Abstract
Cellodextrins, the incomplete hydrolysis products from insoluble cellulose, are accessible as a carbon source to certain members of the human gut microbiota, such as Bifidobacterium breve UCC2003. Transcription of the cldEFGC gene cluster of B. breve UCC2003 was shown to be induced upon growth on cellodextrins, implicating this cluster in the metabolism of these sugars. Phenotypic analysis of a B. breve UCC2003::cldE insertion mutant confirmed that the cld gene cluster is exclusively required for cellodextrin utilization by this commensal. Moreover, our results suggest that transcription of the cld cluster is controlled by a LacI-type regulator encoded by cldR, located immediately upstream of cldE. Gel mobility shift assays using purified CldRHis (produced by the incorporation of a His12-encoding sequence into the 3′ end of the cldC gene) indicate that the cldEFGC promoter is subject to negative control by CldRHis, which binds to two inverted repeats. Analysis by high-performance anion-exchange chromatography with pulsed amperometric detection (HPAEC-PAD) of medium samples obtained during growth of B. breve UCC2003 on a mixture of cellodextrins revealed its ability to utilize cellobiose, cellotriose, cellotetraose, and cellopentaose, with cellotriose apparently representing the preferred substrate. The cldC gene of the cld operon of B. breve UCC2003 is, to the best of our knowledge, the first described bifidobacterial β-glucosidase exhibiting hydrolytic activity toward various cellodextrins.
One of the dominant bacterial groups in the human and mammalian intestinal microbiota is represented by members of the genus Bifidobacterium (47, 48), which are high-GC-content Gram-positive, non-spore-forming, “bifid”-shaped, anaerobic bacteria. Specific bifidobacterial strains have been implicated in promoting host health through one or more beneficial activities, which include prevention of diarrhea, reduction of cholesterol levels, symptom alleviation of inflammatory bowel disease or irritable bowel syndrome, immunomodulation, anticarcinogenicity, easing of lactose intolerance, improving mineral adsorption, and production of vitamins (12, 46). The mechanism(s) of probiotic action is largely unknown and thus merits further investigation of bifidobacteria, which includes research into its genomic content, genetics, biochemistry, and metabolism (52).
Bifidobacterial growth and/or metabolism in the human gastrointestinal tract can be selectively stimulated by various dietary compounds, particularly by so-called prebiotic carbohydrates (22, 35). A prebiotic has been defined as “a selectively fermented ingredient that allows specific changes, both in the composition and/or activity in the gastrointestinal microflora that confers benefits upon host well being and health” (35). According to this definition, the potential prebiotic component must fulfill the following criteria: nondigestible by the host, fermentation by the intestinal microbiota, and selective stimulation of growth and activity of beneficial intestinal bacteria. Recent publications have proposed that fructo-oligosaccharides (FOS), galacto-oligosaccharides (GOS), soybean-derived oligosaccharides (SOS) and xylo-oligosaccharides (XOS) possess properties that match the criteria mentioned above (for a review on this topic, see reference 11). In this context it is important to note that more than 8% of the identified genes on bifidobacterial genomes have been predicted to be involved in carbohydrate metabolism, thus being indicative of extensive carbohydrate-degrading abilities (41, 51). Our model microorganism Bifidobacterium breve UCC2003 has previously been shown to encode a β-fructofuranosidase involved in the partial degradation of FOS (38), an extracellular amylopullanase capable of hydrolyzing α-1,4- and α-1,6-glucosidic linkages in starch, amylose, amylopectin, pullulan, and glycogen (31, 37), an endogalactanase (GalA) responsible for the utilization of galactan (32), and two α-glucosidases that can hydrolyze panose, isomaltose, isomaltotriose, trehalose, and palatinose (34). In addition, a fructose phosphoenolpyruvate phosphotransferase system (PEP-PTS) (24) and a ribose utilization system (33) have been identified in B. breve UCC2003 and characterized.
Humans and other single-stomach animals are incapable of metabolizing cellulose (36). However, human-derived intestinal microorganisms, mainly Bacteroides spp., have been shown to ferment dietary fiber components, including cellulose and hemicellulose (10, 54). Enzymatic hydrolytic degradation of cellulose requires the action of a combination of extracellular endo- and exoglucanases and β-glucosidases (4). In addition, the degradation of cellulose may also be performed by 6-phospho-β-glucosidases (20). β-Glucosidases, including those encoded by noncellulolytic microorganisms, play an important role in the degradation of plant-derived oligosaccharides, such as cellodextrins, by converting them to glucose (4). Bifidobacterium adolescentis Int-57 and B. breve 203 were reported to produce intracellular β-glucosidases with hydrolytic activity toward cellobiose (5, 29, 30). In the current study we describe the identification of a novel cellodextrin utilization (cld) operon of B. breve UCC2003. Furthermore, we present the characterization of the cldC gene, whose protein product is responsible for the hydrolysis of β-1,4 bonds present in cellodextrins, while we also describe how a LacI-type regulator encoded by cldR acts as a likely transcriptional repressor of the cld operon.
MATERIALS AND METHODS
Bacterial strains, plasmids, media, and culture conditions.
Bacterial strains and plasmids used in this study are listed in Table S1 in the supplemental material. Bifidobacteria were cultivated in reinforced clostridium medium (RCM) (Oxoid, Hampshire, England) or modified de Man, Rogosa, and Sharpe (mMRS) medium (7), made from first principles, supplemented with 0.05% (wt/vol) l-cysteine-HCl (Sigma-Aldrich, Steinhein, Germany) and 1% (wt/vol) of a particular carbohydrate solution as the main carbon and energy source. Strains were grown under anaerobic conditions in a modular atmosphere controlled system (Davidson & Hardy Ltd., Dublin, Ireland). Escherichia coli strains were grown at 37°C in Luria-Bertani (LB) medium (40), and Lactococcus lactis strains were grown at 30°C in M17 medium supplemented with 0.5% glucose (GM17). Where appropriate, media were supplemented with 5 μg ml−1 chloramphenicol (Cm), 50 μg ml−1 kanamycin (Km), 100 μg ml−1 erythromycin (Em), or 100 μg ml−1 ampicillin (Amp) for plasmid maintenance.
Cellodextrin preparation.
Cellodextrins were prepared from microcrystalline cellulose (Avicel PH105; 20 μm) using the mixed-acid hydrolysis protocol essentially as described previously (57). Freeze-dried cellodextrin powder was dissolved in deionized water (0.25-g ml−1 final concentration). To remove salts and reduce the amount of glucose, this cellodextrin solution was dialyzed for 24 h against 1 liter of deionized water using cellulose tubing (molecular mass cutoff, 100 Da) (Spectra/Por; Spectrum Europe B.V., Netherlands). The quality and composition of the prepared cellodextrin mixture were analyzed using high-performance anion-exchange chromatography with pulsed amperometric detection (HPAEC-PAD) (see below). The final dialyzed solution was then used for growth experiments.
B. breve UCC2003 consumption of cellodextrin from mMRS.
A dialyzed cellodextrin solution was added to 3× concentrated mMRS medium (to obtain a final sugar concentration of 0.85%). B. breve UCC2003 growth in mMRS supplemented with cellodextrins and 0.05% (wt/vol) l-cysteine-HCl was monitored every hour for 9 h by measuring the optical density at 600 nm (OD600). In addition, HPAEC-PAD analysis (see below) of filtered, cell-free growth medium samples taken at the same time points was performed to analyze cellodextrin consumption.
Bioinformatics.
Sequence data were obtained from the genome annotation of the B. breve UCC2003 sequencing project (S. C. Leahy, M. O'Connell-Motherway, J. A. Moreno Muñoz, G. F. Fitzgerald, D. G. Higgins, and D. van Sinderen, unpublished results). Database searches were performed using the nonredundant sequence database accessible at the National Center for Biotechnology Information website (http://www.ncbi.nlm.nih.gov) with tBlastN, BlastX, and BlastP (1, 2). Sequence analysis was performed using DNASTAR; MapDraw and EditSeq were used for sequence analysis, and MegAlign was used to align multiple protein or DNA sequences (Madison, WI).
DNA manipulations.
Large-scale preparation of chromosomal DNA from Bifidobacterium spp. was performed as described previously (24). Plasmid DNA was obtained from L. lactis NZ9000 using the QIAprep Spin Plasmid Miniprep kit (Qiagen GmbH, Hilden, Germany) as described previously (34).
QRT-PCR.
Differential expression of genes was confirmed by real-time quantitative RT-PCR (QRT-PCR). De novo cDNAs were prepared as described previously (50). All primers were designed using the Universal ProbeLibrary Assay Design Center (Roche Applied Science) based on Primers 3 Plus (49). The resulting primer-probe pairs were tested for efficiency, and only sets yielding an efficiency of 2 using a log fold standard dilution curve evaluated using the Roche 480 software 1.5. were used for subsequent quantitative transcriptional experiments. Primer sequences are presented in Table S2 in the supplemental material. The rnpA gene (encoding RNase P) was used as a housekeeping gene with a presumed constitutive level of transcription to correct for variability in the initial amount of total RNA. Each amplification reaction mixture contained 1 μl of 6.7-fold-diluted cDNA, 10 μl of the 2× FastStart TaqMan Probe Master (Roche), 900 nM each primer, and 250 nM probe mix and was brought to a total volume of 20 μl by the addition of RNase-free water. All QRT-PCRs were performed in triplicate by means of a LightCycler 480 system (Roche) instrument using 384-well plates. Thermal cycling conditions were as recommended by the manufacturer (Roche). The 2−ΔΔCT method (19) was used to calculate relative changes in gene expression determined from real-time quantitative PCR experiments. Pooled cDNA from all samples was used as a reference calibrator for analysis of differential gene expression. Results were calculated from at least two independent RNA extractions. The QRT-PCR expression data, following 2−ΔΔCT, analysis were subjected to a Mann-Whitney t test to compare all groups using GraphPad Prism 4 software (GraphPad Software, CA). Data are represented by mean ± standard error of the mean (SEM). P values of <0.05 were considered significant. The statistical analysis was performed blind to the origin of the data.
Construction of a B. breve UCC2003::cldE insertion mutant.
The procedure by which the cldE insertion mutant was constructed was essentially as described previously (32), the specifics of which are outlined below. An internal 636-bp fragment of cldE (from base 163 to 799 of the 1,307-bp cldE coding region) was amplified by PCR using B. breve UCC2003 chromosomal DNA as a template and primer pair pOCldEFw and pOCldERv (see Table S2 in the supplemental material) and cloned into integration plasmid pORI19 (18) harboring the tetracycline resistance cassette, tetW, cloned from pAM5 (3). Potential mutants harboring pORI19-tet-cldE integrated into the cldE gene were confirmed by colony PCR using primer combinations tetWFw and tetWRv to detect the expected presence of the tetW gene and cldR-Fw (complement to the start of the cldR gene) and pORI19Fv to confirm the presumed chromosomal integration of the pORI19-tet-cldE in the cldE gene.
EMSA.
DNA fragments representing different portions of the cldE promoter region were prepared by PCR using IRD800-labeled primers pairs (MWG Biotech) (see Table S1 in the supplemental material). Electrophoretic mobility shift assays (EMSAs) were performed essentially as described previously (13). In all cases, the binding reactions were carried out in a final volume of 20 μl in the presence of poly[d(I-C)] in binding buffer (20 mM Tris-HCl [pH 8.0], 5 mM MgCl2, 0.5 mM dithiothreitol [DTT], 1 mM EDTA, 100 mM KCl, 10% glycerol). Various amounts of purified CldRHis (produced by the incorporation of a His12-encoding sequence into the 3′ end of the cldC gene) ranging from 0.08 nM to 0.01 μM and probe (0.1 pmol) were mixed on ice and subsequently incubated for 15 min at 37°C. Samples were loaded onto a 6% nondenaturing polyacrylamide (PAA) gel prepared in TAE buffer (40 mM Tris acetate [pH 8.0], 2 mM EDTA) and run in a 0.5× to 2.0× gradient of TAE at 100 V for 60 min in a mini-Protean II system (Bio-Rad Laboratories, Richmond, CA). Signals were detected using the Odyssey infrared imaging system (Li-Cor Biosciences UK Ltd., Cambridge, United Kingdom) and captured using the supplied software Odyssey V3.0.
Primer extension analysis.
Total RNA was isolated from B. breve UCC2003, which was grown in mMRS supplemented with 1% cellobiose or a cellodextrin mixture to early exponential phase, using the Macaloid method (17). RNA samples were treated with RNase-free DNase (Ambion). The 5′ ends of the RNA transcripts were determined by primer extension as previously described (50). The generated cDNA (0.5 μl) was mixed with 0.3 μl loading buffer (Li-Cor Blue stop solution), followed by separation on a 6.5% Li-Cor gel matrix KB Plus and comparison with the products of a sequencing reaction (employing the Thermo Sequenase primer cycle sequencing kit [Amersham]) using the same primer as that employed for the primer extension (see Table S2 in the supplemental material) and using a PCR product encompassing the cldE promoter region as a template. Signal detection was performed by means of a Li-Cor sequencing instrument (Li-Cor Biosciences).
Plasmid construction and overexpression of CldCHis and CldRHis.
The entire coding region of cldC (gene locus Bbr_0109) and cldR (gene locus Bbr_0105) was amplified by PCR with genomic DNA from B. breve UCC2003 serving as a template and primer pairs CldCFw and CldCRv for cldC and CldRFw and CldRRv for cldR (see Table S2 in the supplemental material). For CldC the primers allowed the incorporation of a His12-encoding sequence into the 3′ end of the cldC gene (here designated cldCHis). PCRs were performed using a PTC-200 Peltier thermal cycler (Bio-Sciences, Dublin, Ireland), Taq PCR master mix, and ProofStart DNA polymerase (Qiagen, GmbH, Hilden, Germany). The amplified 1.0-kb cldCHis-encompassing PCR fragment was restricted with SacI and XbaI and ligated into plasmid pNZ8150 (26) cut with the same enzymes, while the amplified cldR-encompassing PCR product was restricted with NcoI and BglII and ligated to similarly digested pQE60. The ligation mixture of the cldCHis-containing PCR fragment and pNZ8150 was introduced into L. lactis NZ9000 by electroporation, and resulting transformants were selected based on Cm resistance. The ligation mixture of the cldRHis-containing PCR fragment and pQE60 was introduced into E. coli XL1-Blue cells by electroporation, with subsequent transformant selection based on Amp and Tc resistance. The expected plasmid content of a number of transformants was verified by sequencing to ensure the genetic integrity of the cloned recombinant cldC and cldR genes. In each case one transformant was selected for further use and designated pNZcldC for CldC overexpression and pQE60CldR for CldR overexpression.
A 400-ml volume of GM17 was inoculated with 8 ml of an overnight culture of L. lactis NZ9000 cells harboring pNZcldC and incubated at 30°C. When the optical density at 600 nm had reached approximately 0.5, expression of the protein was induced by the addition of 800 μl of supernatant from an overnight culture of a nisin A-producing L. lactis NZ9700 culture (8). After 2 h of incubation, cells were harvested, resuspended in lysis buffer (50 mM NaH2PO4, 300 mM NaCl, 10 mM imidazole), and disrupted with glass beads in a Mini Bead Beater (Biospec Products, Bartlesville, OK). Cellular debris was removed by centrifugation. The CldCHis enzyme was purified from the crude cell extract using a nickel-nitrilotriacetic acid column (Qiagen GmbH) according to the manufacturer's instructions (QIAexpressionist 06/2003). Overexpression of CldRHis was achieved as described previously (34). Cell crude extract was subsequently analyzed for the presence of the overexpressed protein by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) as previously described (34).
Determination of hydrolytic activities of CldCHis.
Determination of the hydrolytic activity and substrate specificity of CldCHis was performed using a previously described method (15). Enzymatic assays were performed at 37°C in a total volume of 450 μl of 20 mM MOPS (morpholinepropanesulfonic acid) (pH 7.0) with a 25 mM concentration of a particular carbohydrate substrate. All reactions were initiated by the addition of crude cell extract containing CldCHis (50 μl) and terminated after 17 h of incubation by placing the reaction tubes at 100°C for 5 min followed by cooling on ice. Reaction products were analyzed by HPAEC-PAD or by high-performance thin-layer chromatography (HPTLC).
HPTLC and HPAEC-PAD analyses.
HPTLC analysis was carried out as described previously (34) with some modifications. Following sample spotting, the chromatogram was developed 4-fold using a butanol-acetic acid-water (5:4:1, vol/vol/vol) solvent system in a horizontal developing chamber. The plate was allowed to air dry in a fume hood and was then sprayed evenly with 20% (vol/vol) sulfuric acid in ethanol, air dried again, and finally heated at 120°C for 10 min to visualize the carbohydrate spots.
HPAEC-PAD analysis was performed using a Dionex ICS-3000 system equipped with a CarboPac PA-100 analytical exchange column (250 mm by 4 mm) and a pulsed electrochemical detector in the pulsed amperometric detection (PAD) mode as previously described (34) with the following modifications. The elution was performed at a constant flow rate of 1 ml min−1 at 30°C using the following linear gradient of sodium acetate in 100 mM NaOH: 0 to 5 min, 27.5 mM; 5 to 40 min, 0 to 275 mM; and 40 to 45 min, 275 mM. The injection volume was 10 μl. Reaction products were identified and quantified relative to standard carbohydrates.
Nucleotide sequence accession number.
The nucleotide sequence of the cld cluster has been deposited in the GenBank database under accession number GQ329065.
RESULTS
Growth of Bifidobacterium strains on cellobiose.
We were interested in the ability of bifidobacteria to grow on plant-derived carbohydrates. As part of a larger survey, we investigated if bifidobacteria are capable of growth on cellobiose, the smallest cellulose-derived cellodextrin. For this purpose, growth in modified Rogosa medium (mMRS) supplemented with glucose (as a positive control) or cellobiose was assessed for 36 different bifidobacterial strains, representing 11 bifidobacterial species, by measuring the OD600 following 24 h of anaerobic growth at 37°C (Fig. 1). All bifidobacterial strains grew well on glucose, reaching OD600 values in excess of 1. In contrast, just 4 out of the 36 bifidobacterial strains tested were able to reach an OD600 of higher than 1.0 when grown on cellobiose as the sole carbon source. All four of these strains belong to the B. breve species, including B. breve UCC2003. Eight strains were shown to grow rather poorly on cellobiose, with final OD600 values ranging from 0.3 to 0.7, while the remaining 24 bifidobacterial strains could not or could very poorly metabolize cellobiose (OD600 of <0.3). This indicates that only certain bifidobacterial strains (in particular B. breve strains) can selectively grow on cellobiose and that this sugar possibly represents a selective growth substrate for such strains.
FIG. 1.
Final optical density (OD600) values obtained following 24 h of growth of 36 different bifidobacterial strains in modified MRS containing 0.01 g ml−1 glucose or cellobiose as the sole carbon source. The results presented are mean values and SEMs obtained from three separate experiments.
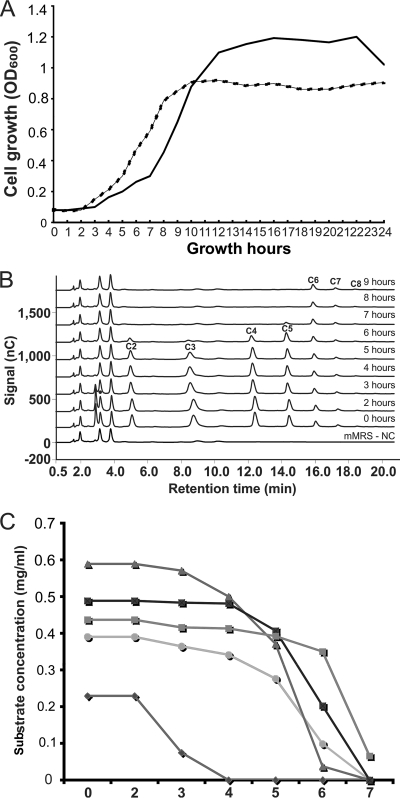
Consumption of cellodextrins from mMRS medium by B. breve UCC2003.
The results described above clearly showed that our model strain B. breve UCC2003 metabolizes cellobiose as its sole carbon source. To expand on this observation, we wanted to analyze the potential of B. breve UCC2003 to utilize higher-molecular-weight cellodextrins, i.e., cellotriose, cellotetraose, etc. For this purpose, a mixture of cellodextrins, obtained through partial acid hydrolysis of cellulose (see Materials and Methods), was included in mMRS as the main carbon source and the growth rate of B. breve UCC2003 was monitored as before, while HPAEC-PAD analysis was performed in order to determine the presence and quantity of each cellodextrin component in the culture supernatant at different time points (Fig. 2). The final OD600 of B. breve UCC2003 grown in mMRS with cellobiose was higher than that with cellodextrins, due to the higher concentration of the carbon source (1% cellobiose versus 0.85% cellodextrins). Nevertheless, this bacterium achieved similar growth rates in mMRS containing the cellodextrin mixture or cellobiose as a sole carbon source (Fig. 2A). It should be noted that the cellodextrin mixture did contain some residual glucose that was consumed by B. breve UCC2003 first before any cellodextrin metabolism was observed (Fig. 2B and C). The HPAEC-PAD analysis clearly demonstrates that B. breve UCC2003 metabolizes various different cellodextrins but does not utilize cellohexaose (C6), celloheptaose (C7), and probably cellodextrins with a higher degree of polymerization (Fig. 2B). Furthermore, from this analysis it appears that cellotriose (C3) represented the preferred substrate, as it was consumed faster than the other cellodextrins present in the growth medium (0.23 mg ml−1 h−1 at between 4 and 6 h of growth). Cellopentaose (C5) was the least preferred substrate, as it was consumed by B. breve UCC2003 at a relatively low rate, 0.03 mg ml−1 h−1, and only when the shorter cellodextrins were nearly exhausted from the medium (Fig. 2C). Cellobiose and cellotetraose were consumed at similar rates (0.12 and 0.14 mg ml−1 h−1, respectively).
FIG. 2.
(A) Growth profile of B. breve UCC2003 in mMRS containing 0.01 g ml−1 cellobiose (solid line) or 0.0085 g ml−1 cellodextrin mixture (broken line). (B) HPAEC-PAD chromatograms monitoring cellodextrin consumption by B. breve UCC2003 from mMRS medium, which was used as a negative-control sample (mMRS-NC). Other chromatograms are labeled with the culture growth time points. A black filled circle indicates the peak that corresponds to glucose. The C2 to C8 peaks indicate cellodextrin molecules with the corresponding degree of polymerization (DP) (ranging from cellobiose [DP = 2] up to cello-octaose [DP = 8]). (C) Cellodextrin consumption by B. breve UCC2003 from mMRS medium as represented by quantified amounts of individual cellodextrins present in the culture medium as calculated from HPAEC-PAD data (shown in panel B). Diamonds, glucose; circles, cellobiose; triangles, cellotriose; dark squares, cellotetraose; gray squares, cellopentaose.
Genome response of B. breve UCC2003 to growth on cellobiose and cellodextrins.
Preliminary microarray experiments had shown that transcription of the adjacent cldE, cldF, cldG, and cldC genes was upregulated when B. breve UCC2003 was grown in cellobiose compared when it was grown in glucose (data not shown). In order to further investigate these preliminary observations, QRT-PCR experiments were performed to measure the transcription levels of these genes when B. breve UCC2003 was grown on cellobiose or cellodextrins and to compare these to their transcription levels when grown on glucose. For this purpose, total RNA was isolated from B. breve UCC2003 cultures grown on cellobiose, cellodextrins, or glucose (see Materials and Methods). The cultures were harvested at the time points that ensured that B. breve UCC2003 was metabolizing cellobiose or cellodextrins (as opposed to the residual glucose present in the cellodextrin preparation). Analysis of the QRT-PCR data obtained from two independent biological replicates indeed clearly showed that the expression of the adjacent cldE, cldF, cldG, and cldC genes was significantly upregulated (fold change of >3.0; P < 0.001) in B. breve UCC2003 cultures grown on cellobiose or cellodextrins relative to cultures grown on glucose (Table 1). These results implicate the cldEFGC gene cluster in cellobiose/cellodextrin metabolism in B. breve UCC2003.
TABLE 1.
Effect of cellobiose or a cellodextrin mixture on the transcription of cld genes from B. breve UCC2003
| Locus tag (gene) | Putative function | Expression ratio quantified by QRT-PCRa |
|
|---|---|---|---|
| Cellobiose | Cellodextrins | ||
| Bbr_0106 (cldE) | Cellodextrin-binding protein | 69.94 | 104.58 |
| Bbr_0107 (cldF) | Cellodextrin transport system permease protein | 74.23 | 97.32 |
| Bbr_0108 (cldG) | Cellodextrin transport system permease protein | 90.56 | 110.54 |
| Bbr_0109 (cldC) | β-Glucosidase | 58.52 | 79.80 |
cDNA templates were derived from RNA samples of B. breve UCC2003 culture grown on cellobiose, cellodextrins, or glucose (as a comparator).
Genetic organization of the putative cellodextrin utilization operon.
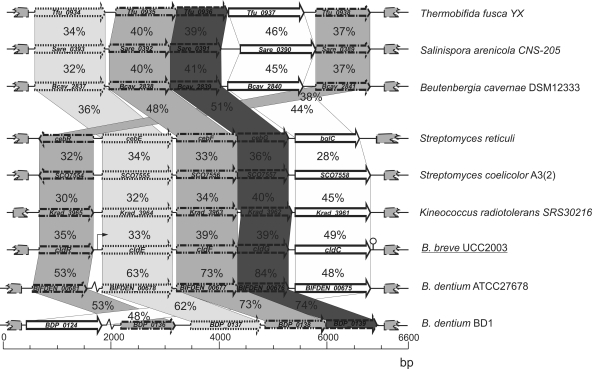
Our presumption, based on the QRT-PCR analysis, that the cld gene cluster is involved in utilization of cellodextrins was supported by the high level of sequence similarity between cldEFGC and predicted and/or proven cellodextrin-specific metabolic genes from a wide variety of bacteria. The genetic organization of the cld gene cluster on the chromosome of B. breve UCC2003 and its comparison to similar clusters found in other bacteria are schematically displayed in Fig. 3. Due to its adjacent location (Fig. 3) and similarity to LacI-type transcriptional repressors, the protein product of the cldR gene was suspected to represent the regulator of the cldEFGC operon of B. breve UCC2003 (see also below). The cldE, cldF, and cldG genes are predicted to encode a putative cellodextrin-binding protein and two cellodextrin permease proteins, respectively, which together are presumed to specify the cellodextrin uptake system of B. breve UCC2003. A β-glucosidase, which is involved in cellodextrin hydrolysis, is encoded by the cldC gene (see below). The gene order of the B. breve UCC2003 cld gene cluster is similar to that of the cld cluster in Streptomyces reticuli (cldREFGbglC) that was previously characterized (42), with the only difference being that the cldR orientation is opposite from that of the cdlEFGC genes. In the case of the Thermobifida fusca bgl operon, which is involved in cellodextrin degradation (45), the gene specifying the transcriptional regulator (celR) in this microorganism is situated downstream of the ABC transporters and β-glucosidase-encoding genes. Comparative genome analysis showed that the B. breve UCC2003 cld operon is most similar to the similarly organized putative cld gene clusters in the Bifidobacterium dentium ATCC 27678 and B. dentium Bd1 genomes (Fig. 3), although the suspected cldR homolog (locus tag, BIFDEN_00681) of B. dentium ATCC27678 and cldC homolog (locus tag, BDP_0124) of B. dentium Bd1 are not linked to the cld operon. A predicted β-glucosidase-encoding gene is present in the genomes of the sequenced B. longum subsp. infantis ATCC 15697 (50% identical to the B. breve UCC2003 CldC protein) and Bifidobacterium animalis subsp. lactis HN019 (49% identical to the CldC protein) (Fig. 3; Table 2 ), but the ABC transporter- and regulator-encoding genes appear to be absent from the immediate vicinity of each of these putative cldC homologs (data not shown). In addition, CldC encoded by B. breve UCC2003 exhibits 48% identity to the β-glucosidase BglC protein encoded by B. breve 203, but the B. breve 203 genome sequence is not available and we therefore cannot comment on the operon organization (29).
FIG. 3.
Comparison of the cld locus of B. breve UCC2003 with corresponding (putative or proven) cellodextrin/cellobiose utilization loci from other bacteria. Each solid arrow indicates an open reading frame (ORF). The length of the arrow is proportional to the length of the predicted ORF, and the gene locus name, which is indicative of its putative function, is indicated within the arrow. Orthologs are marked with the same shade, while the amino acid identity of each predicted protein is indicated as a percentage relative to its equivalent protein encoded by B. breve UCC2003. The bent arrow indicates the B. breve UCC2003 cld promoter; the lollipop sign designates the putative rho-independent terminator.
TABLE 2.
Similarity of CldC from B. breve UCC2003 to proven and annotated β-glucosidases in other bacterial strains
| Bacterial strain | Locus tag or gene | Annotated or proven function | NCBI reference no. | Identity (%) to CldC |
|---|---|---|---|---|
| Micromonospora sp. strain ATCC 39149 | MCAG_00986 | β-Glucosidase | ZP_04604729 | 51 |
| Jonesia denitrificans DSM 20603 | JdenDRAFT_1772 | Broad-specificity cellobiase | ZP_0386834 | 50 |
| B. dentium ATCC 27678 | BIFDEN_00695 | Hypothetical protein BIFDEN_00695 | ZP_02917416 | 50 |
| B. dentium ATCC 27678 | BIFDEN_00675 | Hypothetical protein BIFDEN_00675 | ZP_02917396 | 13 |
| B. dentium BD1 | BDP_0124 | β-Glucosidase | YP_003359633.1 | 48 |
| B. breve 203 | β-d-Glucosidase | BAA19881 | 48 | |
| B. animalis subsp. lactis HN019 | BIFLAC_00409 | β-Glucosidase | ZP_02963645 | 49 |
| B. longum subsp. infantis ATCC 15697 | Blon_1905 | β-Glucosidase | YP_002323355 | 50 |
| B. animalis subsp. lactis DSM 10140 | Balat_0151 | Putative β-glucosidase | YP_002969174 | 47 |
| Kineococcus radiotolerans SRS30216 | Krad_3961 | β-Glucosidase | YP_001363688 | 48 |
| Thermobifida fusca YX | Tfu_0937 | β-Galactosidase | YP_288998 | 44 |
| Streptomyces coelicolor A3(2) | SCO2798 | Cellobiose hydrolase | NP_627028 | 47 |
| Streptomyces reticuli | bglC | β-Glucosidase | AJ009797 | 24 |
| Salinispora arenicola CNS-205 | Sare_0390 | β-Glucosidase | YP_001535310 | 47 |
| Beutenbergia cavernae DSM 12333 | Bcav_2840 | β-Galactosidase | YP_002882847 | 26 |
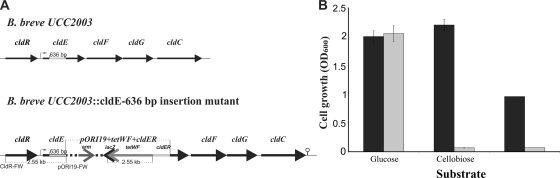
Disruption of the cldE gene in B. breve UCC2003.
In order to establish if disruption of the cldEFGC operon in B. breve UCC2003 would result in loss of this strain's ability to metabolize cellodextrins, a cldE insertion mutant was generated (Fig. 4 A). To investigate the expected phenotype of the B. breve UCC2003::cldE insertion mutant strain, both the wild-type and the insertion mutant strains were analyzed for their ability to grow in mMRS supplemented with cellobiose, the cellodextrin mixture, or glucose (positive control) as the sole carbon source (Fig. 4B).
FIG. 4.
(A) Schematic representation of the cld region of the B. breve UCC2003 and B. breve UCC2003::cldE chromosomes. Chromosomal DNA is represented by a thin line, the cldE gene is represented by a blank arrow, and the internal cldE fragment used for homologous recombination to obtain the insertion mutant is indicated by a solid light gray line. Segments of the integrated plasmid are indicated by solid medium gray (tetW gene), solid (lacZ gene), and boxed black and solid dark gray (erm gene) lines. (B) Growth profiles of wild-type B. breve UCC2003 (dark gray) and the B. breve UCC2003::cldE insertion mutant (light gray) in mMRS supplemented with glucose, cellobiose, or a mixture of cellodextrins.
As expected, and in contrast to the case for the wild type, the B. breve UCC2003::cldE insertion mutant was shown to be incapable of growth on cellobiose or cellodextrins as its sole carbon source. Although the cldE disruption in B. breve UCC2003::cldE is likely to have a polar effect on the transcription of the downstream genes of the cld operon, this nevertheless demonstrates that (elements of) the cld gene cluster is exclusively required for cellodextrin metabolism in B. breve UCC2003.
β-Glucosidic properties of CldCHis.
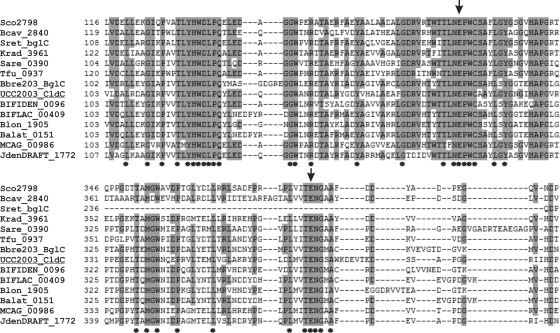
In order to characterize the CldC protein, comparative sequence analysis was performed. The cldC gene (1,401 bp) encodes a protein of 466 amino acids (molecular mass, ∼52.13 kDa). BLASTP and multiple-sequence alignments showed that CldC displays significant similarity to putative or proven β-glucosidases from Micromonospora sp. strain ATCC 39149, Jonesia denitrificans DSM 20603, B. dentium ATCC 27678, B. animalis subsp. lactis HN019, various Streptomyces spp., and T. fusca XY. All these β-glucosidases are members of glycosyl hydrolase family 1 (GH1) (14), and CldC is therefore assigned to this family (Table 2). Multiple-sequence alignment of these putative and/or proven representatives of β-glucosidases and other annotated GH1 members revealed a number of conservative amino acids (Fig. 5). The characteristic conserved glutamate residues of the GH1 family, present in the so-called NEP and ENG motifs, were found in all aligned sequences and correspond to a catalytically active nucleophile and acid-base catalyst, respectively (53).
FIG. 5.
Multiple-sequence alignment between CldC proteins from B. breve UCC2003 and 13 putative or proven β-glucosidases from different bacterial strains using CLUSTAL W (48). The sequence labeled UCC2003_CldC corresponds to CldC encoded by B. breve UCC2003. Other labels indicate locus tags of genes that encode putative GH1 family members from different bacteria (for details, see Table 2). Conserved residues are shaded based on conservation in 60% or more of the sequenced used. Fully conserved residues are indicated by an asterisk below the alignment. The arrows above the sequence point at the conserved glutamate residues that have been identified as an acid-base catalyst (glutamic acid 165 in UCC2003) and an active-site nucleophile (glutamic acid 368 in UCC2003) (57).
In order to demonstrate the suspected β-glucosidase activity of CldC from B. breve UCC2003 and its involvement in cellobiose/cellodextrin degradation, we cloned clcC and overexpressed it as a His-tagged version, designated CldCHis (see Materials and Methods). Crude cell extract containing CldCHis displayed hydrolytic activity against cellobiose, cellodextrins, 2-nitrophenyl-β-d-glucopyranoside, 2-nitrophenyl-β-d-cellobioside, and 4-nitrophenyl-β-d-cellobioside (results not shown). Under the same experimental conditions, we showed that crude cell extract from L. lactis NZ9000 harboring pNZ8150 (negative control) did not exhibit any hydrolytic activity toward the above-mentioned carbohydrates. These results clearly suggest that, consistent with the results from our comparative sequence analysis and the B. breve UCC2003::cldE mutant analysis, CldCHis represents a β-glucosidase being responsible for the observed carbohydrate-degrading activity, although it is theoretically possible that (expression of) CldCHis somehow activates a silent β-glucosidase in its lactococcal expression host. We could not exclude the latter possibility, because for some unknown reason the CldCHis protein completely lost its hydrolytic activity upon purification (results not shown). Under our experimental conditions, the CldCHis-containing cell extract was not capable of hydrolyzing sucrose, lactose, maltose, 1-kestose, and raffinose; however, all cellodextrins present in the reaction mixture (from C2 to C9) were completely converted into glucose (results not shown). The results obtained therefore show that crude extract containing CldCHis is capable of specifically cleaving the β-1,4-linkage present in cellodextrins and that it does not possess hydrolytic activity toward other tested glycosidic linkages.
Transcriptional analysis of the cld gene cluster.
In B. breve UCC2003, the cldE, cldF, cldG, and cldC genes (Fig. 6 A) are presumed to be expressed as a single transcript, based on their similar expression patterns as determined from the microarray analysis and RT-PCR experiments. Additional RT-PCR experiments confirmed the suspected operon structure, as they showed that amplification products were generated from cDNA (generated from total RNA of cellobiose-grown B. breve UCC2003) using various primer combinations that spanned the individual genes of the cld gene cluster (data not shown). Furthermore, the only predicted Rho-independent terminator structure in the DNA region that harbors the very tightly organized cldEFGC genes is present downstream of the cldC gene (Fig. 6C). The transcriptional start site of the presumed cldEFGC operon was determined by primer extension analysis (see Materials and Methods) and was shown to be located 79 bp upstream of the predicted cldE start codon (Fig. 6A and B) and 6 bp downstream of sequences resembling consensus −10 and −35 sequences of a vegetative promoter (Fig. 6A). In addition, the TG sequence upstream of the presumed −10 sequence has previously been shown to act as an enhancer element for transcription in other bacteria (23).
FIG. 6.
(A) Schematic representation of the B. breve UCC2003 cldE promoter region and partial coding sequence. Boldface and underlining indicate the −10 and −35 hexamers (as deduced from the primer extension results shown in panel B) and ribosomal binding site (RBS); the transcriptional start site is indicated by an asterisk. The CldR binding inverted repeat sequences cldEir1 and cldEIR2 are underlined. (B) Primer extension results. CB, cellobiose; CD, cellodextrins. (C) Representation of the B. breve UCC2003 cellodextrinase operon and DNA fragments used in EMSAs for the cldE promoter region. Plus and minus signs indicate whether or not CldR was able to bind to the particular DNA fragment, respectively. (D) Weblogo representation of predicted binding sequences of CldR together with EMSA demonstrating binding of CldR to the C2 fragment.
CldR binds to the cldE promoter region.
The presence of cldR, encoding a putative LacI-type regulator within the cellodextrin gene cluster, suggests that this gene is involved in the transcriptional regulation of the cellodextrin gene cluster, as was obvious from the QRT-PCR data (Table 1). In order to establish if CldR is capable of direct interaction with specific operators within the cldE promoter region, we first cloned the cldR gene in the IPTG (isopropyl-β-d-thiogalactopyranoside)-inducible vector pQE60 with the introduction of a His tag-encoding sequence to facilitate subsequent protein purification. The purified CldR protein was then used to perform electrophoretic mobility shift assays (EMSAs), which clearly demonstrate that the CldR protein can form a complex with IRD700-labeled DNA fragments encompassing a presumed CldR-binding sequence that is located within the cldE promoter region (Fig. 6C). Further delineation of this CldR-binding sequence or CldR box revealed the presence of two inverted repeats, the first located directly downstream of the transcription start site and the second present within the cldE coding sequence (Fig. 6C and D). These CldR box sequences are thus presumed to represent operator sequences for the CldR protein and to act as cis elements in the transcriptional regulation of the cldEFGC operon.
DISCUSSION
Life on Earth is to a significant degree dependent on the generation of plant biomass through photosynthesis. Cellulose, the most abundant component of plant biomass, which in nature is almost exclusively found in plant cell walls (21), cannot be metabolized by humans due to the absence of cellulose-degrading enzymes. Nevertheless, a number of cellulolytic bacteria have been identified in the human gastrointestinal tract and shown to extracellularly hydrolyze cellulose to soluble cellodextrins, which in turn may be utilized by noncellulolytic human gut bacteria with β-glucosidic activity (6, 9, 16, 54). This was further substantiated by a recent study which demonstrated that β-glucosidases are widespread among colonic bacteria (25). Interestingly, bifidobacterial strains are reported to exhibit the highest β-glucosidic activity among the colonic microbiota (6), and several bifidobacterial β-glucosidases involved in the utilization of cellobiose and/or other carbohydrate substances have been described and characterized (5, 29, 30, 39, 56).
The current report describes the genetic identification and characterization of the cellodextrin utilization cluster, cldEFGC and its regulator cldR, carried by B. breve UCC2003. The insertion mutant B. breve UCC2003::cldE confirmed that the cld gene cluster is exclusively responsible for cellodextrin metabolism by B. breve UCC2003. Although B. breve 203 was previously shown to metabolize cellobiose (30), this is, to the best of our knowledge, the first report to demonstrate that particular bifidobacterial strains can utilize a range of cellodextrins and that a specific gene cluster, cldEFGC, is required for its metabolism.
The upregulation of cldEFGC transcription when B. breve UCC2003 was grown in mMRS containing a mixture of cellodextrins was significantly higher than that for growth on cellobiose only. Therefore, these results suggest that cellobiose is less efficient in derepression of the cldR repressor of B. breve UCC2003 cld operon than the cellodextrins. Even though various ruminal cellulolytic bacteria display similar growth behavior, these latter degrade cellodextrins usually by a phospho-dependent reaction by phosphorolytic cleavage, generating glucose-1-phosphate as an end product (44, 55).
Comparative sequence analysis of gene clusters for cellodextrin utilization from other bacteria (e.g., Kineococcus radiotolerans, Streptomyces coelicolor, Beutenbergia cavernae, Salinispora arenicola, and T. fusca) shows that they are generally organized in a similar genetic fashion as the B. breve UCC2003 cld gene cluster, with the exception of B. dentium, where the regulator-encoding gene (in B. dentium ATCC 72678) and putative β-glucosidase-encoding gene (in B. dentium Bd1) appear to be disconnected from the rest of the gene cluster. The results obtained from comparative sequence and genome analyses indicate that just two other bifidobacterial strains, in addition to strain UCC2003, possess the complete cld operon (i.e., strains JCM7017 and NCIMB8815), consistent with the finding that these strains are also capable of cellobiose metabolism. In contrast, B. breve JCM7019 is able to metabolize cellobiose but does not appear to contain the cld cluster, which suggests that this bacterium may use an alternative cellodextrin utilization system which is not homologous to the one identified in B. breve UCC2003. This could be the one found in B. breve 203, which is significantly different in sequence (Table 2) (39).
From the results presented here we have obtained convincing evidence to conclude that the B. breve UCC2003 CldC protein represents a β-glucosidase that exhibits exclusive hydrolytic activity toward cellobiose and higher-molecular-weight cellodextrins, cleaving the β-1→4-glucosidic bond between glucose molecules in these carbohydrates. The presence of β-glucosidases in bifidobacteria has been reported previously for B. breve 203 (29, 30), B. adolescentis Int-57 (5), and Bifidobacterium sp. strain SEN (56). The β-glucosidase from Bifidobacterium sp. strain SEN did not exhibit hydrolytic activity against cellobiose, while both B. breve 203 and B. adolescentis Int-57 produced β-glucosidase activity with the ability to hydrolyze cellobiose. Furthermore, seven different B. bifidum strains, two B. infantis strains, and two B. breve strains were demonstrated to have specific β-glucosidase activities (28). However, none of these enzymes were examined for their ability to degrade cellodextrins with a higher degree of polymerization.
A specific ABC transporter system specified by the cldEFG genes is presumed to be involved in cellodextrin internalization. Interestingly, no gene encoding an ATP-binding protein is present in this gene cluster, suggesting that B. breve UCC2003 may use a general ATP-binding protein whose transcription is not under cellobiose/cellodextrin control. A similar scenario was observed in S. reticuli, which contains the msiK gene, encoding an ABC transporter-type ATP-binding protein involved in both cellobiose and maltose transport (43). Our results (Fig. 2B and C) suggest that the cellodextrin ABC transporter is capable of transporting cellodextrins with a degree of polymerization (DP) ranging from 2 to 5, which is consistent with data from a similar study of Clostridium thermocellus (27). Interestingly, the crude cell extract containing the CldCHis enzyme is capable of hydrolyzing cellodextrins with a higher DP (up to 9 [results not shown]), although they are not utilized by B. breve UCC2003. This observation suggests that the B. breve UCC2003 ABC uptake system for cellodextrins limits the ability of this microorganism to metabolize higher-molecular-weight cellodextrins, although this presumption needs to be confirmed by further experimental work.
Insoluble plant fiber consisting mostly of plant cell wall polysaccharides, including cellulose, is one of the major components of the everyday human diet. The complex microbial community of the human colon is capable of utilization of insoluble cellulose by producing solubilized end products, i.e., cellodextrins, that become accessible to other microorganisms of the human colon microbiota (for a review, see reference 10). This, to the best of our knowledge, represents the first in-depth study describing a bifidobacterial gene cluster responsible for cellodextrin utilization. The ability of B. breve UCC2003 to metabolize cellobiose/cellodextrins may provide a competitive advantage to assist this strain to colonize, adapt to, and persist in the human intestinal environment.
Supplementary Material
Acknowledgments
The Alimentary Pharmabiotic Centre is a research center funded by Science Foundation Ireland (SFI) through the Irish Government's National Development Plan. The authors and their work were supported by SFI (grant no. 02/CE/B124 and 07/CE/B1368).
We are grateful to Therese Uniacke from the Department of Food and Nutritional Sciences, University College Cork, Ireland, for technical assistance.
Footnotes
Published ahead of print on 7 January 2011.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., et al. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alvarez-Martin, P., M. O'Connell-Motherway, D. Van Sinderen, and B. Mayo. 2007. Functional analysis of the pBC1 replicon from Bifidobacterium catenulatum L48. Appl. Microbiol. Biotechnol. 76:1395-1402. [DOI] [PubMed] [Google Scholar]
- 4.An, C. L., et al. 2004. Analysis of bgl operon structure and characterization of beta-glucosidase from Pectobacterium carotovorum subsp. carotovorum LY34. Biosci. Biotechnol. Biochem. 68:2270-2278. [DOI] [PubMed] [Google Scholar]
- 5.Choi, Y. J., C. J. Kim, and G. E. Ji. 1996. A partially purified beta-glucosidase from Bifidobacterium adolescentis converts cycasin to a mutagenic compound. Lett. Appl. Microbiol. 22:145-148. [DOI] [PubMed] [Google Scholar]
- 6.Dabek, M., S. I. McCrae, V. J. Stevens, S. H. Duncan, and P. Louis. 2008. Distribution of beta-glucosidase and beta-glucuronidase activity and of beta-glucuronidase gene gus in human colonic bacteria. FEMS Microbiol. Ecol. 66:487-495. [DOI] [PubMed] [Google Scholar]
- 7.De Man, J. C., A. Rogosa, and M. E. Sharpe. 1960. A medium for the cultivation of lactobacilli. J. Appl. Bacteriol. 23:130-135. [Google Scholar]
- 8.de Ruyter, P. G., O. P. Kuipers, and W. M. de Vos. 1996. Controlled gene expression systems for Lactococcus lactis with the food-grade inducer nisin. Appl. Environ. Microbiol. 62:3662-3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Flint, H. J., and E. A. Bayer. 2008. Plant cell wall breakdown by anaerobic microorganisms from the mammalian digestive tract. Ann. N. Y. Acad. Sci. 1125:280-288. [DOI] [PubMed] [Google Scholar]
- 10.Flint, H. J., E. A. Bayer, M. T. Rincon, R. Lamed, and B. A. White. 2008. Polysaccharide utilization by gut bacteria: potential for new insights from genomic analysis. Nat. Rev. Microbiol. 6:121-131. [DOI] [PubMed] [Google Scholar]
- 11.Gibson, G. R. 2008. Prebiotics as gut microflora management tools. J. Clin. Gastroenterol. 42:S75-S79. [DOI] [PubMed] [Google Scholar]
- 12.Guarner, F. 2006. Enteric flora in health and disease. Digestion 73:5-12. [DOI] [PubMed] [Google Scholar]
- 13.Hamoen, L. W., A. F. Van Werkhoven, J. J. Bijlsma, D. Dubnau, and G. Venema. 1998. The competence transcription factor of Bacillus subtilis recognizes short A/T-rich sequences arranged in a unique, flexible pattern along the DNA helix. Genes Dev. 12:1539-1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Henrissat, B. 1991. A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem. J. 280:309-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jeoh, T., J. O. Baker, M. K. Ali, M. E. Himmel, and W. S. Adney. 2005. Beta-d-glucosidase reaction kinetics from isothermal titration microcalorimetry. Anal. Biochem. 347:244-253. [DOI] [PubMed] [Google Scholar]
- 16.Kopecny, J., J. Hajer, and J. Mrazek. 2004. Detection of cellulolytic bacteria from the human colon. Folia Microbiol. (Praha) 49:175-177. [DOI] [PubMed] [Google Scholar]
- 17.Kuipers, O. P., M. M. Beerthuyzen, R. J. Siezen, and W. M. de Vos. 1993. Characterization of the nisin gene cluster nisABTCIPR of Lactococcus lactis. Requirement of expression of the nisA and nisI genes for development of immunity. Eur. J. Biochem. 216:281-291. [DOI] [PubMed] [Google Scholar]
- 18.Law, J., et al. 1995. A system to generate chromosomal mutations in Lactococcus lactis which allows fast analysis of targeted genes. J. Bacteriol. 177:7011-7018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Livak, K. J., and T. D. Schmittgen. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods 25:402-408. [DOI] [PubMed] [Google Scholar]
- 20.Lou, J., K. A. Dawson, and H. J. Strobel. 1996. Role of phosphorolytic cleavage in cellobiose and cellodextrin metabolism by the ruminal bacterium Prevotella ruminicola. Appl. Environ. Microbiol. 62:1770-1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lynd, L. R., P. J. Weimer, W. H. van Zyl, and I. S. Pretorius. 2002. Microbial cellulose utilization: fundamentals and biotechnology. Microbiol. Mol. Biol. Rev. 66:506-577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Macfarlane, G. T., H. Steed, and S. Macfarlane. 2008. Bacterial metabolism and health-related effects of galacto-oligosaccharides and other prebiotics. J. Appl. Microbiol. 104:305-344. [DOI] [PubMed] [Google Scholar]
- 23.Madsen, S. M., T. Hindre, J. P. Le Pennec, H. Israelsen, and A. Dufour. 2005. Two acid-inducible promoters from Lactococcus lactis require the cis-acting ACiD-box and the transcription regulator RcfB. Mol. Microbiol. 56:735-746. [DOI] [PubMed] [Google Scholar]
- 24.Maze, A., M. O'Connell-Motherway, G. F. Fitzgerald, J. Deutscher, and D. Van Sinderen. 2007. Identification and characterization of a fructose phosphotransferase system in Bifidobacterium breve UCC2003. Appl. Environ. Microbiol. 73:545-553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McBain, A. J., and G. T. Macfarlane. 1998. Ecological and physiological studies on large intestinal bacteria in relation to production of hydrolytic and reductive enzymes involved in formation of genotoxic metabolites. J. Med. Microbiol. 47:407-416. [DOI] [PubMed] [Google Scholar]
- 26.Mierau, I., and M. Kleerebezem. 2005. 10 years of the nisin-controlled gene expression system (NICE) in Lactococcus lactis. Appl. Microbiol. Biotechnol. 68:705-717. [DOI] [PubMed] [Google Scholar]
- 27.Nataf, Y., et al. 2009. Cellodextrin and laminaribiose ABC transporters in Clostridium thermocellum. J. Bacteriol. 191:203-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Noriega, L., M. Gueimonde, B. Sanchez, A. Margolles, and C. G. de los Reyes-Gavilan. 2004. Effect of the adaptation to high bile salts concentrations on glycosidic activity, survival at low pH and cross-resistance to bile salts in Bifidobacterium. Int. J. Food Microbiol. 94:79-86. [DOI] [PubMed] [Google Scholar]
- 29.Nunoura, N., et al. 1996. Cloning and nucleotide sequence of the beta-d-glucosidase gene from Bifidobacterium breve clb, and expression of beta-d-glucosidase activity in Escherichia coli. Biosci. Biotechnol. Biochem. 60:2011-2018. [DOI] [PubMed] [Google Scholar]
- 30.Nunoura, N., K. Ohdan, T. Yano, K. Yamamoto, and H. Kumagai. 1996. Purification and characterization of beta-d-glucosidase (beta-d-fucosidase) from Bifidobacterium breve clb acclimated to cellobiose. Biosci. Biotechnol. Biochem. 60:188-193. [DOI] [PubMed] [Google Scholar]
- 31.O'Connell-Motherway, M., et al. 2008. Characterisation of ApuB, an extracellular type II amylopullulanase from Bifidobacterium breve UCC2003. Appl. Environ. Microbiol. 74:6271-6279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.O'Connell-Motherway, M., J. O'Driscoll, G. F. Fitzgerald, and D. Van Sinderen. 2008. Overcoming the restriction barrier to plasmid transformation and targeted mutagenesis in Bifidobacterium breve UCC2003. Microb. Biotechnol. 2:321-332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pokusaeva, K., et al. 2009. Ribose utilization by the human commensal Bifidobacterium breve UCC2003. Microb. Biotechnol. 3:311-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pokusaeva, K., M. O'Connell-Motherway, A. Zomer, G. F. Fitzgerald, and D. Van Sinderen. 2009. Characterization of two novel alpha-glucosidases from Bifidobacterium breve UCC2003. Appl. Environ. Microbiol. 75:1135-1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roberfroid, M. 2007. Prebiotics: the concept revisited. J. Nutr. 137:830S-837S. [DOI] [PubMed] [Google Scholar]
- 36.Russell, J. B., R. E. Muck, and P. J. Weimer. 2009. Quantitative analysis of cellulose degradation and growth of cellulolytic bacteria in the rumen. FEMS Microbiol. Ecol. 67:183-197. [DOI] [PubMed] [Google Scholar]
- 37.Ryan, S. M., G. F. Fitzgerald, and D. Van Sinderen. 2006. Screening for and identification of starch-, amylopectin-, and pullulan-degrading activities in bifidobacterial strains. Appl. Environ. Microbiol. 72:5289-5296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ryan, S. M., G. F. Fitzgerald, and D. Van Sinderen. 2005. Transcriptional regulation and characterization of a novel beta-fructofuranosidase-encoding gene from Bifidobacterium breve UCC2003. Appl. Environ. Microbiol. 71:3475-3482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sakai, K., T. Tachiki, H. Kumagai, and T. Tochikura. 1986. Isolation and characterization of two β-d-glucosidases from Bifidobacterium breve 203. Agric. Biol. Chem. 50:2287-2293. [Google Scholar]
- 40.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 41.Schell, M. A., et al. 2002. The genome sequence of Bifidobacterium longum reflects its adaptation to the human gastrointestinal tract. Proc. Natl. Acad. Sci. U. S. A. 99:14422-14427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schlosser, A., J. Jantos, K. Hackmann, and H. Schrempf. 1999. Characterization of the binding protein-dependent cellobiose and cellotriose transport system of the cellulose degrader Streptomyces reticuli. Appl. Environ. Microbiol. 65:2636-2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schlosser, A., T. Kampers, and H. Schrempf. 1997. The Streptomyces ATP-binding component MsiK assists in cellobiose and maltose transport. J. Bacteriol. 179:2092-2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shi, Y., and P. J. Weimer. 1996. Utilization of individual cellodextrins by three predominant ruminal cellulolytic bacteria. Appl. Environ. Microbiol. 62:1084-1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Spiridonov, N. A., and D. B. Wilson. 2001. Cloning and biochemical characterization of BglC, a beta-glucosidase from the cellulolytic actinomycete Thermobifida fusca. Curr. Microbiol. 42:295-301. [DOI] [PubMed] [Google Scholar]
- 46.Tuohy, K. M., H. M. Probert, C. W. Smejkal, and G. R. Gibson. 2003. Using probiotics and prebiotics to improve gut health. Drug Discov. Today 8:692-700. [DOI] [PubMed] [Google Scholar]
- 47.Turroni, F., A. Ribbera, E. Foroni, D. van Sinderen, and M. Ventura. 2008. Human gut microbiota and bifidobacteria: from composition to functionality. Antonie Van Leeuwenhoek 94:35-50. [DOI] [PubMed] [Google Scholar]
- 48.Turroni, F., D. Van Sinderen, and M. Ventura. 2009. Bifidobacteria: from ecology to genomics. Front. Biosci. 14:4673-4684. [DOI] [PubMed] [Google Scholar]
- 49.Untergasser, A., et al. 2007. Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Res. 35:W71-W74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ventura, M., C. Canchaya, V. Meylan, T. R. Klaenhammer, and R. Zink. 2003. Analysis, characterization, and loci of the tuf genes in Lactobacillus and Bifidobacterium species and their direct application for species identification. Appl. Environ. Microbiol. 69:6908-6922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ventura, M., et al. 2007. From bacterial genome to functionality; case bifidobacteria. Int. J. Food Microbiol. 120:2-12. [DOI] [PubMed] [Google Scholar]
- 52.Ventura, M., et al. 2009. Genome-scale analyses of health-promoting bacteria: probiogenomics. Nat. Rev. Microbiol. 7:61-71. [DOI] [PubMed] [Google Scholar]
- 53.Voorhorst, W. G., R. I. Eggen, E. J. Luesink, and W. M. de Vos. 1995. Characterization of the celB gene coding for beta-glucosidase from the hyperthermophilic archaeon Pyrococcus furiosus and its expression and site-directed mutation in Escherichia coli. J. Bacteriol. 177:7105-7111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wedekind, K. J., H. R. Mansfield, and L. Montgomery. 1988. Enumeration and isolation of cellulolytic and hemicellulolytic bacteria from human feces. Appl. Environ. Microbiol. 54:1530-1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wells, J. E., J. B. Russell, Y. Shi, and P. J. Weimer. 1995. Cellodextrin efflux by the cellulolytic ruminal bacterium Fibrobacter succinogenes and its potential role in the growth of nonadherent bacteria. Appl. Environ. Microbiol. 61:1757-1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang, L., T. Akao, K. Kobashi, and M. Hattori. 1996. Purification and characterization of a novel sennoside-hydrolyzing beta-glucosidase from Bifidobacterium sp. strain SEN, a human intestinal anaerobe. Biol. Pharm. Bull. 19:705-709. [DOI] [PubMed] [Google Scholar]
- 57.Zhang, Y. H., and L. R. Lynd. 2003. Cellodextrin preparation by mixed-acid hydrolysis and chromatographic separation. Anal. Biochem. 322:225-232. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.