using Monte Carlo permutation tests (4,999 permutations) (39). Both PCA and the VP analysis were first performed on the complete data set. As habitat appeared to be the dominant component structuring the protozoan communities (see Results), separate analyses of the data of each habitat were subsequently performed.
RESULTS
Bacterial status of the broiler flocks.
All farms were Campylobacter positive for at least one rearing period. Campylobacter jejuni was isolated from cecal droppings in farm B (broiler houses X1 and X2) on sampling occasions 3 and 6; Campylobacter coli was isolated from farm A (broiler houses X1 and X2) and farm C (broiler house X1) during the first rearing period. Salmonella-positive birds were found only in farm A (broiler house X1) on sampling occasion 3 (Fig. 1).
Free-living protozoan phylotype diversity.
A total of 62 eukaryotic phylotypes were distinguished. Most excised bands showed high similarities (>97%) to known sequences. In total, 38 unique sequences were recovered, belonging to the Amoebozoa (2 sequences), Chromalveolata (13), Excavata (2), Opisthokonta—fungi (9), Opisthokonta—Metazoa (1), Opisthokonta—others (1), Archaeplastida-Plantae (5), and Rhizaria (1) and unknown affiliations (4). Seventeen sequences were affiliated with free-living protozoa (6 ciliates, 8 flagellates, and 3 amoebae), with most identified free-living protozoa (64.7%) belonging to the Chromalveolata (Table 1). The most common protozoan phylotypes (present in more than half of the samples) had affinities with the Spumella-like flagellate JBM/S11, Colpoda spp., a Glissomonad sp., and Naegleria spp. Almost one-fourth of the protozoan phylotypes (23.5%) represented protozoan genera known to host pathogens, like Vannella, Platyamoeba, Cyclidium, and Naegleria (23, 29, 34).
TABLE 1.
BLAST matches of free-living protozoan sequences obtained from DGGE bands
| Phylotype no. | Abbreviation | Closest relative(s) | GenBank accession no. | % similarity (no. of matching bp/total no. of bp) | Group |
|
|---|---|---|---|---|---|---|
| Taxonomic | Functional | |||||
| 16 | UROGLEN | Uroglena sp. CCMP2768 | EF165132 | 95.9 (140/146) | Chromalveolata | Flagellate |
| 19 | LOXOPH1 | Loxophyllum sp. GD-070419 | EU242511 | 97.1 (136/140) | Chromalveolata | Ciliate |
| 20 | LOXOPH2 | Loxophyllum spp.a | 97.6 (122/125) | Chromalveolata | Ciliate | |
| 21 | VAN | Vannella sp. CAZ6/1 | AY929914 | 90.8 (157/173) | Amoebozoa | Amoeba |
| 22 | PLATY | Platyamoeba placida | AY294150 | 92.8 (141/152) | Amoebozoa | Amoeba |
| 23 | ENCHELYS | Enchelys polynucleata | DQ411861 | 99.3 (141/142) | Chromalveolata | Ciliate |
| 25 | COLP | Colpoda spp.a | 98.1 (158/161) | Chromalveolata | Ciliate | |
| 26 | PARAPHYS | Paraphysomonas vestita | Z28335 | 86.8 (132/152) | Chromalveolata | Flagellate |
| 27 | GLISSOM | Glissomonad sp. Panama 103 | EU709272 | 98.1 (152/155) | Rhizaria | Flagellate |
| 29 | SPUMEL | Spumella-like flagellate JBM/S11 | EF043285 | 100 (146/146) | Chromalveolata | Flagellate |
| 30 | VORTICEL | Vorticella spp.a | 98.6 (145/147) | Chromalveolata | Ciliate | |
| 32 | SPUM/PAR | Spumella-like flagellate JBAS37 | AY651094 | 99.3 (149/150) | Chromalveolata | Flagellate |
| Paraphysomonas vestita | AF109325 | 99.3 (149/150) | ||||
| 34 | CYCL GL | Cyclidium glaucoma | EU032356 | 98.7 (155/157) | Chromalveolata | Ciliate |
| 35 | NAEGLE | Naegleria spp.a | 99.4 (159/160) | Excavata | Amoeba | |
| 37 | SPHAERO | Sphaeroeca WS 3-Uni05 | AJ867611 | 87.2 (136/156) | Opisthokonta | Flagellate |
| 48 | STRAMEN | Stramenopile E2-85 | EU528042 | 97.3 (142/146) | Chromalveolata | Flagellate |
| 58 | RHYN NAS | Rhynchomonas nasuta HFCC348 | DQ46552 | 100 (166/166) | Excavata | Flagellate |
Several species within one genus all gave the same similarity results.
Samples possessed an average protozoan phylotype diversity of 5.4 ± 2.6 (minimum, 0; maximum, 12). The average phylotype diversity (across all farms) was lower in miscellaneous samples than in those from the other habitats, as follows: 4.7 ± 2.4 versus 5.8 ± 3.0 (pipeline samples) and 5.7 ± 2.2 (water samples). There also were differences between habitats and farms: the average phylotype diversity was highest in the pipeline samples from farm A (6.3 ± 1.0) and in the water (10.0 ± 3.9) and miscellaneous (6.1 ± 1.8) samples from farm C. The average phylotype diversity was lowest in the pipeline samples from farm C (3.7 ± 1.9) and in the water (5.2 ± 1.7) and miscellaneous (4.1 ± 2.8) samples from farm B.
Spatial distribution and temporal patterns in free-living protozoan communities.
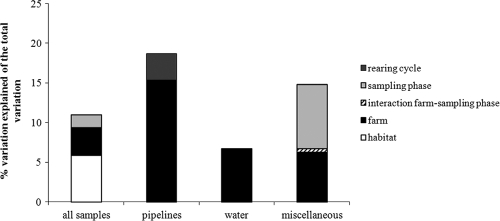
On the basis of preliminary ordinations, five outlier samples were identified and removed from the complete data set prior to PCA and variation partitioning (VP) of the data set. VP of the complete data set (131 samples and 17 protozoan phylotypes) showed that only 11% of the variation in the protozoan data can be significantly explained by the introduced dummy variables for habitat (ca. 6%), farm (3.5%), and sampling phase (1.6%) (see Fig. 3). No overlap in the variation explained by any of the four environmental components was found. Variation in the species data could not be significantly related to dummy variables for rearing cycles, indicating that no significant change in the protozoan community composition occurred from one rearing cycle to another.
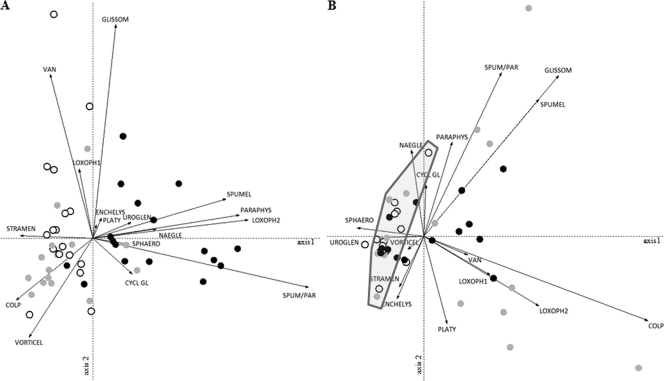
Because of the important effect of habitat on the variation in free-living protozoan communities and in order to assess whether significant changes in composition occurred between rearing cycles within the different habitat types, separate PCAs (Fig. 2) and VP analyses (Fig. 3) of the three habitat types were performed. The VP analyses showed that 18.6%, 6.7%, and 14.8% of the variation in the protozoan data obtained from the pipeline, water, and miscellaneous habitats, respectively, can be explained by the introduced variables. The farm from which samples were collected contributed importantly in all three habitat types; significant differences in community composition between the farms persisted throughout the whole sampling period. The sampling phase contributed to almost half of the explained variation in the miscellaneous habitat, suggesting that significant change in community composition occurred within the rearing cycles in this habitat but not in the other habitats (note, however, that the down period in the water habitat could never be sampled). In the pipeline habitat only, a slight (3.3%) but significant change between the rearing cycles was observed. The PCAs confirmed that pronounced differences existed between the farms, as shown for the pipeline and miscellaneous habitats in Fig. 2. The protozoan communities in the pipelines obtained from farms A (spread out along the positive side of the first axis in Fig. 2A) were different from those in the pipelines obtained from farms B and C (which both cluster more tightly on the negative side of the first axis in Fig. 2A). The communities obtained from farm A were characterized mainly by the presence of chromalveolate flagellates (Spumella-like and Paraphysomonas flagellates) and the ciliate Loxophyllum (LOXOPH2), and those obtained from farms B and C were characterized mainly by an unidentified chromalveolate flagellate (STRAMEN), the ciliates Vorticella (VORTICEL) and Colpoda (COLP), and the amoeba Vannella (VAN) (Fig. 2A). Variation in protozoan species composition in the miscellaneous habitat (Fig. 2B) was most pronounced in farm C and, to a lesser degree, in farm A, while the samples obtained from farm B were more similar to one another and clustered closely together on the negative side of the first axis. Samples on this side of the first axis were negatively characterized mainly by the absence of chromalveolate and rhizarian flagellates (except a Uroglena sp.) and the ciliates Loxophyllum (LOXOPH1 and -2) and Colpoda (COLP). All miscellaneous samples taken during sampling phase 3 in all three farms (Fig. 2B) clustered strongly together and are also characterized by the absence of many protozoans, indicating that during each rearing cycle, a significant change in community composition took place in this habitat between the down period and the other sampling phases. Spumella-like organisms and a Glissomonad sp. were found mainly in miscellaneous samples during the down periods (sampling phase 1) and sampling phase 2 (Fig. 2B).
FIG. 2.
PCA correlation biplots (axes 1 and 2) showing samples (symbols) and free-living protozoans (arrows) (labels are shown in Table 1) from analyses of the pipeline (A) and miscellaneous (B) samples. Farms A, B, and C are represented by black, white, and gray circles, respectively. (B) The gray polygon encircles the samples taken during sampling phase 3 from the three farms.
FIG. 3.
Contribution of spatially (habitat, farm) and temporally (rearing cycle, sampling phase) structured variation to the total variation in free-living protozoan community composition for the whole data set (all samples) and for each habitat separately. Only significant variables were included in the analysis (see the text for more details).
Persistence of free-living protozoa.
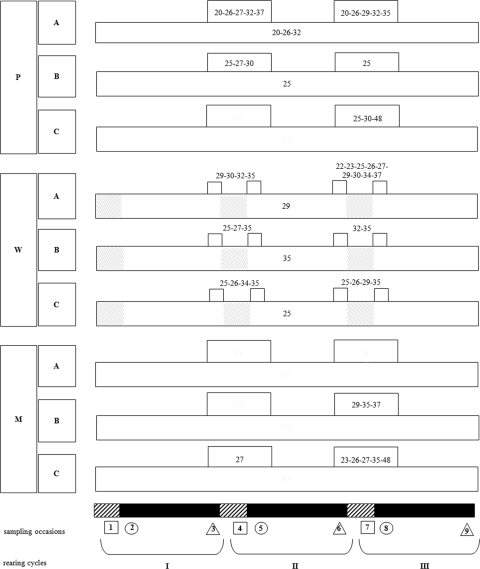
Protozoan phylotypes were defined as persistent if they persisted in the broiler house environment from one rearing cycle to another (Fig. 4). A small number of protozoans was detected on all consecutive sampling occasions in some farms and/or habitats. Loxophyllum spp., Paraphysomonas vestita, and Spumella-like flagellate JBAS37/Paraphysomonas vestita were found in all pipeline samples from farm A, while Colpoda was present in all pipeline samples from farm B. Spumella-like flagellate JBM/S11, Naegleria spp., and Colpoda spp. could be detected in all water samples from farms A, B, and C, respectively (Fig. 4). No phylotypes persisted during the whole sampling campaign in the miscellaneous habitat. Other protozoans persisted between at least two consecutive rearing cycles, i.e., they were detected in sampling phase 3 of one rearing cycle, during the down period (sampling phase 1), and in sampling phase 2 of the next cycle (note, however, that because the down period for the water habitat could never be sampled, protozoan persistence in this habitat pertains to a presence in sampling phase 3 of one cycle and sampling phase 2 of the next) (Fig. 4). Examples of these include a Glissomonad sp., Paraphysomonas vestita, and Naegleria spp. (all habitats); Spumella-like flagellate JBAS37/Paraphysomonas vestita and Colpoda spp. (pipeline and water); Loxophyllum spp. (pipeline); and Cyclidium glaucoma (water) (Fig. 4).
FIG. 4.
Persistence of protozoan phylotypes during the whole sampling campaign and across down periods. Pipeline, water, and miscellaneous habitats are represented by P, W, and M, respectively; A, B, and C refer to the three farms. Gray boxes indicate that the water habitat could not be sampled during the down periods (see Materials and Methods). Free-living protozoa are represented by their phylotype numbers (Table 1). Long, horizontal boxes represent phylotypes detected on all consecutive sampling occasions, while short, horizontal boxes contain phylotypes which persisted between two consecutive rearing cycles.
DISCUSSION
Transmission of pathogenic bacteria via protozoan vectors from one broiler flock to the next requires persistence of these vectors in between rearing cycles. This could be inside the rearing area of the broiler house or in the protozoan pool in the immediate surroundings (such as in the water supply, litter, or feed). Until today, no information about the occurrence, composition, and persistence of protozoan communities in various habitats (pipeline, water, and miscellaneous samples) across consecutive rearing cycles was available. Data obtained during the present study provide strong evidence for the long-term (ca. 6-month) persistence of free-living protozoa in broiler house environments across consecutive rearing cycles. Protozoan communities show almost no change across rearing cycles and remain highly habitat and farm specific.
Chromalveolata are the most frequently encountered protozoan group, which confirms published data (7). Most organisms belong to the Chrysophyceae (especially flagellates related to Spumella) or are ciliates (e.g., Colpoda and Cyclidium). Many small flagellates (such as Spumella and the cercozoan Glissomonadida) are common bacterivores in soil and freshwater environments (12, 19, 42, 43). Through the formation of a gelatinous matrix that enhances bacterial growth (42), certain Spumella species are especially apt at colonizing environments where food is initially scarce. It is also worth noting that the amoeba Naegleria is one of the dominant organisms in all three habitats, as this genus comprises several human (Naegleria fowleri) and animal (e.g., Naegleria australiensis and Naegleria italica) pathogens which can cause central nervous system infections (37). Further, Naegleria is known to harbor bacterial pathogens (34). The diversity estimates in the present study are almost certainly an underestimate of true protozoan diversity in these environments, due to the inability to sequence all phylotypes and to potential DNA extraction and/or PCR biases (16, 38).
The pipeline, water, and miscellaneous habitats harbor more or less distinct protozoan communities. There is considerable overlap in community composition between the water samples and samples from the other habitat types, because water communities are affected by both the water supply from the anteroom and the aerial habitats inside the broiler houses. The present study shows that this overriding effect of habitat on the protozoan community composition is maintained across consecutive rearing cycles. In addition, the different farms are also characterized by specific communities, and those differences persist across rearing cycles as well. Variation between farms is most pronounced in the pipeline communities. This is probably due to the fact that different water sources are used, from the chlorinated main supply water in farm C to local groundwater in farms A and B. In the miscellaneous habitat, the high variability in the samples obtained from farm C may be related to the fact that untreated straw from the field was used as bedding material rather than commercial wood shavings, as used in farms A and B.
Within a specific habitat and farm, there are no pronounced changes in protozoan community structure with time. Significant temporal change observed in the miscellaneous habitat is uniquely related to change within and not between rearing cycles. Significant (albeit small) temporal variation between rearing cycles was observed only in the pipeline habitat, which may be related to external, possibly seasonal changes in the water supply communities. The within-rearing-cycle change in the miscellaneous habitat is due mainly to an overall decrease in diversity toward the end of each cycle (sampling phase 3). This appears to be counterintuitive, as one would expect higher diversity after early colonization stages. This decrease, however, may be due mainly to an increase in the diversity of nonprotozoan eukaryotes (especially Fungi) (data not shown). A high abundance of these organisms may interfere with the amplification of (rarer) protozoan DNA, resulting in lower apparent protozoan diversity.
The present study, for the first time, provides a strong indication that commercial poultry farms are inhabited by farm-specific (endemic) protozoan communities which persist in the broiler house environment half year round. This indication is based on the facts that (i) protozoa were recovered from samples taken during the down periods between rearing cycles and that (ii) unlike natural communities (4, 24), virtually no change in the protozoan community composition between consecutive rearing cycles is observed. If the protozoa in the broiler houses would only be newly recruited from the outside environment, a seasonal signal (and hence more variation between rearing cycles) in the data would be expected. Most of the protozoan phylotypes detected during the down periods (e.g., Spumella-like flagellate, Paraphysomonas vestita, a Glissomonad sp., Vorticella spp., Naegleria spp., Platyamoeba placida, and Colpoda spp.) belong to groups which are capable of forming cysts under unfavorable conditions (14, 19, 27, 42) and as such might survive the cleaning and disinfection procedures applied during the down periods.
Several persistent protozoa, like Platyamoeba placida, Cyclidium species, and Naegleria species, are known vectors or reservoirs for bacterial pathogens (23, 34), but for most protozoa detected during this study, this information is still lacking. In vitro experiments (under simulated broiler house conditions) (8) recently revealed that Campylobacter jejuni was able to survive for up to 14 days (the advisable duration of a down period) (17) in association with Acanthamoeba castellanii, a genus previously observed in the broiler houses (7) but not without this organism. The presence of free-living protozoa can as such in theory enable the survival of pathogenic bacteria in between rearing periods. The logical next step will be to demonstrate the presence of Campylobacter or other pathogens inside persistent protozoans (and/or their cysts) in broiler house samples taken during the down periods. We tested several methods to detect the presence of Campylobacter in protozoa in samples, but these were as yet not successful. First, fixatives used for fluorescent in situ hybridization (FISH) analyses distorted the protozoa (including their internal organelles) (J. Baré, unpublished data). Second, the antibiotic gentamicin, which is utilized in studies of protozoa-bacteria associations (to make sure only internalized bacteria are detected) (20, 33), was not effective against the Campylobacter strains used but did affect the protozoon (Acanthamoeba castellanii) (8). In order to unequivocally establish internalization of Campylobacter in protozoa obtained from natural samples, an effective protocol for extracting the protozoa from the samples and killing free-living or externally associated bacteria should be designed.
The results obtained indicate that the biosecurity measures and cleaning and disinfection procedures used are not effective against (potential pathogenic) free-living protozoa, as organisms persisted during the down periods. If protozoa are involved in the transfer of pathogenic bacteria from one broiler flock to the next, the currently used approaches cannot prevent bacterial cross-contamination between rearing cycles via protozoa. Therefore, cleaning and disinfection procedures should be thoroughly evaluated for their effectiveness against protozoan trophozoites, cysts, and associated bacterial pathogens. The fact that even the main supply water used in farm C contained protozoa suggests that standard water chlorination procedures are not effective against all free-living protozoa present. Improvement of biosecurity measures, like for instance the avoidance of the use of natural straw as bedding material or the use of a hygiene barrier at the entrance of the broiler houses, should further reduce the entrance of organisms.
Acknowledgments
This work was supported by a Ph.D. grant from the Institute for the Promotion of Innovation through Science and Technology in Flanders (grant IWT-SB/53141; IWT Vlaanderen, Brussels, Belgium) to J.B. and by the Research Fund of Ghent University (grant O1J18206; BOF, Ghent, Belgium).
We thank Andy Vierstraete for his help with sequencing. Many thanks to the farmers for their valuable cooperation during this study and to three anonymous reviewers for their valuable comments on the manuscript.
Footnotes
Published ahead of print on 14 January 2011.
REFERENCES
- 1.Adl, S. M., et al. 2005. The new higher level classification of eukaryotes with emphasis on the taxonomy of protists. J. Eukaryot. Microbiol. 52:399-451. [DOI] [PubMed] [Google Scholar]
- 2.Akya, A., A. Pointon, and C. Thomas. 2009. Viability of Listeria monocytogenes in co-culture with Acanthamoeba spp. FEMS Microbiol. Ecol. 70:20-29. [DOI] [PubMed] [Google Scholar]
- 3.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 4.Andrushchyshyn, O. P., K. P. Wilson, and D. D. Williams. 2007. Ciliate communities in shallow groundwater: seasonal and spatial characteristics. Freshwater Biol. 52:1745-1761. [Google Scholar]
- 5.Axelsson-Olsson, D., J. Waldenström, T. Broman, B. Olsen, and M. Holmberg. 2005. Protozoan Acanthamoeba polyphaga as a potential reservoir for Campylobacter jejuni. Appl. Environ. Microbiol. 71:987-992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Axelsson-Olsson, D., et al. 2010. Amoebae and algae can prolong the survival of Campylobacter species in co-culture. Exp. Parasitol. 126:59-64. [DOI] [PubMed] [Google Scholar]
- 7.Baré, J., et al. 2009. Diversity and habitat specificity of free-living protozoa in commercial poultry houses. Appl. Environ. Microbiol. 75:1417-1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baré, J., et al. 2010. Influence of temperature, oxygen and bacterial strain identity on the association of Campylobacter jejuni with Acanthamoeba castellanii. FEMS Microbiol. Ecol. 74:371-381. [DOI] [PubMed] [Google Scholar]
- 9.Borcard, D., P. Legendre, and P. Rapeau. 1992. Partialling out the spatial component of ecological variation. Ecology 73:1045-1055. [Google Scholar]
- 10.Brandl, M. T., B. M. Rosenthal, A. F. Haxo, and S. G. Berk. 2005. Enhanced survival of Salmonella enterica in vesicles released by a soilborne Tetrahymena species. Appl. Environ. Microbiol. 71:1562-1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cirillo, J. D., S. Falkow, L. S. Tompkins, and L. E. Bermudez. 1997. Interaction of Mycobacterium avium with environmental amoebae enhances virulence. Infect. Immun. 65:3759-3767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fenchel, T. 1987. Ecology of protozoa. The biology of free-living phagotrophic protists. Science Tech Publishers, Inc., Berlin, Germany.
- 13.Finlay, B. J., et al. 2000. Estimating the growth potential of the soil protozoan community. Protist 151:69-80. [DOI] [PubMed] [Google Scholar]
- 14.Foissner, W., H. Blatterer, H. Berger, and F. Kohmann. 1991. Taxonomische und ökologische Revision der Ciliaten des Saprobiensystems. Band I: Cyrthophorida, Oligotrichida, Hypotrichida, Colpodea. Bayerisches Landesamt für Wasserwirtschaft, Munich, Germany.
- 15.Gaze, W. 2003. Interactions between Salmonella typhimurium and Acanthamoeba polyphaga, and observation of a new method of intracellular growth within food vacuoles. Microb. Ecol. 36:358-369. [DOI] [PubMed] [Google Scholar]
- 16.Goldschmidt, P., et al. 2008. Resistance of Acanthamoeba to classic DNA extraction methods used for the diagnosis of corneal infections. Br. J. Ophthalmol. 92:112-115. [DOI] [PubMed] [Google Scholar]
- 17.Hald, B., A. Wedderkopp, and M. Madsen. 2000. Thermophilic Campylobacter spp. in Danish broiler production: a cross-sectional survey and a retrospective analysis of risk factors for occurrence in broiler flocks. Avian Pathol. 29:123-131. [DOI] [PubMed] [Google Scholar]
- 18.Heikkinen, R. K., M. Luoto, M. Kuussaari, and J. Poyry. 2005. New insights into butterfly-environment relationships using partitioning methods. Proc. R. Soc. B Biol. Sci. 272:2203-2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Howe, A. T., D. Bass, K. Vickerman, E. E. Chao, and T. Cavalier-Smith. 2009. Phylogeny, taxonomy, and astounding genetic diversity of Glissomonadida ord. nov., the dominant gliding zooflagellates in soil (Protozoa: Cercozoa). Protist 160:159-189. [DOI] [PubMed] [Google Scholar]
- 20.Huws, S. A., R. J. Morley, M. V. Jones, M. R. Brown, and A. W. Smith. 2008. Interactions of some common pathogenic bacteria with Acanthamoeba polyphaga. FEMS Microbiol. Lett. 282:258-265. [DOI] [PubMed] [Google Scholar]
- 21.Jongman, R. H. G., C. J. F. ter Braak, and O. F. R. Van Tongeren. 1995. Data analysis in community and landscape ecology. Cambridge University Press, Cambridge, United Kingdom.
- 22.Jules, M., and C. Buchrieser. 2007. Legionella pneumophila adaptation to intracellular life and the host response: clues from genomics and transcriptomics. FEBS Lett. 581:2829-2838. [DOI] [PubMed] [Google Scholar]
- 23.King, C. H., E. B. Shotts, Jr., R. E. Wooley, and K. G. Porter. 1988. Survival of coliforms and bacterial pathogens within protozoa during chlorination. Appl. Environ. Microbiol. 54:3023-3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kiss, A. K., E. Acs, K. T. Kiss, and J. K. Torok. 2009. Structure and seasonal dynamics of the protozoan community (heterotrophic flagellates, ciliates, amoeboid protozoa) in the plankton of a large river (River Danube, Hungary). Eur. J. Protistol. 45:121-138. [DOI] [PubMed] [Google Scholar]
- 25.Lamothe, J., S. Thyssen, and M. A. Valvano. 2004. Burkholderia cepacia complex isolates survive intracellularly without replication within acidic vacuoles of Acanthamoeba polyphaga. Cell. Microbiol. 6:1127-1138. [DOI] [PubMed] [Google Scholar]
- 26.Newell, D. G., and C. Fearnley. 2003. Sources of Campylobacter colonization in broiler chickens. Appl. Environ. Microbiol. 69:4343-4351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Page, F. C. 1988. A new key to freshwater and soil Gymnamoebae. Freshwater Biological Association, Ambleside, United Kingdom.
- 28.Rasschaert, G., K. Houf, J. Van Hende, and L. De Zutter. 2007. Investigation of the concurrent colonization with Campylobacter and Salmonella in poultry flocks and assessment of the sampling site for status determination at slaughter. Vet. Microbiol. 123:104-109. [DOI] [PubMed] [Google Scholar]
- 29.Scheid, P. 2007. Mechanism of intrusion of a microspordian-like organism into the nucleus of host amoebae (Vannella sp.) isolated from a keratitis patient. Parasitol. Res. 101:1097-1102. [DOI] [PubMed] [Google Scholar]
- 30.Snelling, W. J., J. P. McKenna, D. M. Lecky, and J. S. Dooley. 2005. Survival of Campylobacter jejuni in waterborne protozoa. Appl. Environ. Microbiol. 71:5560-5571. (Author's correction, 71:7631.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Snelling, W. J., J. E. Moore, J. P. McKenna, D. M. Lecky, and J. S. Dooley. 2006. Bacterial-protozoa interactions; an update on the role these phenomena play towards human illness. Microbes Infect. 8:578-587. [DOI] [PubMed] [Google Scholar]
- 32.ter Braak, C. J. F., and P. Smilauer. 1998. Canoco reference manual and user's guide to Canoco for Windows: software for canonical community ordination (version 4). Microcomputer Power, Ithaca, NY.
- 33.Tezcan-Merdol, D., et al. 2004. Uptake and replication of Salmonella enterica in Acanthamoeba rhysodes. Appl. Environ. Microbiol. 70:3706-3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thomas, V., G. McDonnel, S. P. Denyer, and J.-Y. Maillard. 2010. Free-living amoebae and their intracellular pathogenic microorganisms: risk for water quality. FEMS Microbiol. Rev. 34:231-259. [DOI] [PubMed] [Google Scholar]
- 35.Valster, R. M., B. A. Wullings, G. Bakker, H. Smidt, and D. van der Kooij. 2009. Free-living protozoa in two unchlorinated drinking water supplies, identified by phylogenic analysis of 18S rRNA gene sequences. Appl. Environ. Microbiol. 75:4736-4746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van Hannen, E. J., M. P. van Agterveld, H. J. Gons, and H. J. Laanbroek. 1998. Revealing genetic diversity of eukaryotic microorganisms in aquatic environments by denaturing gradient gel electrophoresis. J. Phycol. 34:206-213. [Google Scholar]
- 37.Visvesvara, G. S., H. Moura, and F. L. Schuster. 2007. Pathogenic and opportunistic free-living amoebae: Acanthamoeba spp., Balamuthia mandrillaris, Naegleria fowleri, and Sappinia diploidea. FEMS Immunol. Med. Microbiol. 50:1-26. [DOI] [PubMed] [Google Scholar]
- 38.von Wintzingerode, F., U. B. Göbel, and E. Stackebrandt. 1997. Determination of microbial diversity in environmental samples: pitfalls of PCR-based rRNA analysis. FEMS Microbiol. Rev. 21:213-229. [DOI] [PubMed] [Google Scholar]
- 39.Vyverman, W., et al. 2007. Historical processes constrain patterns in global diatom diversity. Ecology 88:1924-1931. [DOI] [PubMed] [Google Scholar]
- 40.Wildschutte, H., D. M. Wolfe, A. Tamewitz, and J. G. Lawrence. 2004. Protozoan predation, diversifying selection, and the evolution of antigenic diversity in Salmonella. Proc. Natl. Acad. Sci. U. S. A. 101:10644-10649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wildschutte, H., and J. G. Lawrence. 2007. Differential Salmonella survival against communities of intestinal amoebae. Microbiology 153:1781-1789. [DOI] [PubMed] [Google Scholar]
- 42.Yubuki, N., T. Nakayama, and I. Inouye. 2008. A unique life cycle and perennation in a colorless chrysophyte Spumella sp. J. Phycol. 44:164-172. [DOI] [PubMed] [Google Scholar]
- 43.Zwart, K. B., and J. F. Darbyshire. 1992. Growth and nitrogenous excretion of a common soil flagellate Spumella sp.—a laboratory experiment. J. Soil Sci. 43:145-157. [Google Scholar]
- 44.Zweifel, C., K. D. Scheu, M. Keel, F. Renggli, and R. Stephan. 2008. Occurrence and genotypes of Campylobacter in broiler flocks, other farm animals, and the environment during several rearing periods on selected poultry farms. Int. J. Food Microbiol. 125:182-187. [DOI] [PubMed] [Google Scholar]