Abstract
We report here the construction, characterization, and application of a bacterial bioreporter for fructose and sucrose that was designed to monitor the availability of these sugars to microbial colonizers of the phyllosphere. Plasmid pPfruB-gfp[AAV] carries the Escherichia coli fruB promoter upstream from the gfp[AAV] allele that codes for an unstable variant of green fluorescent protein (GFP). In Erwinia herbicola, this plasmid brings about the accumulation of GFP fluorescence in response to both fructose and sucrose. Cells of E. herbicola (pPfruB-gfp[AAV]) were sprayed onto bean plants, recovered from leaves at various time intervals after inoculation, and analyzed individually for GFP content by quantitative analysis of digital microscope images. We observed a positive correlation between single-cell GFP accumulation and ribosomal content as determined by fluorescence in situ hybridization, indicating that foliar growth of E. herbicola occurred at the expense of fructose and/or sucrose. One hour after inoculation, nearly all bioreporter cells appeared to be actively engaged in fructose consumption. This fraction dropped to approximately 11% after 7 h and to ≈1% a day after inoculation. This pattern suggests a highly heterogeneous availability of fructose to individual E. herbicola cells as they colonize the phyllosphere. We estimated that individual cells were exposed to local initial fructose abundances ranging from less than 0.15 pg fructose to more than 4.6 pg.
The phyllosphere (1), or plant leaf surface, is a habitat for many microorganisms. The most commonly found epiphytic residents are bacteria and fungi. The communities that these microbes form on leaves can vary dramatically from one leaf to another and undergo constant change in both size and composition (2–6). Microbes may arrive to or depart from a leaf surface through the action of rain, wind, or insects (7–10). On arrival, new immigrants are challenged with the harsh conditions of the leaf environment (11, 12), including highly fluctuating water availability, exposure to UV radiation from sunlight, and limited access to nutrient resources. To cope with these conditions and to survive the leaf environment, microorganisms have adopted different strategies (13) such as the production of pigments to protect against the effects of UV radiation (14) or the secretion of polysaccharides to prevent desiccation when water becomes scarce (15, 16).
Many leaf-associated microbes are capable of growth, i.e., multiplying, by exploitation of the few resources that the leaf surface offers. Exogenous nutrients may be available fortuitously in the form of pollen, honeydew, dust, air pollution, or microbial debris (17, 18). Occasionally, plant sap may ooze from wounds inflicted by insect feeding (17) or frost damage (12). But even healthy plants passively leak small amounts of metabolites such as carbohydrates, amino acids, and organic acids to the leaf surface (19, 20). The amounts that are leaked depend on many factors, including the leachate itself, the plant species, leaf characteristics such as wettability, waxiness, and age, and duration and intensity of rain or dew (20).
It has been demonstrated that leached compounds are used for growth by the microflora residing on a leaf (21–23). Photoassimilates like sucrose, fructose, and glucose, which are found in abundances of 0.2–2.0 μg per leaf on uninhabited bean leaf surfaces, were readily consumed and converted into biomass by the bacterium Pseudomonas fluorescens A506 (24). Bacterial and fungal colonization of the phyllosphere does not occur evenly across the leaf (25, 26), suggesting that resources are not evenly available or exploitable. Microscopic analyses have revealed that bacteria are more likely to be found clustered in crevices between epidermal cells, near the base of trichomes, in the proximity of stomates, and along veins (27–29). The factors that influence the availability of nutrients within or beyond these preferred sites remain elusive. Also, there is no information on leaf nutrient availability at the scale that is most relevant to its microbial colonizers. It seems unlikely that nutrient abundance as averaged over an entire leaf surface is of any relevance to an individual bacterial colonizer. Instead, a bacterium probably perceives its local environment in multiples of its own dimensions, i.e., on a micrometer scale.
The aim of this research was to quantify the availability of nutrients to individual microbial residents in the phyllosphere. We targeted sugars as the model nutrient, as they generally are recognized as the most abundant carbon source available in the phyllosphere (24). Our strategy involved the use of reporter gene technology in which the well-characterized bacterial epiphyte Erwinia herbicola 299R (Eh299R) (30) harbored a sugar-responsive element that was fused to a gene for green fluorescent protein (GFP). The GFP content of single cells thus becomes a measure for local sugar availability. We opted for the promoter region of the fruBKA operon from Escherichia coli (31) to drive expression of GFP. This gene cluster codes for the metabolism of fructose and its expression is controlled by the catabolite repressor/activator or Cra protein (FruR) (32) in response to fructose 1-phosphate, the first intermediate in the fructose metabolic pathway (33). Because E. coli and E. herbicola are closely related bacteria, we anticipated that the fruB promoter would function properly in Eh299R. It is important to note that Eh299R can metabolize sugars such as fructose, so that its ability to report the presence of fructose is a function not only of local fructose abundance but also of fructose consumption. Instead of the original GFP from Aequorea victoria (34), we used variant GFP[AAV] (35), which matures faster and yields a brighter fluorescence and also has a reduced stability, which allows for real-time monitoring of gene expression (36, 37). Analysis of individual Eh299R fructose bioreporters that were exposed to leaf surfaces for different periods of time disclosed a substantial heterogeneity in the availability of this sugar, both spatially (i.e., at different sites along the leaf surface) and temporally (i.e., at different times during leaf colonization).
Materials and Methods
Bacterial Strains and Culture Conditions.
Eh299R (30) was host to the fructose reporter construct pPfruB-gfp[AAV] (see below). E. coli DH5α (38) was used for routine cloning of plasmids and as a source for the fruB promoter (see below). Bacterial cultures were grown at 28°C (Eh299R) or 37°C (DH5α) in LB broth, on solid LB medium, or in liquid M9 minimal medium (38) supplemented with 0.2% casaminoacids, 0.4% galactose, and fructose at final concentrations as indicated in the text. Where appropriate, ampicillin, kanamycin, and rifampicin were added to final concentrations of 100, 50, and 100 μg/ml, respectively.
Fructose Reporter Construct.
Plasmid pPfruB-gfp[AAV] was constructed as follows. The fruB promoter (PfruB) was amplified as a 0.5-kb DNA fragment from E. coli DH5α genomic DNA in a PCR using primers FRUB-1 (5′-GGGAGTTATGCATGCTGGTCG-3′) and FRUB-2 (5′-CGCGCCAGCATGCCATTG-ACG-3′). After insertion into pCR2.1 (Invitrogen) and verification of its nucleotide sequence, PfruB was reisolated as an XhoI–KpnI fragment, and inserted upstream of the promoterless gfp[AAV] gene on SalI–KpnI double-digested promoter-probe vector pPROBE-gfp[AAV] (39). The resulting plasmid pPfruB-gfp[AAV] was introduced into Eh299R by triparental mating using E. coli DH5α (pRK2073) (40) as a helper strain. As a control plasmid to pPfruB-gfp[AAV], we constructed pPnptII-gfp[AAV] by inserting into pPROBE-gfp[AAV] a 131-bp HindIII-BamHI DNA fragment containing the constitutive nptII promoter from Tn5.
Fructose Induction Experiments.
We analyzed cultures of Eh299R (pPfruB-gfp[AAV]) for GFP content of individual cells by epifluorescence microscopy. Cells were collected by centrifugation or by filtration on a 0.1-μm Durapore filter (Millipore), fixed according to the protocol of Akkermans et al. (41), spotted on Polysine microscope slides (Erie Scientific, Portsmouth, NH), air-dried, mounted with Aqua-Poly/Mount (Polysciences), and analyzed microscopically (see below).
Plant Experiments and Fluorescence in Situ Hybridization (FISH).
Eh299R cells harboring pPfruB-gfp[AAV] or pPnptII-gfp[AAV] were grown to early midlog phase (optical density at 600 nm between 0.65 and 0.85) on M9 medium containing galactose, and then diluted in sterile water to a density of approximately 5 × 106 cells per ml. These bacterial suspensions were sprayed onto the aerial parts of 10-day-old bean plants (Phaseolus vulgaris cv. Bush Blue Lake 274) at the primary leaf stage. Inoculated plants were placed at room temperature (24–28°C) in a sealed chamber that was kept at a high relative humidity throughout the experiment. We collected bacteria from the surface of three individual leaves at 1, 3, 7, and 24 h after inoculation. To do so, leaves were detached and transferred to a tube containing 20 ml washing buffer (42). After sonication for 7 min and vortexing for 15 sec, a 50-μl aliquot was taken from the wash solution for plate counting on LB agar containing rifampicin and kanamycin. The cells in the remainder of the wash solution were collected by filtration and prepared as described above. One aliquot of fixed cells was spotted and analyzed directly for single-cell GFP content as described below. Another aliquot was first subjected to a FISH procedure using a tetramethylrhodamine (TAMRA)-labeled oligonucleotide probe specific for Eh299R 16S rRNA, as described by Brandl et al. (43).
Microscopy and Quantitative Image Analysis.
Cells were viewed at a ×1,000 total magnification on a Zeiss Axiophot epifluorescence microscope by using phase-contrast objectives. To allow for the visualization of fluorescence from GFP or TAMRA, the transmitted light was turned off and samples were illuminated with a Hg/Ne arc lamp using filter combinations that are specific for the detection of GFP (BP470/40, LP495, HQ525/50) or TAMRA (BP546/6, FT580, LP590). Microscopy views were digitally captured as black-and-white images by using a charged-couple device camera (Princeton Instruments, Trenton, NJ). Captured images were analyzed by using iplab software (Scanalytics, Fairfax, VA). In short, the periphery of each individual cell in a phase-contrast image was marked by an overlay. All overlays then were transferred from the phase-contrast image to the corresponding GFP and/or TAMRA image. The software computed for each overlay a mean pixel intensity (MPI), i.e., the average grayscale value for all of the pixels within that overlay. These values were corrected for nonspecific background fluorescence (background noise), which was defined as the lowest MPI value from among a randomly sampled collection of background areas in the fluorescent image. The difference between the highest and lowest background MPI values set the limit of detection: cells with an MPI that fell below this value were considered to be not significantly different from background noise. Background-corrected MPI values were interpreted as a measure for single-cell GFP content and further analyzed in Microsoft excel, either in an untransformed form, or after a cube-root transformation (MPI1/3). Where appropriate, average MPI1/3 values were back-transformed as (MPI1/3)3.
We also observed Eh299R bacteria expressing GFP in situ, i.e., directly on the leaf surface. For this purpose, individual 15-mm diameter leaf discs were punched from different parts of detached leaves and scanned for fluorescent bacterial cells at a ×100–400 total magnification. Phase-contrast and GFP fluorescent images were captured as above, and saved as images in Corel photo-paint (Corel, Salinas, CA). The black-and-white GFP fluorescent image was pseudocolored green and overlaid onto the phase-contrast image to give the final picture.
Results
Fructose-Induced GFP Expression in Eh299R (pPfruB-gfp[AAV]).
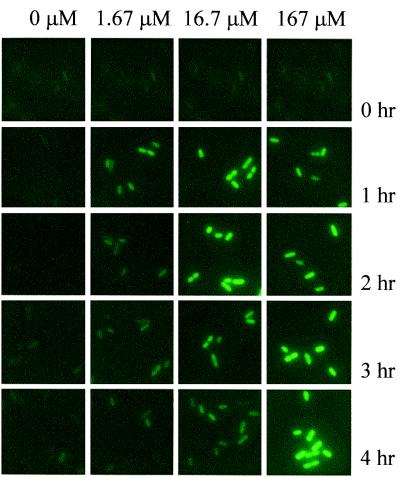
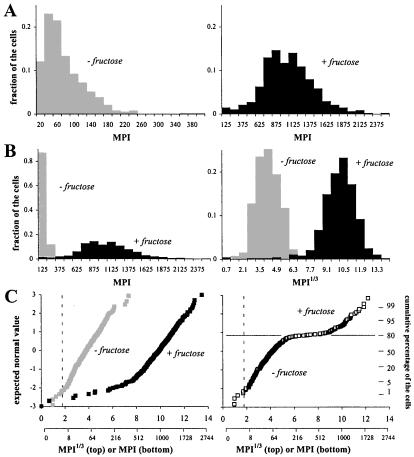
Single cells of fructose bioreporter Eh299R (pPfruB-gfp[AAV]) showed a low apparent GFP content during growth on galactose: they were barely visible with epifluorescence microscopy (Fig. 1, column marked 0 μM). When cells from the same culture were exposed to fructose however, they became green fluorescent (Fig. 1, columns marked 1.67, 16.7, and 167 μM), indicating that the E. coli fruB promoter was functional in Eh299R. GFP appeared to be evenly present within individual bioreporter cells (Fig. 1). This allowed us to express single-cell GFP content as the mean brightness, or intensity, of all of the pixels that comprise a given cell in a digitally captured image. Among uninduced cells of Eh299R (pPfruB-gfp[AAV]) (represented by the cells in Fig. 1, t = 0 h), GFP fluorescence averaged around an MPI of 67 ± 53 (n = 391). The frequency distribution of single-cell GFP content was quite right-hand skewed (Fig. 2A Left). With a mean MPI of 989 ± 379, fructose-induced cells (n = 478) (represented by the cells in Fig. 1, t = 4 h, 167 μM) were on average about 14 times brighter than uninduced cells, and their distribution profile was also right-hand skewed, but considerably less so (Fig. 2A Right).
Figure 1.
Accumulation of GFP fluorescence in single cells of Eh299R carrying plasmid pPfruB-gfp[AAV]. At t = 0, a galactose-grown culture (with an optical density of 1.0) was diluted 120-fold into the same medium supplemented with fructose at initial concentrations of 0, 1.67, 16.7, or 167 μM.
Figure 2.
Distribution profiles of single-cell GFP content in Eh299R (pPfruB-gfp[AAV]) cells from cultures that were supplemented with 0 or 167 μM fructose. See text for details. Note that due to the nature of the cube-root transformation, the MPI1/3 axis disproportionally overrepresents low values. An auxiliary axis shows corresponding MPI values. The broken lines in C mark the limit of GFP detection: MPI1/3 values above this limit were significantly different from background fluorescence (see Materials and Methods).
The variance in GFP fluorescence when expressed as MPI values was considerably larger among induced cells than among uninduced cells, making it hard to compare the two populations on the same scale (Fig. 2B Left). To standardize this variance, we subjected MPI values to a cube-root transformation, i.e., MPI1/3. This normalized the data and produced symmetric frequency distributions, which allowed the two populations to be compared in a single histogram (Fig. 2B Right). Also, the two populations appeared as straight and parallel lines in a normal probability plot (Fig. 2C Left). The two lines crossed the z = 0 axis at their corresponding population average MPI1/3 of 3.8 (uninduced) and 9.8 (induced), respectively. The slope of both lines is determined by the intrapopulation variability in single-cell GFP content. It is important to note here that this variability is independent of whether or not the fruB promoter was activated. Any deviation from this slope would be an indication for heterogeneity in fruB promoter activity, and thus fructose exposure, within the population. In the induced population, about 5% of the cells appeared dimmer than expected. These cells probably had lost the plasmid pPfruB-gfp[AAV], which complied with our observation that approximately 4% of the induced cells were sensitive to kanamycin (not shown).
Presentation of MPI1/3 data in a normal probability plot is especially useful when a population is expected to include two or more subpopulations with different exposure to fructose, for example on a leaf where fructose availabilities might vary. To illustrate this, we considered a hypothetical population made up of uninduced and induced cells that were unequally mixed in a ratio of 4 to 1. In a normal probability plot, this population was described by two parallel lines connected by a horizontal line at the expected normal value z = 0.85, which divides the mixed population into 20% bright and 80% dim cells (Fig. 2C Right).
GFP Accumulation in Eh299R (pPfruB-gfp[AAV]) in Response to Different Fructose Availabilities.
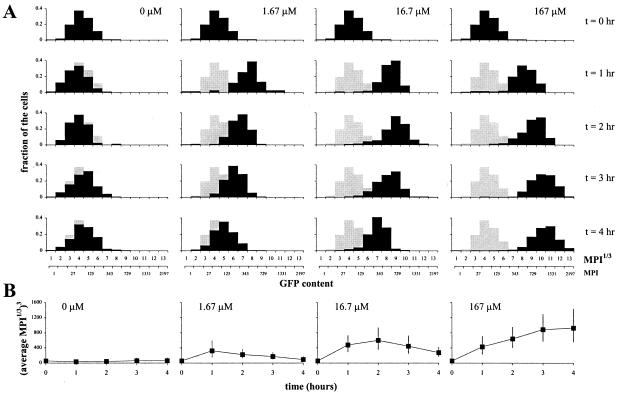
To determine the sensitivity of the bioreporter and the rate of GFP accumulation, we analyzed individual cells from growing cultures of Eh299R (pPfruB-gfp[AAV]) that were exposed to increasing fructose concentrations for different time periods (representative cells from these cultures are shown in Fig. 1). To display population dynamics of GFP accumulation, we represented single-cell GFP fluorescence as MPI1/3 in a histogram format (Fig. 3A). Also, for each time point and each initial fructose concentration, an average MPI1/3 was calculated, and its back-transformed value (average MPI1/3)3 was plotted as a function of time (Fig. 3B). With the two lowest concentrations of fructose, we observed a transient accumulation of fluorescence (Figs. 1 and 3); after an initial increase for 1 and 2 h, respectively, GFP content gradually declined again to uninduced levels. From initial cell densities, observed growth rates, and an estimated consumption of 0.3 pg fructose per individual cell doubling (as determined from our observation that M9 medium plus 0.4% fructose supports a final population of approximately 1.3⋅1010 Eh299R cells per ml), we calculated that initial fructose abundances of 1.67 and 16.7 μM would be consumed by the bacteria within approximately 15 min and 1.5 h, respectively. This explains our observation that the response of Eh299R (pPfruB-gfp[AAV]) to the presence of fructose was transient and further shows that GFP accumulation is concomitant with fructose consumption. In the presence of an initial fructose concentration of 167 μM, which would take the bacteria an estimated 4.2 h to consume completely, GFP continued to accumulate for the entire duration of the experiment (Figs. 1 and 3). At even higher concentrations of fructose (1.67 and 16.7 mM), distributions of single-cell GFP fluorescence were indistinguishable (not shown) from the one at 167 μM. This suggests that the activation of the PfruB-gfp[AAV] fusion became saturated at around 167 μM fructose. In fact, we suspect that activation of the fruB promoter in Eh299R (PfruB-gfp[AAV]) is already maximal at 1.67 μM fructose, because the frequency distributions at t = 1 h looked very similar for all concentrations of fructose that were tested. This does not rule out that the fruB promoter is induced dose-dependently at lower concentrations of fructose.
Figure 3.
Dynamics of single-cell GFP accumulation in growing cultures of Eh299R (pPfruB-gfp[AAV]) in response to different fructose concentrations. See the legend to Fig. 1 for induction conditions. The gray distribution curves in A represent the population at t = 0 and serve as a reference.
From the results of Fig. 3, we concluded that there are three features of the fructose bioreporter that are important for a meaningful interpretation of GFP data from environments with unknown fructose availabilities. First, due to intrapopulation variance in GFP signal in response to a uniform fructose availability, there may be overlap among populations of cells that experience different fructose concentrations. In other words, the dimmest cell in a (completely or partially) induced population might be as bright as the brightest cell in an uninduced population, without the latter ever being exposed to fructose. Second, GFP content merely presents a snapshot of a cell's activity. Depending on the availability of fructose at the time of the snapshot, GFP content may subsequently increase or decrease. And third, induction of the PfruB-gfp[AAV] fusion by fructose appears to be an all-or-none process that is independent of fructose concentration, at least above 1.67 μM. The implications of these features are that a single cell's GFP fluorescence must be interpreted (i) in the context of other cells within the same population, (ii) as a function of time, and (iii) as evidence of active fructose consumption rather than a measure for available fructose concentration.
Besides the sensitivity of the bioreporter, we determined its specificity. In addition to fructose, the disaccharide sucrose elicited accumulation of fluorescence although to a lesser extent (about 2-fold) than fructose alone (not shown). On uptake, sucrose is phosphorylated to sucrose 6-phosphate and subsequently hydrolyzed to glucose 6-phosphate plus fructose; intracellular conversion of the latter to fructose 1-phosphate would trigger activation of the PfruB-gfp fusion. Glucose, like galactose, was not an inducer (not shown). However, when added in concentrations that were equal to or higher than those of fructose, GFP accumulation rates in response to fructose were reduced approximately by half (not shown). These observations should caution against a direct comparison of GFP accumulation rates from the fructose induction experiments (Fig. 3) with those from bioreporter cells that are recovered from environments like the leaf surface in which sucrose and glucose concentrations are essentially unknown. Yet, the more important observation is that even in the presence of glucose or when fructose is available as sucrose, GFP accumulation remains indicative of fructose metabolism.
Fructose Consumption and Metabolic Activity of Eh299R (pPfruB-gfp[AAV]) on Bean Leaf Surfaces.
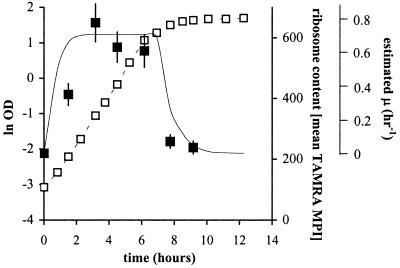
Bean leaves were inoculated by spraying with a suspension of galactose-grown cells, and at different times after inoculation, total epiphytic communities were collected from individual leaves and analyzed for single-cell GFP content using epifluorescence microscopy. To distinguish Eh299R from other bacterial leaf residents, we subjected the cells to a FISH protocol using a TAMRA-labeled probe that was specifically designed to target the 16S ribosomal RNA of strain Eh299R (43). This probe does not target bacteria that are indigenous to greenhouse-grown bean plant leaves (43). An additional advantage of the FISH procedure was that from the intensity of TAMRA fluorescence we were able to infer the metabolic state of individual Eh299R cells based on the relationship between growth rate and ribosome content (44, 45). This is illustrated for a growing culture of Eh299R (pPfruB-gfp[AAV]) in Fig. 4 where average single-cell TAMRA fluorescence is shown in relation to the growth stage and growth rate of the culture. Clearly, actively growing cells revealed a significantly higher ribosome content (average TAMRA MPI = 550–650) than stationary phase cells (average TAMRA MPI 200).
Figure 4.
Correlation between ribosome content and growth rate of a Eh299R (pPfruB-gfp[AAV]) culture in minimal medium on galactose. Shown as a function of time are cell density (□) and average single-cell TAMRA-fluorescence intensity (■). The solid curve represents the change in growth rate as estimated from the slope of the growth curve (μmax = 0.71 h−1).
In preliminary experiments, we had noticed that the FISH procedure slightly reduced GFP fluorescence in single cells. Accordingly, GFP fluorescence intensities of hybridized cells would not be directly comparable to those from the fructose induction experiment (Fig. 3). Therefore, each batch of cells that was recovered from the leaf surface was divided in two subsets. One subset was hybridized with the 16S rRNA probe and analyzed for both GFP and TAMRA fluorescence (Fig. 5) to establish a correlation between fructose consumption and active metabolism. The second subset was not hybridized and analyzed for GFP only (Fig. 6A) so that GFP accumulation rates could be compared with those observed in Fig. 3. We present here the results of a single plant experiment that are typical for the results we obtained from several independent repetitions. Typically, plate counts revealed that foliar populations of Eh299R (pPfruB-gfp[AAV]) increased approximately 10-fold between 1 and 7 h after inoculation, from an average of 1.0 × 106 to 1.1 × 107 colony-forming units/g leaf, respectively. This corresponds to an initial average growth rate of 0.4 h−1. Population sizes did not change significantly during the remainder of the experiment. This in planta behavior was similar to that of wild-type strain Eh299R (not shown), indicating that the production of GFP[AAV] had no adverse effect on the ability of Eh299R (pPfruB-gfp[AAV]) to colonize bean leaves.
Figure 5.
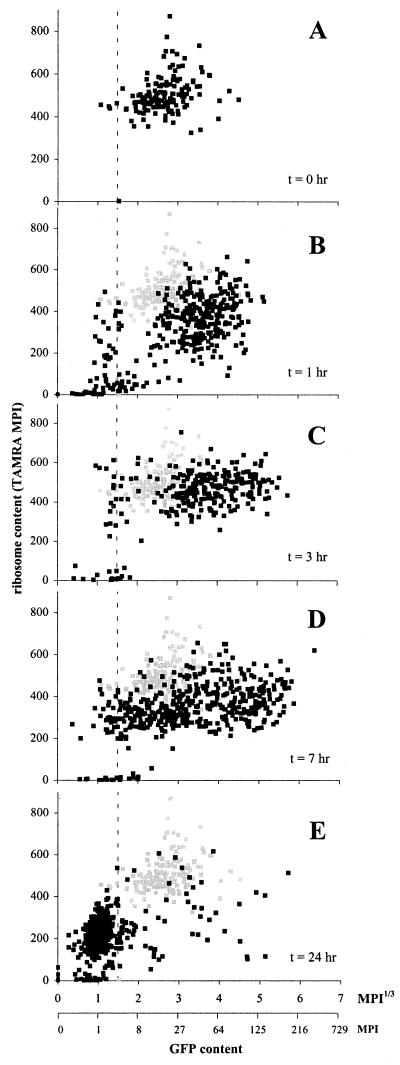
Scatter plot of GFP and TAMRA fluorescence for individual cells from epiphytic populations of Eh299R (pPfruB-gfp[AAV]) that were recovered from leaves 1, 3, 7, and 24 h after inoculation (t = 0). The gray scatter in B–E refers to the inoculum population. Because we observed no major differences in the distribution patterns among three leaves taken at each time point, we grouped, analyzed, and presented the data as if they were obtained from a single leaf. Broken lines indicate the limit of GFP detection.
Figure 6.
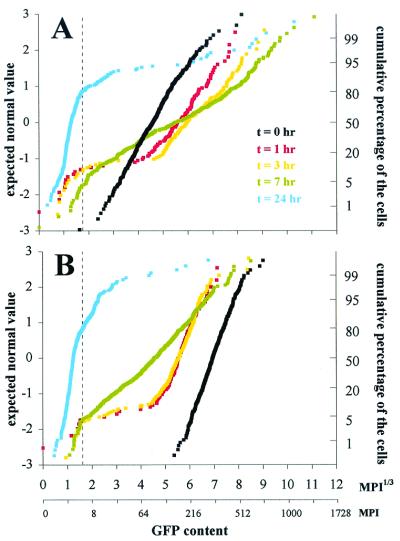
Normal probability plots of GFP fluorescence in single cells from epiphytic populations of Eh299R harboring pPfruB-gfp[AAV] (A) or pPnptII-gfp[AAV] (B). See text for details. Broken lines indicate the limit of GFP detection.
A simultaneous analysis of GFP and ribosomal content in single bioreporter cells from the leaf surface revealed that most Eh299R (pPfruB-gfp[AAV]) cells in the phyllosphere that were metabolically active were engaged in fructose consumption. In the inoculum (Fig. 5A), TAMRA fluorescence varied between 300 and 900, and GFP content averaged at a MPI of 20 (average MPI1/3 = 2.7). One hour after inoculation, 81% of all recovered cells had a ribosome content above 200 TAMRA units (Fig. 5B). Of these cells (all Eh299R), 96% contained GFP levels that were, on average, higher than those found within the inoculum, suggesting that these cells had started the consumption of fructose and/or sucrose. The other 4% of the Eh299R cells, however, showed no GFP fluorescence. We suspect that they no longer carried the plasmid pPfruB-gfp[AAV], but we cannot rule out that they were metabolizing a substrate other than fructose and sucrose. Of the 19% of the cells with a TAMRA signal below 200 units, about half had GFP levels above background, suggesting they were representative of dormant Eh299R cells exposed to very low initial fructose abundances. Only 3% of all recovered cells represented indigenous leaf residents, as judged from their complete lack of GFP and TAMRA fluorescence. Three hours after inoculation, 94% of all cells could be identified as metabolically active Eh299R cells (Fig. 5C). The average TAMRA signal closely but not entirely approached that of the inoculum, suggesting that the cells had become adapted to their new environment. The majority of these cells also showed increased GFP content, indicating active fructose catabolism. The portion of Eh299R cells seemingly without a plasmid remained constant at 5%. By 7 h, ribosomal content had dropped for most, but not all, of the Eh299R population (Fig. 5D), and at the same time, GFP content varied dramatically among cells. As a trend, however, cells with a low GFP content (MPI1/3 < 3) also contained less ribosomal RNA (TAMRA MPI < 400). By 24 h (Fig. 5E), the vast majority of Eh299R cells no longer contained significant levels of GFP and showed a low ribosomal content that was comparable to that of stationary phase cells in culture (Fig. 4). Still, 1–2% of all cells gave a GFP signal corresponding to MPI > 50 (MPI1/3 > 3.7) and a TAMRA intensity over 300, suggesting that these cells were still actively growing at the expense of fructose and/or sucrose. Also, ≈1% of all Eh299R cells had elevated GFP content (MPI > 50) but low ribosome (TAMRA MPI < 200) content. Perhaps these cells still had access to sugar, but became limited in another nutrient requirement such as nitrogen.
Quantitative Analysis of GFP Content and Accumulation Rate in Foliar Populations of Eh299R (pPfruB-gfp[AAV]).
For a more quantitative assessment of the dynamics of GFP accumulation on the leaf surface in comparison to fructose-induced cultures, we analyzed a subset of the foliar Eh299R (pPfruB-gfp[AAV]) population for GFP content without prior hybridization with the 16S rRNA probe. The data are presented in a normal probability plot (Fig. 6A). If all cells on the leaf had access to the same amount of fructose, we would expect to see the entire population move away from the inoculum as a straight and parallel line toward a higher average GFP content and shift back again after all fructose had been consumed. Although this appeared to be true for most cells during the first couple of hours on the leaf, it was no longer the case in later stages of leaf colonization (Fig. 6A). A large part of the curve at t = 7 appeared flatter than before, which indicates that in the 4-hr period after t = 3 many but not all cells stopped fructose-induced accumulation of GFP, and some did so earlier than others. As leaf colonization proceeded, a progressively smaller fraction of cells continued to accumulate GFP (Fig. 6A). We estimated from Fig. 6A that this fraction was reduced from approximately 84% of the total population at 1 h after inoculation to 11% after 7 h. To estimate heterogeneity in fructose availability, we exploited the observation that the time during which a reporter cell continues to accumulate GFP is indicative of the initial availability of fructose (Fig. 3). With an average foliar growth rate of 0.4 h−1, cells doubled on average 0.6 and 4.0 times in 1 and 7 h, respectively. Assuming that 0.3 pg of fructose is consumed during every cell doubling, this corresponds to initial single-cell fructose exposures of 0.15 and 4.6 pg. This represents a 30-fold range in initial sugar abundance to individual cells. At 24 h after inoculation, a small fraction of the cells still appeared to be induced (Fig. 6A). Under the same assumptions as above, the progenitors of these cells would have been exposed to 4.4 ng of fructose. At 48 h after inoculation, none of the approximately 1,000 analyzed cells were green fluorescent (not shown), indicating that less than 0.1% of the entire population was exposed to any significant amounts of fructose or sucrose.
The rate at which GFP accumulated in Eh299R bioreporter cells in the phyllosphere (Fig. 6A) was significantly lower than was observed for cells in culture (Fig. 3B). Possibly, the cells on the leaf were reporting sucrose, not fructose. Alternatively, the presence of glucose, the third major sugar in the phyllosphere, might have interfered with the sensing of available fructose. A third explanation, which is compatible with the other two, is that exposure to the leaf environment caused a slight reduction in all cellular activities, including that of promoters. This was confirmed by analysis of the in planta GFP expression pattern of Eh299R cells harboring pPnptII-gfp[AAV]. This plasmid is identical to pPfruB-gfp[AAV] except that the fruB promoter was replaced with the nptII promoter, which is constitutive and not responsive to fructose (not shown). The GFP content of Eh299R (pPnptII-gfp[AAV]) cells in the inoculum averaged around MPI1/3 = 7.0 (MPI = 343). In a normal probability plot (Fig. 6B), the curve was slightly steeper than that of the pPfruB-gfp[AAV] inoculum, indicating that nptII promoter activity varies less than that of PfruB. One and three hours after inoculation on bean plants, GFP content of Eh299R (pPnptII-gfp[AAV]) cells was reduced considerably compared with the inoculum population (Fig. 6B). The pPnptII-gfp[AAV] fusion probably acts as a kind of metabolic indicator, and its output should correlate loosely with ribosomal content. Indeed, at 7 h, there was great heterogeneity in GFP content among Eh299R (pPnptII-gfp[AAV]) cells, just as there was an increase in heterogeneity in the TAMRA signal among Eh299R cells on the leaf surface (Fig. 5D). At 24 h, most Eh299R (pPnptII-gfp[AAV]) cells were no longer active and contained no significant levels of GFP.
Direct Observations of Eh299R (PfruB-gfp[AAV]) in the Phyllosphere.
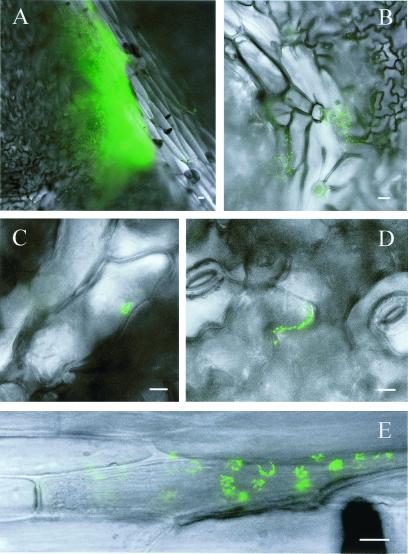
Our approach using Eh299R (pPfruB-gfp[AAV]) to quantify fructose consumption during leaf colonization presents itself with one obvious disadvantage: by removing bacteria from the leaf for analysis, any information regarding the spatial dimensions of fructose utilization was lost. We tried to gain a sense for the spatial element by scanning different parts of the leaf surface for the presence of green fluorescent bacteria (Fig. 7). Many fluorescent bacteria on the leaf could be found in water drops (Fig. 7A). Within such drops, we often observed clusters of green cells that appeared to be kept together by some kind of loose matrix. Such structures were commonly associated with trichomes (e.g., Fig. 7B) or veins. Consistently, we saw clusters of bacteria near vein cells (e.g., Fig. 7C), in cracks between larger veins, in crevices between epidermal cells (e.g., Fig. 7D), at the tip and base of trichomes, and near stomates. At 24 h after inoculation, we recorded many instances of green fluorescent cells associated with discolored plant cells in small veins (e.g., Fig. 7E) or in the epidermal cell layer. It is tempting to assume that these plant cells were compromised in the integrity of their membranes, cell walls, or in the cuticular layer above them, and that as a result they were leaking nutrients. It remains unclear whether the bacteria played an active role in this process.
Figure 7.
In situ observations of fructose consumption by Eh299R (pPfruB-gfp[AAV]) in the phyllosphere. Descriptions are given in the text. The bar in each photograph represents a distance of 10 μm.
From the images in Fig. 7 it cannot be appreciated whether there were Eh299R cells that were not exposed to sugars because these would appear dim. However, vast overexposure of the image in Fig. 7D revealed the presence of dim cells as close as 10 μm from the cluster of bright green cells (not shown). This suggests that sugar availability has a very local component indeed. We are presently using fructose bioreporter cells that harbor a second stable red fluorescent marker, which allows visualization of all bioreporter cells, whether they are reporting the consumption of fructose or not.
Discussion
Based on our observations with fructose bioreporter Eh299R (pPfruB-gfp[AAV]), we propose the following model for the role of local sugar availability in the colonization of bean plant leaves. On arrival to a previously uncolonized leaf surface, an individual cell has a good chance of finding itself in a place where it is presented with an amount of sugar that is sufficient to allow for a short and transient adaptation to its new environment. There usually remains ample sugar, predominantly in the form of fructose and/or sucrose, to then multiply and start colonization of the local area. As this fate is shared by most immigrants in other places on the same leaf, the total leaf population rapidly increases during the initial stage of leaf colonization. However, due to heterogeneity in initial availability, sugar pools are depleted earlier in some localities than in others. The majority of these pools are relatively small in size: almost all initial colonizers were exposed to at least 0.15 pg fructose equivalents, but only a few (roughly 11% or less) had access to abundances close to or exceeding 4.6 pg. As increasingly more cells run out of resources and cease to multiply, growth of the total population slows down and eventually halts. This suggests that sugar pools on previously uncolonized bean leaves are discontinuous, i.e., they are not replenished by the plant at rates that support continual fast growth of the bacteria.
What caused the differences in initial sugar availability is not clear. A first factor is local variation in sugar abundances. Most of the sugar that the reporter cells encountered on arrival had probably accumulated on the leaf surface before the immigration to the leaf i.e., during the 10 days that the plants developed in the greenhouse. Generally, these plants carry low indigenous epiphytic populations, so that most of the sugar remained unutilized and continued to build up to amounts that were large enough to support the establishment of 107 Eh299R bacteria per leaf. Differences in sugar accumulation probably can be related to microscopic structure of the leaf surface. Stomates, epidermal cells, and trichomes are dissimilarly layered with waxes and are therefore suspected of differential leakage (46). Photoassimilates may be found in higher abundances in the vicinity of veins where they are transported in concentrations as high as 0.3 to 0.9 M (47). Furthermore, the presence of water can stimulate leaching (20). Consequently, sites or structures that are more likely to retain water or dew, such as veins, trichomes, and crevices, also are expected to harbor more nutrients. Many of these are locations in which we observed bacteria engaged in fructose consumption (Fig. 7).
A second factor to be considered is the spatial pattern in which the cells first landed on the leaf. Our spray-inoculation did not necessarily deliver the bacteria evenly across the surface. Some arrived close together whereas others were more scattered. In the former case, a cell's share of the available sugar pool will be inversely proportional to the number of coimmigrants to the same site. If a cell can claim a spot for itself, however, it could profit from the available fructose for a longer time and produce substantial progeny. Not much is known about how cells immigrate to leaves outside the laboratory, whether they arrive as single cells or in aggregates. Apparently, many airborne bacteria occur in groups of more than one (48). As argued above, such a pattern of immigration might expose groups of cells to different nutrient availabilities than solitary immigrants.
Faced with a condition that does not favor microbial growth or survival, Eh299R bioreporter cells would probably fail to report even if they were exposed to copious amounts of fructose. Examples of such adverse conditions are water stress and the presence of inhibitory substances like toxins. Local areas on the leaf may differ in relative humidity or presence of toxins, and therefore may be differentially conducive to fructose reporting. However, we should immediately point out that this property of the bioreporter is essential when one is interested, as we are, in the biological relevance of sugar availability. Cells that have access to a resource such as fructose but are hindered from using it for whatever reason will not consume the resource, will not multiply, and will therefore not contribute to growth of the population. In other words, the sugar may be available but is not exploitable. As microbes differ in their resistance to adverse conditions, so might their ability to use available sugar.
We plan to use the Eh299R (pPfruB-gfp[AAV]) reporter system in determining the factors that influence the availability of nutrients. One interesting application of the fructose bioreporter is to assess whether and how bacteria modify their local surroundings on the leaf surface. Like many other leaf-associated bacteria, E. herbicola is capable of synthesis of the plant hormone 3-indole acetic acid (IAA), or auxin (30). It has been suggested that this trait helps the bacterium by inducing the plant to release nutrients (49). The green fluorescent bacteria in Fig. 7E might well represent an example of such IAA-induced leakage. Clearly, the Eh299R (pPfruB-gfp[AAV]) fructose bioreporter system offers tremendous potential for the study of microbe-microbe and host-microbe interactions in the phyllosphere and possibly other habitats, awarding us with an exciting opportunity to view such interactions from the unique perspective of the microbes themselves.
Acknowledgments
We thank Steve Ruzin and Denise Schichnes at the College of Natural Resources Biological Imaging Center for their support with epifluorescence microscopy. We are indebted to Maria Marco, Joyce Loper, and Jan Roelof van der Meer for helpful comments on the manuscript. Thanks also to Dr. Bridget Coughlin at PNAS for her suggestions. This research was funded in part by United States Department of Agriculture National Research Initiative Grant 96–35303-3377 and Grant DE-FG03–86ER13518 from the U.S. Department of Energy.
Abbreviations
- GFP
green fluorescent protein
- FISH
fluorescence in situ hybridization
- TAMRA
tetramethylrhodamine
- MPI
mean pixel intensity
Footnotes
This contribution is part of the special series of Inaugural Articles by members of the National Academy of Sciences elected on April 27, 1999.
References
- 1.Ruinen J. Plant Soil. 1961;15:81–109. [Google Scholar]
- 2.Ellis R J, Thompson I P, Bailey M J. FEMS Microbiol Ecol. 1999;28:345–356. [Google Scholar]
- 3.Hirano S S, Upper C D. Appl Environ Microbiol. 1989;55:623–630. doi: 10.1128/aem.55.3.623-630.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kinkel L L, Wilson M, Lindow S E. Microb Ecol. 2000;39:1–11. doi: 10.1007/s002489900182. [DOI] [PubMed] [Google Scholar]
- 5.Kinkel L L. Annu Rev Phytopathol. 1997;35:327–347. doi: 10.1146/annurev.phyto.35.1.327. [DOI] [PubMed] [Google Scholar]
- 6.Lindow S E. In: Aerial Plant Surface Microbiology. Morris C E, Nicot P C, Nguyen-The C, editors. New York: Plenum; 1996. pp. 155–168. [Google Scholar]
- 7.Hirano S S, Baker L S, Upper C D. Appl Environ Microbiol. 1996;62:2560–2566. doi: 10.1128/aem.62.7.2560-2566.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lilley A K, Hails R S, Cory J S, Bailey M J. FEMS Microbiol Ecol. 1997;24:151–157. [Google Scholar]
- 9.Lindemann J, Upper C D. Appl Environ Microbiol. 1985;50:1229–1232. doi: 10.1128/aem.50.5.1229-1232.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Walker J C, Patel P N. Phytopathology. 1964;54:140–141. [Google Scholar]
- 11.Beattie G A, Lindow S E. Annu Rev Phytopathol. 1995;33:145–172. doi: 10.1146/annurev.py.33.090195.001045. [DOI] [PubMed] [Google Scholar]
- 12.Hirano S S, Upper C D. Microbiol Mol Biol Rev. 2000;64:624–653. doi: 10.1128/mmbr.64.3.624-653.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beattie G A, Lindow S E. Phytopathology. 1999;89:353–359. doi: 10.1094/PHYTO.1999.89.5.353. [DOI] [PubMed] [Google Scholar]
- 14.Sundin G W, Jacobs J L. Microb Ecol. 1999;38:27–38. doi: 10.1007/s002489900152. [DOI] [PubMed] [Google Scholar]
- 15.Ophir T, Gutnick D L. Appl Environ Microbiol. 1994;60:740–745. doi: 10.1128/aem.60.2.740-745.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu J, Penaloza-Vazquez A, Chakrabarty A M, Bender C L. Mol Microbiol. 1999;33:712–720. doi: 10.1046/j.1365-2958.1999.01516.x. [DOI] [PubMed] [Google Scholar]
- 17.Stadler B, Mueller T. Can J Forest Res. 2000;30:631–638. [Google Scholar]
- 18.Warren R C. Neth J Plant Pathol. 1972;78:89–98. [Google Scholar]
- 19.Derridj S. In: Aerial Plant Surface Microbiology. Morris C E, Nicot P C, Nguyen-The C, editors. New York: Plenum; 1996. pp. 25–42. [Google Scholar]
- 20.Tukey H B., Jr Annu Rev Plant Physiol. 1970;21:305–324. [Google Scholar]
- 21.Dik A J, Fokkema N J, van Pelt J A. Neth J Plant Pathol. 1991;97:209–232. [Google Scholar]
- 22.Rodger G, Blakeman J P. Trans Br Mycol Soc. 1984;82:45–51. [Google Scholar]
- 23.Wilson M, Savka M A, Hwang I, Farrand S K, Lindow S E. Appl Environ Microbiol. 1995;61:2151–2158. doi: 10.1128/aem.61.6.2151-2158.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mercier J, Lindow S E. Appl Environ Microbiol. 2000;66:369–374. doi: 10.1128/aem.66.1.369-374.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kinkel L, Wilson M, Lindow S E. Microb Ecol. 1995;29:283–297. doi: 10.1007/BF00164891. [DOI] [PubMed] [Google Scholar]
- 26.Weller D M, Saettler A W. Phytopathology. 1980;70:500–506. [Google Scholar]
- 27.Davis C L, Brlansky R H. Appl Environ Microbiol. 1991;57:3052–3055. doi: 10.1128/aem.57.10.3052-3055.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mansvelt E L, Hattingh M J. Can J Bot. 1987;65:2517–2522. [Google Scholar]
- 29.Morris C E, Monier J-M, Jacques M-A. Appl Environ Microbiol. 1997;63:1570–1576. doi: 10.1128/aem.63.4.1570-1576.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brandl M T, Lindow S E. Appl Environ Microbiol. 1996;62:4121–4128. doi: 10.1128/aem.62.11.4121-4128.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reizer J, Reizer A, Kornberg H L, Saier M H. FEMS Microbiol Lett. 1994;118:159–161. doi: 10.1111/j.1574-6968.1994.tb06819.x. [DOI] [PubMed] [Google Scholar]
- 32.Saier M H, Ramseier T M. J Bacteriol. 1996;178:3411–3417. doi: 10.1128/jb.178.12.3411-3417.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ramseier T M. Res Microbiol. 1996;147:489–493. doi: 10.1016/0923-2508(96)84003-4. [DOI] [PubMed] [Google Scholar]
- 34.Tsien R Y. Annu Rev Biochem. 1998;67:509–544. doi: 10.1146/annurev.biochem.67.1.509. [DOI] [PubMed] [Google Scholar]
- 35.Andersen J B, Sternberg C, Kongsbak-Poulsen L, Petersen-Bjorn S, Givskov M, Molin S. Appl Environ Microbiol. 1998;64:2240–2246. doi: 10.1128/aem.64.6.2240-2246.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ramos C, Molbak L, Molin S. Appl Environ Microbiol. 2000;66:801–809. doi: 10.1128/aem.66.2.801-809.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sternberg C, Christensen B B, Johansen T, Nielsen T, Andersen J B, Givskov M, Molin S. Appl Environ Microbiol. 1999;65:4108–4117. doi: 10.1128/aem.65.9.4108-4117.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sambrook J, Maniatis T, Fritsch E F. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 39.Miller W G, Leveau J H J, Lindow S E. Mol Plant–Microbe Interact. 2000;13:1243–1250. doi: 10.1094/MPMI.2000.13.11.1243. [DOI] [PubMed] [Google Scholar]
- 40.Better M, Helinski D R. J Bacteriol. 1983;155:311–316. doi: 10.1128/jb.155.1.311-316.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Akkermans A D L, van Elsas J D, de Bruijn F J. Molecular Microbial Ecology Manual. Dordrecht, The Netherlands: Kluwer; 1996. [Google Scholar]
- 42.O'Brien R D, Lindow S E. Phytopathology. 1989;79:619–627. [Google Scholar]
- 43.Brandl M T, Quiñones B, Lindow S E. Proc Natl Acad Sci USA. 2001;98:3454–3459. doi: 10.1073/pnas.061014498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Neidhardt F C, Ingraham J L, Schaechter M. Physiology of the Bacterial Cell: A Molecular Approach. Sunderland, MA: Sinauer; 1990. [Google Scholar]
- 45.Ruimy R, Breittmayer V, Boivin V, Christen R. FEMS Microbiol Ecol. 1994;15:207–213. [Google Scholar]
- 46.Schoenherr J, Bauer P. In: Aerial Plant Surface Microbiology. Morris C E, Nicot P C, Nguyen-The C, editors. New York: Plenum; 1996. pp. 1–24. [Google Scholar]
- 47.Taiz L, Zeiger E. Plant Physiology. Sunderland, MA: Sinauer; 1998. [Google Scholar]
- 48.Lighthart B. FEMS Microbiol Ecol. 1997;23:263–274. [Google Scholar]
- 49.Brandl M T, Lindow S E. Appl Environ Microbiol. 1998;64:3256–3263. doi: 10.1128/aem.64.9.3256-3263.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]