Abstract
We report the biochemical characterization of a novel haloalkane dehalogenase, DatA, isolated from the plant pathogen Agrobacterium tumefaciens C58. DatA possesses a peculiar pair of halide-stabilizing residues, Asn-Tyr, which have not been reported to play this role in other known haloalkane dehalogenases. DatA has a number of other unique characteristics, including substrate-dependent and cooperative kinetics, a dimeric structure, and excellent enantioselectivity toward racemic mixtures of chiral brominated alkanes and esters.
Haloalkane dehalogenases (HLDs) (EC 3.8.1.5) are enzymes that catalyze the dehalogenation of alkyl halides, and they are found in a wide range of bacteria (3). They promote the hydrolysis of a broad range of halogenated compounds, cleaving the carbon-halogen bond to generate the corresponding alcohol, a halide, and a proton (6). A number of halogenated compounds are environmentally toxic industrial by-products, and it has been suggested that haloalkane dehalogenases may be useful catalysts for their biodegradation, with potential applications in bioremediation. In biocatalysis, there is a long-standing interest in these enzymes, particularly for the production of optically pure alcohols. Therefore, the identification of dehalogenating enzymes with appropriate selectivity patterns is very important in terms of their industrial utility (6-7, 18, 20-21).
Agrobacterium tumefaciens causes the plant disease crown gall by transferring a discrete set of genes located on the tumor-inducing (Ti) plasmid into the plant's cells. These genes are then integrated and expressed, resulting in the disease (23). The open reading frame atu6064 (AE009425), which we refer to as datA, is located on the Ti plasmid of Agrobacterium tumefaciens C58. A recent phylogenetic analysis suggested that the sequence of DatA from Agrobacterium tumefaciens C58 belongs to the HLD-II subfamily (sequence identities of 31% to 35%; see Table S1 in the supplemental material). HLDs of this subfamily have a characteristic catalytic pentad composed of Asp-His-Glu and Asn-Trp residues (3). Interestingly, sequence alignment revealed that DatA differs from all of the other known HLDs in that one of its halide-stabilizing residues is a tyrosine rather than the tryptophan observed in other members of the family. The halide-stabilizing residues play a key role in the binding of a halogenated substrate and in the stabilization of the transition state of carbon-halogen bond-breaking transition (6).
The start codon of datA has been predicted differently by the two groups who determined independently the entire genome sequence of C58 (see Fig. S1 in the supplemental material). Therefore, we predicted another start codon of datA on the basis of comparison with other HLDs and the existence of the most probable Shine-Dalgarno (SD) sequence upstream of our predicted start codon. An optimized synthetic gene coding for DatA with a hexahistidine tag at the C terminus was subcloned into the pAQN vector (15). DatA was overexpressed under the control of the tac promoter in Escherichia coli BL21(DE3) Arctic Express (Stratagene, La Jolla, CA) overnight at 12°C. The overexpression was induced by isopropyl-β-d-thiogalactopyranoside added to a final concentration 0.5 mM. The produced DatA was purified to homogeneity by Q-Sepharose ion-exchange chromatography (GE Healthcare, Waukesha, WI) and affinity chromatography with Ni-nitrilotriacetic acid resin (Qiagen, Hilden, Germany), with a yield of 30 mg of purified protein per liter of culture.
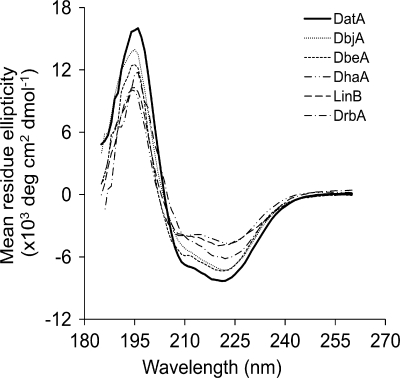
Circular dichroism spectroscopy in the far-UV region was used to assess the folding and stability of DatA. The spectrum of pure DatA contains two negative peaks at 208 and 222 nm and one positive peak at 195 nm (Fig. 1) and is similar to other characterized HLDs possessing an α/β-hydrolase fold (9-10, 16-17, 20). The differences between the depths of the minima for DatA are due to differences in the abundances of its helical structures. The thermal stability of DatA was measured by heating a solution of the protein from 22 to 80°C at rate of 1°C min−1 and monitoring the changes in its ellipticity at 222 nm. The melting temperature of DatA was found to be 48.3 ± 0.2°C, which is comparable to the melting temperatures of other HLDs (9-10, 16). 1,3-Dibromopropane was used to examine the effect of temperature and pH on the activity of DatA. The activity was accessed in the temperature range from 20 to 60°C, with the peak activity at 40°C, and in the range of pH from 5 to 11, with the maximal activity at pH 9.8. These values are similar to those observed with other HLDs (8-11, 14, 21).
FIG. 1.
Far-UV circular dichroism spectra of DatA and five different biochemically characterized HLDs: DbjA (13), DbeA (R. Chaloupkova, T. Prudnikova, P. Rezacova, Z. Prokop, T. Mozga, Y. Sato, M. Kuty, Y. Nagata, I. K. Smatanova, and J. Damborsky, unpublished data), DhaA (11), LinB (9), and DrbA (7). The protein concentration used for measurement was 0.2 mg ml−1.
The native structure of DatA was examined by gel filtration with a Superdex TM200 10/300 GL column (GE Healthcare, Waukesha, WI), using an elution buffer consisting of 50 mM Tris-HCl at pH 7.5 in the presence of 150 mM NaCl, and by native electrophoresis using the same buffer with or without 150 mM NaCl. DatA, with an estimated molecular mass of 34 kDa (see Fig. S3A in the supplemental material), exists as a monomer under high-NaCl salt conditions (see Fig. S2 and Fig. S3B) but is dimeric under low-salt conditions (see Fig. S3C). This suggests that ionic interactions are involved in the association of the protein, similar to the case with esterase (4), cyclomaltodextrinase (13), amidase (19), and formyltransferase (22).
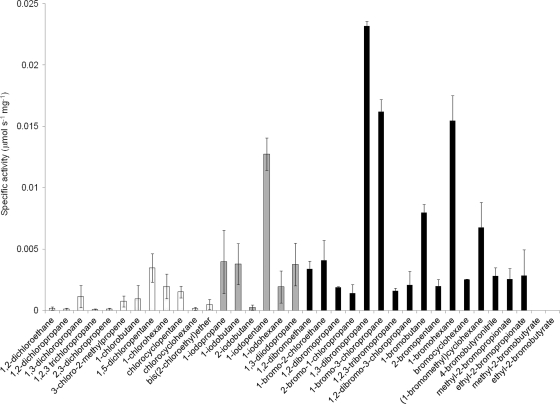
The specificity of DatA was studied using 36 different halogenated substrates. It was found to be highly active toward brominated and iodinated compounds but poorly active toward most of the chlorinated compounds. The highest activity (0.024 μmol s−1 mg−1 of protein) was observed with 1,3-dibromopropane (Fig. 2). Comparison of the functional properties of DatA with characterized HLDs indicated that DatA possesses substrate specificity similar to those of DmbC and DbeA; the activity levels with various halogenated substrates are similar to those of DmbC and DrbA (T. Koudelakova, E. Chovancova, J. Brezovsky, M. Monincova, A. Fortova, J. Jarkovsky, and J. Damborsky, unpublished data). Generally low activity of DatA and preference for brominated and iodinated substrates can be related to the Asn-Tyr pair of halide-stabilizing residues and their spatial arrangement with the catalytic triad (3). A lower stabilization efficiency of the Asn-Tyr pair may result in weakened binding of the substrate and less-efficient stabilization of the transition state (1, 12). DatA showed no activity with known substrates of epoxide hydrolases, which are evolutionarily closely related to HLDs and employ Tyr in the activation of the epoxide substrate.
FIG. 2.
Substrate specificity profile of DatA with the set of 36 chlorinated (in white), iodinated (in gray), and brominated (in black) substrates. Specific activity of DatA was measured using the Iwasaki method (5). The error bars indicate standard deviations from at least triplicate experiments.
A detailed kinetic analysis was performed with six selected substrates, consisting of brominated, chlorinated, and iodinated analogs of 1-halohexane and 1,3-dihalopropane. Kinetics of DatA with 1,3-dibromopropane (K0.5 [concentration of substrate at half-maximal velocity that binds at the given Hill coefficient {nH}] = 2.15 ± 0.15 mM; kcat = 1.47 ± 0.08 s−1) and 1-bromohexane (K0.5 = 0.14 ± 0.09 mM; kcat = 1.7 ± 0.1 s−1) follow single hyperbolic relationship with a Hill coefficient (nH value) equal to 1. Interestingly, sigmoidal kinetics were observed with 1,3-diiodopropane and 1-chlorohexane, suggesting that these species react with DatA via a mechanism involving cooperative substrate binding. In keeping with this suggestion, Hill coefficients (nH values) of 3.5 ± 1.6 and 1.27 ± 0.23, respectively, were observed with these two substrates. Other unusual kinetics of DatA are a combination of a cooperative mechanism with substrate inhibition observed in the case of 1-iodohexane, with a K0.5 value of 0.63 ± 0.25 and a substrate inhibition constant (Ksi) of 1.7 ± 0.1 mM. The Hill coefficients nH and mH for this substrate were 2.4 ± 1.0 and 11.6 ± 6.0, respectively (Table 1). A structural analysis of DatA is in progress to provide additional information on this interesting phenomenon. The kinetics of DatA with 1,3-dichloropropane could not be accurately measured. The reaction velocity of DatA toward this substrate increased linearly between 0 and 4 mM, indicating that the K0.5 value was significantly higher than the solubility limit (4 mM) of the substrate (Table 1). Taken together, these results indicate that the kinetic mechanism by which DatA achieves haloalkane hydrolysis is strongly substrate dependent.
TABLE 1.
| Substrate | K0.5 (mM) | kcat (s−1) | kcat/K0.5 (s−1·mM−1) | nHe | mHf | Ksi (mM) |
|---|---|---|---|---|---|---|
| 1,3-Dibromopropanec | 2.15 ± 0.15 | 1.47 ± 0.08 | 0.69 | 1.0 | —g | — |
| 1,3-Dichloropropanec | >4 | >0.008 | < 0.002 | 1.0 | — | — |
| 1,3-Diiodopropanec | 0.20 ± 0.03 | 1.2 ± 0.1 | 6.0 | 3.5 ± 1.6 | — | — |
| 1-Bromohexanec | 0.14 ± 0.09 | 1.7 ± 0.1 | 12.20 | 1.0 | — | — |
| 1-Chlorohexanec | 0.07 ± 0.02 | 0.039 ± 0.004 | 0.58 | 1.27 ± 0.23 | — | — |
| 1-Iodohexaned | 0.63 ± 0.25 | 0.26 ± 0.09 | 0.42 | 2.4 ± 1.0 | 11.6 ± 6.0 | 1.7 ± 0.1 |
All measurements were performed in 100 mM glycine buffer with pH 8.6 at 37°C.
Plus or minus standard error of the fit.
All parameters were calculated using the Hill equation: 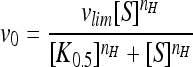 (where S is the substrate concentration and vlim is the limiting rate at the saturating substrate concentration).
(where S is the substrate concentration and vlim is the limiting rate at the saturating substrate concentration).
All parameters were calculated using the Hill equation with substrate inhibition: 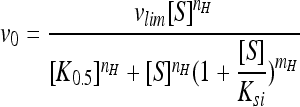 .
.
Hill coefficient.
Hill coefficient in inhibitory mode.
—, not applicable for the scheme used in a fit.
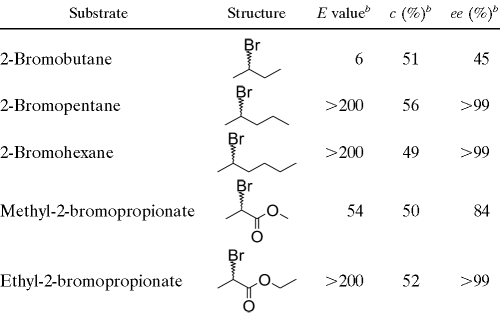
The enantioselectivity of DatA was assessed by determining the kinetic resolution of racemic brominated alkanes and esters. These data make it possible to quantify the ability of the dehalogenase to discriminate between two enantiomers present at equal concentrations (14-15). Based on kinetic resolution of racemic mixtures, DatA exhibited a high ability to discriminate between the R and S enantiomers of 2-bromopentane, 2-bromohexane, and ethyl 2-bromopropionate (E > 200 [E represents the enantiomeric ratio]). Low to medium selectivities were observed with 2-bromobutane (E = 6) and methyl 2-bromopropionate (E = 54), while no activity was detected with methyl and ethyl 2-bromobutyrate (Table 2). To the best of our knowledge, DatA is the most enantioselective of all the HLDs reported to date toward bromoalkane substrates and thus may be of considerable interest in biocatalysis (6, 18, 20).
TABLE 2.
Enantioselectivity of DatA with racemic β-bromoalkanes and α-bromoestersa
All measurements were performed in 50 mM Tris-sulfate buffer (pH 8.2) at room temperature.
b The enantiomeric ratio (E value) is a quantitative measure of enzyme stereospecificity and its relationship with substrate enantiomeric excess (ee) and degree of conversion (c) has been described previously (2).
Nucleotide sequence accession number.
The nucleotide sequence of the datA gene was deposited in the EMBL/GenBank/DDBJ database under accession number AB478945.
Supplementary Material
Acknowledgments
We thank Eva Chovancova (Masaryk University, Brno, Czech Republic) for preparation of Table S1 in the supplemental material.
This work was supported by The Ministry of Education, Youth, and Sports of the Czech Republic (grants LC06010 to T.K. and MSM0021622412 to Z.P.), by project CETOCOEN from the European Regional Development Fund (CZ.1.05/2.1.00/01.0001 to R.C.), by the Grant Agency of the Czech Academy of Sciences (IAA401630901 to J.D.), and by Grant-in-Aids from The Ministry of Education, Culture, Sports, Science, and Technology and The Ministry of Agriculture, Forestry, and Fisheries, Japan (to Y.N.).
Footnotes
Published ahead of print on 30 December 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Bohac, M., et al. 2002. Halide-stabilising residues of haloalkane dehalogenases studied by quantum mechanic calculations and site-directed mutagenesis. Biochemistry 41:14272-14280. [DOI] [PubMed] [Google Scholar]
- 2.Chen, C. S., Y. Fujimoto, G. Girdaukas, and C. J. Sih. 1982. Quantitative analyses of biochemical kinetic resolutions of enantiomers. J. Am. Chem. Soc. 104:7294-7299. [Google Scholar]
- 3.Chovancova, E., J. Kosinski, J. M. Bujnicki, and J. Damborsky. 2007. Phylogenetic analysis of haloalkane dehalogenases. Proteins 67:305-316. [DOI] [PubMed] [Google Scholar]
- 4.Ferrer, M., et al. 2005. Microbial enzymes mined from the Urania deep-sea hypersaline anoxic basin. Chem. Biol. 12:895-904. [DOI] [PubMed] [Google Scholar]
- 5.Iwasaki, I., S. Utsumi, and T. Ozawa. 1952. New colorimetric determination of chloride using mercuric thiocyanate and ferric ion. Bull. Chem. Soc. Jpn. 25:226. [Google Scholar]
- 6.Janssen, D. B. 2004. Evolving haloalkane dehalogenase. Curr. Opin. Chem. Biol. 8:150-159. [DOI] [PubMed] [Google Scholar]
- 7.Janssen, D. B. 2007. Biocatalysis by dehalogenating enzymes. Adv. Appl. Microbiol. 61:233-252. [DOI] [PubMed] [Google Scholar]
- 8.Jesenska, A., et al. 2002. Cloning and expression of the haloalkane dehalogenase gene dhmA from Mycobacterium avium N85 and preliminary characterization of DhmA. Appl. Environ. Microbiol. 68:3724-3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jesenska, A., et al. 2005. Cloning, biochemical properties, and distribution of mycobacterial haloalkane dehalogenases. Appl. Environ. Microbiol. 71:6736-6745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jesenska, A., et al. 2009. Biochemical characterization of haloalkane dehalogenase DrbA and DmbC, representative of a novel subfamily. Appl. Environ. Microbiol. 75:5157-5160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Keuning, S., D. B. Janssen, and B. Witholt. 1985. Purification and characterization of hydrolytic haloalkane dehalogenase from Xanthobacter autotrophicus GJ10. J. Bacteriol. 163:635-639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krooshof, G. H., et al. 1998. Kinetic analysis and X-ray structure of haloalkane dehalogenase with a modified halide-binding site. Biochemistry 37:15013-15023. [DOI] [PubMed] [Google Scholar]
- 13.Lee, H.-S., et al. 2006. Dissociation/association properties of a dodecameric cyclomaltodextrinase: effect of pH and salt concentration on the oligomeric state. FEBS J. 273:109-121. [DOI] [PubMed] [Google Scholar]
- 14.Nagata, Y., et al. 1997. Purification and characterization of haloalkane dehalogenase of a new substrate class from a γ-hexachlorohexane-degrading bacterium, Sphingomonas paucimobilis UT26. Appl. Environ. Microbiol. 63:3707-3710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nagata, Y., K. Hynkova, J. Damborsky, and M. Takagi. 1999. Construction and characterization of histidine-tagged haloalkane dehalogenase (LinB) of a new substrate class from a γ-hexachlorocyclohexane-degrading bacterium, Sphingomonas paucimobilis UT26. Protein Expr. Purif. 17:299-304. [DOI] [PubMed] [Google Scholar]
- 16.Nakamura, T., et al. 2006. Expression of glycosylated haloalkane dehalogenase LinB in Pichia pastoris. Protein Expr. Purif. 46:85-91. [DOI] [PubMed] [Google Scholar]
- 17.Pavlova, M., et al. 2009. Redesigning dehalogenase access tunnels as a strategy for degrading an anthropogenic substrate. Nat. Chem. Biol. 5:727-733. [DOI] [PubMed] [Google Scholar]
- 18.Pieters, R. J., L. J. H. Spelberg, R. M. Kellogg, and D. B. Janssen. 2001. The enantioselectivity of haloalkane dehalogenases. Tetrahedron Lett. 42:469-471. [Google Scholar]
- 19.Politi, L., et al. 2009. pH-, temperature- and ion-dependent oligomerization of Sulfolobus solfataricus recombinant amidase: a study with site-specific mutants. Archaea 2:221-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Prokop, Z., et al. 2010. Enantioselectivity of haloalkane dehalogenases and its modulation by surface loop engineering. Angew. Chem. Int. Ed. 49:6111-6115. [DOI] [PubMed] [Google Scholar]
- 21.Sato, Y., et al. 2005. Two rhizobial strains, Mesorhizobium loti MAFF303099 and Bradyrhizobium japonicum USDA110, encode haloalkane dehalogenases with novel structures and substrate specificity. Appl. Environ. Microbiol. 71:4372-4379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shima, S., R. K. Thauer, and U. Ermler. 2004. Hyperthermophilic and salt-dependent formyltransferase from Methanopyrus kandleri. Biochem. Soc. Trans. 32:269-272. [DOI] [PubMed] [Google Scholar]
- 23.Wood, D. W., et al. 2001. The genome of natural genetic engineer Agrobacterium tumefaciens C58. Science 294:2317-2323. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.