Abstract
Autotrophic CO2 fixation represents the most important biosynthetic process in biology. Besides the well-known Calvin-Benson cycle, five other totally different autotrophic mechanisms are known today. This minireview discusses the factors determining their distribution. As will be made clear, the observed diversity reflects the variety of the organisms and the ecological niches existing in nature.
Autotrophic CO2 fixation represents the most important biosynthetic process in nature, being responsible for the net fixation of 7 × 1016 g carbon annually, corresponding to the conservation of 2.8 × 1018 kJ of energy (107). The photosynthetic path of carbon in algae and plants was elucidated in the laboratory of Melvin Calvin in the 1940s and 1950s; the discovered reductive pentose phosphate (Calvin-Benson [CB]) cycle was immediately proposed to be the universal autotrophic carbon dioxide assimilation pathway, and the presence of its key enzyme, ribulose-1,5-bisphosphate carboxylase, was regarded as a synonym for autotrophy. This idea fit perfectly with the central biological dogma of that time, the biochemical unity of life (74). However, already in 1966 the second autotrophic CO2 fixation cycle (the reductive citric acid cycle) had been discovered in the laboratory of Daniel Arnon (33). Today, six autotrophic CO2 fixation mechanisms are known, raising the question of why so many pathways are necessary. In this review, the factors determining their distribution are discussed. As will be made clear, the observed diversity reflects the variety of the organisms and the ecological niches existing in nature. First, the general aspects of autotrophic CO2 fixation will be discussed; then the known pathways will be introduced in the order of their discovery, highlighting their main characteristic features; finally, the main factors determining their distribution will be summarized. For those interested in a more detailed discussion of different aspects of autotrophy, several highly valuable reviews published in the last few years can be recommended, covering the function, structure, and evolution of ribulose-1,5-bisphosphate carboxylase (7, 101-103, 111) and carboxysomes (61, 126); the reductive acetyl coenzyme A (acetyl-CoA) pathway (25, 78, 80) and the reductive citric acid cycle (2); CO2 fixation in Archaea (14) and in deep-sea vent chemoautotrophs (73); and autotrophy in various oceanic ecosystems (47).
GENERAL ASPECTS OF AUTOTROPHIC CO2 FIXATION
In general terms, the assimilation of CO2 (oxidation state of +4) into cellular carbon (average oxidation state of 0, as in carbohydrates) requires four reducing equivalents. An input of energy is also required for the reductive conversion of CO2 to cell carbon and is provided by ATP hydrolysis. Anaerobes often use low-potential electron donors like reduced ferredoxin for CO2 fixation, whereas aerobes usually rely on NAD(P)H as a reductant. Since reduced ferredoxin bears more energy than NADPH (ferredoxin, E0′ ≈ −400 mV; NADPH, E0′ = −320 mV), aerobic pathways usually require more ATP equivalents than anaerobic ones. A carboxylating enzyme links either CO2 or HCO3− with an organic acceptor molecule, which must be regenerated in the following steps of the pathway. These inorganic carbon species are related by the pH-dependent equilibrium CO2 + H2O ↔ H2CO3 ↔ H+ + HCO3− ↔ 2H+ + CO32−, with apparent pKa [HCO3−/ CO2] = 6.3. Therefore, the bicarbonate concentration under slightly alkaline conditions (e.g., in marine water) is much higher than the concentration of dissolved CO2, and the usage of bicarbonate instead of CO2 may be advantageous; the solubility of CO2 is also affected by the salt concentration and temperature. The product of the autotrophic pathway that is drained off should be a central metabolite, from which carbohydrates, proteins, nucleic acids, and lipids can be derived. However, these macromolecules are made from different building blocks, and their content varies in different organisms, making one CO2 fixation product/pathway preferable to another.
The enzymes catalyzing mechanistically difficult steps in a given pathway (e.g., ribulose-1,5-bisphosphate carboxylase, CO dehydrogenase/acetyl-CoA synthase) evolved just once during the evolution, are conserved, and are often regarded as its key enzymes. They may be present in large amounts in the cell. Their synthesis itself devours a lot of energy and may actually determine the energy costs of the pathway. In contrast, noncharacteristic enzymes belong to large enzyme families (carboxylic acid-CoA ligases, enoyl-CoA hydratases, different dehydrogenases, etc.). The corresponding genes may easily be recruited from the gene pools and mutated in order to perform the required function.
ROUTE 1: THE CALVIN-BENSON REDUCTIVE PENTOSE PHOSPHATE CYCLE
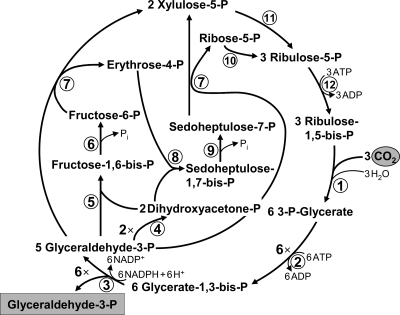
The reductive pentose phosphate cycle is the quantitatively most important mechanism of autotrophic CO2 fixation in nature. It is centered around carbohydrates and can primarily be found in organisms synthesizing large ammounts of sugars (i.e., plants). High detachment of the CB cycle from the rest of the central carbon metabolism allows its effective regulation. The success of the CB cycle was probably governed by the full robustness of its enzymes to molecular oxygen. The key enzymes are ribulose-1,5-bisphosphate carboxylase/oxygenase (RubisCO; 121) and phosphoribulokinase. In addition, the sedoheptulose bisphosphatase reaction can also be regarded as specific for the cycle. RubisCO catalyzes the electrophilic addition of CO2 to the C5 sugar ribulose-1,5-bisphosphate in its enediolate form, giving rise to an unstable C6 intermediate (23). The latter is subsequently spontaneously hydrolyzed into two molecules of 3-phosphoglycerate, which are further reduced with NADPH to glyceraldehyde-3-phosphate. This reduction is catalyzed by two gluconeogenic reactions and driven by ATP hydrolysis. The regeneration part of the cycle comprises the interconversion of triose phosphates via various sugar phosphates to ribulose-5-phosphate, which is phosphorylated by phosphoribulokinase to ribulose-1,5-bisphosphate, closing the cycle (Fig. 1). The cycle requires nine ATP equivalents and six NADPHs for the synthesis of one glyceraldehyde-3-phosphate molecule.
FIG. 1.
The reductive pentose phosphate (Calvin-Benson) cycle. Enzymes: 1, ribulose-1,5-bisphosphate carboxylase/oxygenase; 2, 3-phosphoglycerate kinase; 3, glyceraldehyde-3-phosphate dehydrogenase; 4, triose-phosphate isomerase; 5, fructose-bisphosphate aldolase; 6, fructose-bisphosphate phosphatase; 7, transketolase; 8, sedoheptulose-bisphosphate aldolase; 9, sedoheptulose-bisphosphate phosphatase; 10, ribose-phosphate isomerase; 11, ribulose-phosphate epimerase; and 12, phosphoribulokinase.
The CB cycle operates in some thermophiles but never in hyperthermophiles—its upper temperature limit appears to be at ∼70 to 75°C. This may be explained by the heat instability of some intermediates of the cycle, mainly of glyceraldehyde-3-phosphate, producing toxic methylglyoxal at high temperature (50, 75). The CB cycle operates in plants, algae, cyanobacteria, and many aerobic or facultative aerobic proteobacteria belonging to the alpha, beta, and gamma subgroups. It was also shown for Sulfobacillus spp., iron and sulfur-oxidizing members of the firmicutes (19, 129), some mycobacteria (65), and green non-sulfur bacteria of the genus Oscillochloris (phylum Chloroflexi) (12, 51). The CB cycle not only functions in autotrophic CO2 fixation but is also required as an electron sink for (anaerobic) photoheterotrophic growth of some purple bacteria (e.g., Rhodobacter, Rhodospirillum, and Rhodopseudomonas) on substrates, which are more reduced than the average cell carbon (54, 118).
The key CB cycle enzyme, RubisCO, is the most abundant protein in the world (29), as it can comprise up to 50% of the total soluble protein in chloroplasts of a C3 plant or in bacteria using this cycle (85, 121). This fact is a consequence of the notorious catalytic inefficiency of RubisCO—i.e., low affinity for CO2, slow catalytic turnover rate (kcat = 1 to 12 s−1) (7), and a wasteful oxygenase side reaction responsible for photorespiration, resulting in a futile cleavage of the substrate to form toxic phosphoglycolate (1) as a side product. The assimilation (or detoxification) of phosphoglycolate starts with its conversion to glyoxylate, which can only be returned to the metabolism by wasting fixed carbon (1 CO2 per 2 glyoxylate molecules). The presence of three different routes for glyoxylate assimilation in cyanobacteria emphasizes the importance of this scavenging pathway in the metabolism (27) (Fig. 2). Although the precise loss of carbon caused by the photorespiration in bacteria is not known, it was estimated to be 21% of the net CO2 fixation for C3 plants (21).
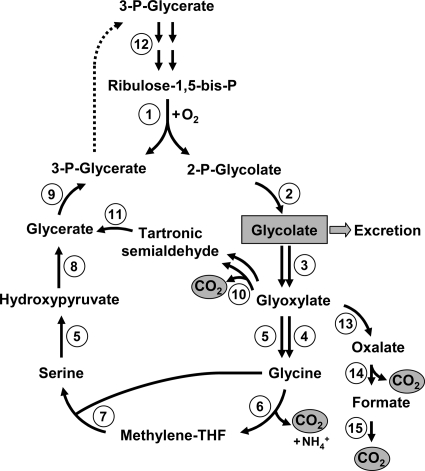
FIG. 2.
Photorespiration. RubisCO-catalyzed oxygenation of ribulose-1,5-bisphosphate (enzyme 1) leads to the formation of 3-phosphoglycerate and 2-phosphoglycolate. 3-Phosphoglycerate returns to the CB cycle, whereas 2-phosphoglycolate is converted to glycolate by 2-phosphoglycolate phosphatase (enzyme 2). Glycolate can either be excreted or oxidized to glyoxylate by glycolate dehydrogenase or glycolate oxidase (enzyme 3). In the classical pathway functioning in, e.g., plants, glyoxylate is converted to glycine by glutamate-glyoxylate aminotransferase (enzyme 4) and serine-glyoxylate transaminase (enzyme 5). One molecule of glycine is then oxidized by the glycine decarboxylase complex (enzymes 6) to methylene-tetrahydrofolate (THF), which reacts with the second glycine molecule in the reaction, catalyzed by serine hydroxymethyltransferase (enzyme 7). The resulting serine is transaminated to hydroxypyruvate (enzyme 5) and then converted to 3-phosphoglycerate by hydroxypyruvate reductase (enzyme 8) and glycerate kinase (enzyme 9). An alternative pathway of glyoxylate assimilation is the condensation of two glyoxylate molecules giving rise to tartronic semialdehyde (glyoxylate carboligase [enzyme 10]). Tartronic semialdehyde reductase reaction (enzyme 11) leads to glycerate, and the latter is converted to 3-phosphoglycerate. Ribulose-1,5-bisphosphate regeneration proceeds via the CB cycle reactions (step 12). In some cases, glyoxylate can also be completely oxidized to CO2 in reactions catalyzed by hydroxyacid dehydrogenase (enzyme 13), oxalate decarboxylase (enzyme 14), and formate dehydrogenase (enzyme 15). All of these pathways can be found in cyanobacteria (27).
These considerations can help us to understand the requirement of concentrating mechanisms for inorganic carbon in organisms using the CB cycle. These mechanisms involve transmembrane pumps actively concentrating bicarbonate inside the cell (for a review, see references 8 and 77). Furthermore, in order to increase the CO2 concentration in the vicinity of RubisCO and decrease photorespiration, some prokaryotes evolved special carbon-fixing organelle-like microcompartments called carboxysomes. Carboxysomes are filled with RubisCO. They are present in cyanobacteria, where a saturated oxygen concentration occurs, and in some (aerobic chemoautotrophic) proteobacteria (7, 94, 95). The shell of these bacterial microcompartments is easily permeable for the negatively charged RubisCO substrates/products (ribulose-1,5bisphosphate and bicarbonate, 3-phosphoglycerate) but less for oxygen, which allows depletion of O2 from the RubisCO active site (60). Besides RubisCO, carboxysomes contain carbonic anhydrase, which converts bicarbonate to CO2, the actual RubisCO substrate (76).
Crassulacean acid metabolism and C4 photosynthesis functioning in many higher plants may also be regarded as carbon-concentrating mechanisms (59). The primary carboxylating enzyme in these pathways is phosphoenolpyruvate (PEP) carboxylase, which is much faster than RubisCO, uses bicarbonate instead of CO2 as a substrate and has a much lower Km value for the inorganic carbon (56). Although these mechanisms require almost twice as much ATP as the “classical” CB cycle, ATP supply is not the major problem for phototrophs. Their growth is rather limited by the supply of water, phosphorus, nitrogen, or iron.
Forms of RubisCO.
Photorespiration in an oxygen-containing atmosphere is probably inevitable, because RubisCO is unable to strictly discriminate between the two fairly similar CO2 and O2 molecules (68). Futhermore, even the fastest RubisCOs adapted to anoxic conditions have a low catalytic efficiency (63). Nevertheless, RubisCO properties are shaped by selective pressure, and all RubisCOs seem to be optimized to their environment (7, 106). The improvement of a given property (e.g., of Km to CO2, kcat, or decrease of the photorespiration rate) comes at some cost to other properties. In other words, for the optimized enzyme, an increase of the maximal velocity leads to a decrease in the affinity for the substrate(s). Furthermore, increase of the carboxylation/oxygenation ratio can be exerted at the expense of the overall reaction rate, and more discriminatory RubisCOs adapted to a lower CO2/O2 ratio are slower (106). As a consequence of these factors, different forms of the enzyme with different catalytic properties evolved, and some organisms express different RubisCO isoenzymes, depending on the growth conditions. The preference of RubisCO for CO2 versus O2 is represented by the specificity factor Ω (Ω = VcKo/VoKc), which is the ratio of the catalytic efficiency (Vmax/Km ) for the carboxylase (Vc/Kc) and oxygenase (Vo/Ko) reactions.
Up to now, four forms of RubisCO had been recognized, with forms I to III being bona fide ribulose-1,5-bisphosphate carboxylases (101). The fastest of all are the enzymes of Archaea belonging to form III (kcat = 23 s−1 for Archaeoglobus enzyme), which have, however, unusually high affinity to oxygen (Ω = 4) (63). Form III RubisCO is not known to participate in autotrophy, and its function in metabolism is at issue (14, 86, 101). RubisCOs participating in autotrophic CO2 assimilation via the CB cycle belong to form I or II. Form I consists of large (L) and small (S) subunits and has an L8S8 structure, whereas form II contains only the large subunit (L2). The small subunits influence the catalytic efficiency and specificity of RubisCO (96), and the form I enzyme usually has lower Vmax and kcat but also higher specificity to CO2, than the form II RubisCO (7, 43, 53, 106). Consequently, the specificity factor Ω for form II enzymes (Ω = 10 to 15) is much lower than that for form I (Ω = 25 to 75 in bacteria, ∼80 in higher plants, and 100 to 240 in nongreen algae) (100).
Purple non-sulfur bacteria possessing RubisCO form II (i.e., Rhodobacter, Rhodospirillum, and Rhodopseudomonas) preferentially grow photoheterotrophically under anoxic conditions (7, 64, 101). In addition, many of them have form I enzyme, which is used in response to carbon limitation (54). Only the form II enzyme is present in Magnetospirillum strains; accordingly, they grow autotrophically under microoxic conditions, where the oxygenase side reaction of the form II is not of great importance (37). The obligately autotrophic hydrogen-oxidizing gammaproteobacterium Hydrogenovibrio marinus possesses three sets of RubisCO: i.e., one copy of the form II enzyme and two of form I—cytoplasmatic and carboxysome associated. They are expressed depending on the inorganic carbon supply, with form II RubisCO exclusively expressed under very high CO2 conditions (15%), carboxysomal form I at 0.15% CO2, and cytoplasmatic form I in between (note, however, that all three forms are synthesized at ambient air CO2 concentration) (127). Inorganic carbon limitation can also be a result of alkaliphilic growth conditions, where inorganic carbon exists preferentially in the form of bicarbonate, which is not a RubisCO substrate. The occurrence of different RubisCO forms in the phylogenetic group Hydrogenovibrio/Thiomicrospira/Thioalkalimicrobium illustrates the importance of this factor. Hydrogenovibrio and related neutrophilic species with three RubisCO genes may probably represent an ancestral status for this group (110); alkalitolerant Thiomicrospira pelophila lost the cytoplasmatic form I RubisCO, whereas obligate alkaliphilc species possess only the carboxysome-associated enzyme, which is adapted to support better growth at low CO2 concentration (110).
ROUTE 2: THE REDUCTIVE CITRIC ACID CYCLE (ARNON-BUCHANAN CYCLE)
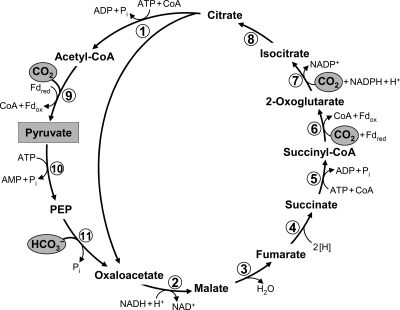
The reductive citric acid cycle (or reductive tricarboxylic acid [rTCA] cycle) was first proposed for the green sulfur bacterium Chlorobium limicola (Chlorobi) (33, 36, 52). Later it was found in anaerobic or microaerobic members of various other phyla such as Aquificae (11, 49, 92), Proteobacteria (especially of the delta and epsilon subdivisions) (20, 48, 70, 89, 104, 122), and Nitrospirae (e.g., Nitrospira and Leptospirillum) (38, 66, 69). This cycle may in fact be widespread among anaerobic or microaerobic bacteria, but until now it had not been found in the archaeal domain (14, 81). It reverses the reactions of the oxidative citric acid cycle (Krebs cycle) and forms acetyl-CoA from two CO2s (Fig. 3). Three reactions of the conventional Krebs cycle are considered to be irreversible and have to be substituted by other reactions/enzymes to reverse the cycle: succinate dehydrogenase is replaced by fumarate reductase, NAD+-dependent 2-oxoglutarate dehydrogenase by ferredoxin-dependent 2-oxoglutarate synthase, and citrate synthase by ATP-citrate lyase (33, 34, 52). These three enzymes are usually regarded as the characteristic enzymes of the cycle.
FIG. 3.
The reductive citric acid (Arnon-Buchanan) cycle as it functions in green sulfur bacteria (33, 52). The pathway of acetyl-CoA assimilation to pyruvate, phosphoenolpyruvate (PEP), and oxaloacetate is shown as well. For deviations from this variant of the cycle, see the text. Enzymes: 1, ATP-citrate lyase; 2, malate dehydrogenase; 3, fumarate hydratase; 4, fumarate reductase (natural electron donor is not known); 5, succinyl-CoA synthetase; 6, ferredoxin (Fd)-dependent 2-oxoglutarate synthase; 7, isocitrate dehydrogenase; 8, aconitate hydratase; 9, Fd-dependent pyruvate synthase; 10, PEP synthase; 11, PEP carboxylase.
The primary CO2 fixation product of the cycle, acetyl-CoA, must be further converted to other central intermediates of the carbon metabolism: i.e., to pyruvate/phosphoenolpyruvate (PEP), oxaloacetate, and 2-oxoglutarate. Therefore, bacteria using the rTCA cycle require additional enzymes for acetyl-CoA assimilation. Acetyl-CoA is reductively carboxylated to pyruvate by ferredoxin-dependent pyruvate synthase. Pyruvate can be further converted to PEP. Oxaloacetate is synthesized in pyruvate or PEP carboxylase reactions (33, 92) (Fig. 3).
Although most of the species using this cycle are mesophiles, representatives of the Aquificae are thermophiles growing best at 70°C or above (Aquifex aeolicus up to 95°C). The most heat-labile intermediate of the cycle is succinyl-CoA. Its hydrolysis does not result in the formation of any toxic intermediate and just leads to succinate and CoA, but it eventually results in a loss of energy, becoming stronger with temperature increase. In the classical Chlorobium variant of the cycle (Fig. 3), the reactions involved in further conversion of succinyl-CoA (up to oxaloacetate and acetyl-CoA) work preferably in the reverse direction. Consequently, heat-labile succinyl-CoA (24) forms a large pool and cannot be effectively removed and trapped in stabile intermediates of the pathway. Aquificae (as studied in Hydrogenobacter thermophilus) solve this problem by investing additional ATP in the conversion of 2-oxoglutarate to isocitrate by the combined action of an irreversible biotin-dependent 2-oxoglutarate carboxylase and a nondecarboxylating isocitrate dehydrogenase (3, 6). Together with low Km values of 2-oxoglutarate synthase for succinyl-CoA (125), this probably makes the process reasonably effective and irreversible at elevated temperatures. Other (mesophilic) bacteria using the rTCA cycle seem to possess normal reversible (decarboxylating) isocitrate dehydrogenase for the conversion of 2-oxoglutarate to isocitrate (57).
In some species, there are further minor modifications of the cycle. Citrate cleavage can be catalyzed by two enzymes, citryl-CoA synthetase and citryl-CoA lyase, rather than by ATP-citrate lyase. Citryl-CoA synthetase and citryl-CoA lyase are, however, phylogenetically related to ATP-citrate lyase (4, 5, 49). They were found in representatives of Aquificaceae (but not in other Aquificae) (4, 5, 49) and proposed for Leptospirillum (66). Furthermore, some proteobacteria (e.g., Magnetococcus sp. strain MC-1) probably have a novel type of ATP-citrate lyase (47, 70, 91).
The rTCA cycle uses both reduced ferredoxin and NAD(P)H as electron donors. Although the native electron donor for fumarate reductase in Chlorobium is not known, it was shown to be NADH in Hydrogenobacter (72). The cycle requires (at least in Chlorobium) only two ATP equivalents to form pyruvate (and three additional ATPs to convert it to triosephosphates) and is therefore less energy-consuming than the CB cycle. Interestingly, the usage of the rTCA cycle fits perfectly with the lifestyle of green sulfur bacteria. These bacteria possess a FeS photosynthetic reaction center type I capable of direct reduction of ferredoxin (18). Furthermore, they are adapted to low light intensities (10, 113), and ATP supply may therefore be a factor limiting their growth. This is in a striking contrast to, e.g., purple bacteria, which use the CB cycle (and the quinone reaction center type II, which produces the weak reductant [i.e., reduced quinone]) and are adapted to high light conditions.
Desulfobacter hydrogenophilus uses the rTCA cycle (with ATP-citrate lyase and 2-oxoglutarate:ferredoxin oxidoreductase), not only for autotrophic CO2 fixation, but also in the reversed (oxidative) direction for acetyl-CoA oxidation (89, 109). This underlines the absence of the irreversible steps in the cycle. Interestingly, in acetate oxidation in D. hydrogenophilus, succinyl-CoA is used for substrate activation by a CoA-transferase, and ATP synthesis during condensation of acetyl-CoA and oxaloacetate catalyzed by ATP-citrate lyase is the only substrate phosphorylation step (89, 109).
Although the rTCA cycle was first discovered in Chlorobium, it was most intensively studied in the thermophilic aerobic hydrogen-oxidizing bacterium Hydrogenobacter thermophilus (Aquificae) (2-6, 72, 92, 124, 125). How can aerobic organisms make use of enzymes requiring reduced ferredoxin for their activity, growing at up to 40% of oxygen in the gas phase? The solubility of oxygen at the optimum growth temperature of Hydrogenobacter (70°C) is reduced, especially in the cytoplasm, which is protected by active respiration. Furthermore, the organism possesses special biochemical adaptations to oxic conditions, such as two isoforms of 2-oxoglutarate:ferredoxin oxidoreductase with different sensitivities to molecular oxygen (124). Under oxic conditions, a five-subunit form of the enzyme (that is O2 tolerant) is preferentially synthesized. However, its robustness comes at the expense of a >5-times-lower specific activity, compared to the two-subunit anaerobic 2-oxoglutarate:ferredoxin oxidoreductase (125). Therefore, this aerobic izoenzyme may probably constitute a substantial part of the cellular soluble protein under oxic conditions. Interestingly, a similar five-subunit 2-oxoglutarate:ferredoxin oxidoreductase was identified in the genomes of aerobic Nitrospira and Leptospirillum spp. (66, 69). In addition, some still unknown mechanisms may contribute to the O2 robustness of the cycle in aerobes; however, this protection may significantly increase the energy demands of the cycle. Growth of Nitrospira and Leptospirillum in biofilms and flocs could offer additional protection from ambient O2 (69). Note that the mechanism for ferredoxin reduction in these aerobic organisms is not known.
ROUTE 3: THE REDUCTIVE ACETYL-COA (WOOD-LJUNGDAHL) PATHWAY
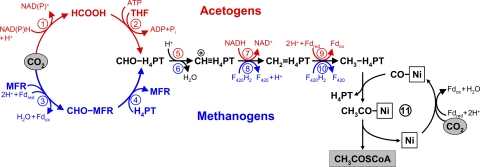
The Wood-Ljungdahl pathway is a noncyclic pathway that results in the fixation of two CO2 molecules to form acetyl-CoA, with a coenzyme and an enzyme metal center as CO2 acceptors (67, 78, 80, 123). One molecule of CO2 is reduced to the level of a methyl group, which is bound to a tetrahydropterin coenzyme (Fig. 4). Another CO2 molecule is reduced to carbon monoxide bound to nickel in the reaction center of CO dehydrogenase, which also acts as an acetyl-CoA synthase. It accepts the methyl group from the methylated tetrahydropterin via a methylated corrinoid protein, combines it with CO to form an enzyme-bound Ni-acetyl group, and releases this group with coenzyme A to form acetyl-CoA. This key enzyme (CO dehydrogenase/acetyl-CoA synthase) may represent a substantial part of soluble cell protein (e.g., 6 to 9% of soluble cell protein together with the corrinoid protein in Moorella thermoacetica) (83). CO dehydrogenase/acetyl-CoA synthase has common roots in all prokaryotes using this pathway (79), in contrast to the enzymes involved in the formation of methyltetrahydropterin from CO2.
FIG. 4.
The reductive acetyl-CoA (Wood-Ljungdahl) pathway. One CO2 molecule is reduced to carbon monoxide bound to a nickel atom in the active center of CO dehydrogenase, and another is reduced to a methyl group bound to the carrier tetrahydropterin; subsequent methyl transfer to nickel-bound CO leads to acetyl-CoA synthesis. The upper part (red) shows the variant of the pathway functioning in acetogens, and the lower part (blue) depicts the pathway in methanogens. Fd, ferredoxin; THF, tetrahydrofolate; H4TPT, tetrahydropterin (THF in acetogens and tetrahydromethanopterin or tetrahydrosarcinopterin in methanogens); MFR, methanofuran; F420, coenzyme F420. Enzymes: 1, formate dehydrogenase; 2; formyl-THF synthetase; 3, formyl-MFR dehydrogenase; 4, formyl-MFR:tetrahydromethanopterin formyltransferase; 5, methenyl-THF cyclohydrolase; 6, methenyl-tetrahydromethanopterin cyclohydrolase; 7, methylene-THF dehydrogenase; 8, methylene-tetrahydromethanopterin dehydrogenase; 9, methylene-THF reductase; 10, methylene-tetrahydromethanopterin reductase; 11, CO dehydrogenase/acetyl-CoA synthase.
This pathway is preferred by prokaryotes living close to the thermodynamic limit, like acetogenic bacteria and methanogenic archaea. Methanogens and acetogens use the reductive acetyl-CoA pathway, not only for CO2 fixation but also for energy conservation via generation of an electrochemical gradient (16, 80, 108). In the process of energy conservation during autotrophic growth or fermentation, acetogens generate acetate, whereas methanogens withdraw methyltetrahydromethanopterine from the pathway and reduce it via methyl coenzyme M to methane. The reductive acetyl-CoA pathway functions also in anaerobic ammonia-oxidizing planctomycetes (98), sulfate-reducing bacteria (Desulfobacterium sp., Deltaproteobacteria) (88), and in autotrophic Archaeoglobales (Euryarchaeota) growing by means of anaerobic respiration (114, 115).
The assimilation of acetyl-CoA synthesized in the reductive acetyl-CoA pathway proceeds via pyruvate synthase, as described above for the rTCA cycle. However, the pathways of 2-oxoglutarate synthesis may differ. Many organisms use an incomplete oxidative (horseshoe) citric acid cycle, in which 2-oxoglutarate oxidation enzyme is lacking (26, 28, 114, 115, 120). Others form 2-oxoglutarate via the reductive branch of the citric acid cycle, involving ferredoxin-dependent 2-oxoglutarate synthase (35).
There are many variants of the pathway that differ in the use of coenzymes and electron carriers. The variant of the Wood-Ljungdahl pathway functioning in methanogens does not require any additonal ATP input, but chemiosmotical energy is used for the reduction of ferredoxin and for the reduction of CO2 to the level of formylmethanofuran. In bacteria, usage of NADPH instead of reduced ferredoxin for the reduction of CO2 in the methyl branch of the pathway dictates the necessity of spending an additional ATP equivalent for the formation of formyltetrahydrofolate (Fig. 4).
The pathway can also be used for the assimilation of a variety of C1 compounds like CO, formaldehyde, methanol, methylamine, methylmercaptane, and methyl groups of aromatic O-methyl ethers/esters. Since CO2 reduction to acetate through the Wood-Ljungdahl pathway does not dissipate ATP, it can be exploited for channeling electrons produced during fermentations to CO2 (acetogenesis). Acetoclastic methanogenic archaea disproportionate acetate through the pathway into CH4 plus CO2, and hydrogenotrophic methanogens use its methyl branch for methane formation (accompanied with energy production), in addition to autotrophic CO2 fixation. In many organisms, the Wood-Ljungdahl pathway may be reversed for the complete oxidation of acetyl-CoA, and sometimes even in organisms which can use it for acetyl-CoA production in the course of autotrophic CO2 fixation (42, 88).
The reductive acetyl-CoA pathway functions in psychrophiles as well as hyperthermophiles. Its currently known upper temperature limit is the same as the maximum for possible cell proliferation (Methanopyrus kandleri; 122°C) (105).
The functioning of the pathway requires strict anoxic conditions, since some of its enzymes, notably CO dehydrogenase/acetyl-CoA synthase, are highly oxygen sensitive. Since organisms possessing the pathway can be isolated from environments with fluctuating oxygen tensions like soils, they had to evolve some strategies to deal with oxidative stress. Among these are the synthesis of enzymes of oxidative response (catalase, peroxidase, etc.), switching to other electron acceptors, and symbiotic relationships with O2-consuming partners (25, 93). This, however, does not render oxygen-tolerant organisms into true aerobes. A related problem of the acetyl-CoA pathway is the high requirement for metals (Mo or W, Co, Ni, and Fe), which are water soluble preferentially in the reduced oxidation state (i.e., under anoxic conditions). Furthermore, the pathway makes extensive use of coenzymes (tetrahydropterin and cobalamin); for example, methanogens synthesize up to ∼2 mg/g dry mass cobamides (99). Therefore, the demand for metals, cofactors, and anaerobiosis restricts the pathway to a limited number of ecological niches, despite its high energetic efficiency.
ROUTE 4: THE HYDROXYPROPIONATE (FUCHS-HOLO) BI-CYCLE (3-HYDROXYPROPIONATE CYCLE)
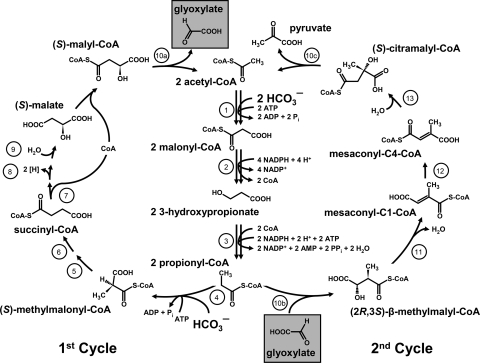
The 3-hydroxypropionate cycle was discovered in Chloroflexus aurantiacus by Helge Holo, who designed the first draft of the pathway (45), and Georg Fuchs, whose laboratory closed and completely elucidated it (44, 97, 130), and is named here after its discoverers as the “Fuchs-Holo (bi-)cycle.” Originally, glyoxylate was proposed as the primary CO2 fixation product in Chloroflexus (97). However, glyoxylate is not a central precursor metabolite, and its conversion to the cellular building blocks requires a second cycle (44). Therefore, the cycle turned out to be a bi-cycle (Fig. 5; 130). In the first glyoxylate synthesis cycle, acetyl-CoA is carboxylated to malonyl-CoA, which is further reduced to propionyl-CoA via 3-hydroxypropionate as a free characteristic intermediate. Subsequent carboxylation of propionyl-CoA followed by carbon rearrangement yields succinyl-CoA. Succinyl-CoA is converted to (S)-malyl-CoA, the second characteristic intermediate of the cycle. (S)-Malyl-CoA cleavage regenerates the starting molecule acetyl-CoA and releases glyoxylate as a first carbon fixation product (97). A second glyoxylate assimilation cycle starts with glyoxylate addition to propionyl-CoA, forming β-methylmalyl-CoA, which in turn is converted via mesaconyl-CoA to citramalyl-CoA. The latter is cleaved into pyruvate and acetyl-CoA. Acetyl-CoA conversion to propionyl-CoA occurs as described above, closing the second cycle (44, 130).
FIG. 5.
The 3-hydroxypropionate (Fuchs-Holo) bi-cycle (130). Enzymes: 1, acetyl-CoA carboxylase; 2, malonyl-CoA reductase; 3, propionyl-CoA synthase; 4, propionyl-CoA carboxylase; 5, methylmalonyl-CoA epimerase; 6, methylmalonyl-CoA mutase; 7, succinyl-CoA:(S)-malate-CoA transferase; 8, succinate dehydrogenase; 9, fumarate hydratase; 10a, -b, and -c, trifunctional (S)-malyl-CoA (a)/β-methylmalyl-CoA (b)/(S)-citramalyl-CoA lyase (c); 11, mesaconyl-C1-CoA hydratase; 12, mesaconyl-CoA C1-C4 CoA transferase; 13, mesaconyl-C4-CoA hydratase. (Reprinted from reference 130 with permission.)
The energy costs of the 3-hydroxypropionate bi-cycle are high: it requires seven ATP equivalents for the synthesis of pyruvate and three additional ATPs for triose phosphate. However, its key carboxylase(s), biotin-dependent acetyl-CoA/propionyl-CoA carboxylase, is virtually irreversible and uses bicarbonate as an active inorganic carbon species. This may be especially advantageous under neutrophilic and alkaliphilic conditions (i.e., in the natural environment for the organisms using this pathway).
The 3-hydroxypropionate bi-cycle allows coassimilation of numerous compounds (e.g., fermentation products like acetate, propionate, and succinate—substrates that are metabolized via acetyl-CoA or propionyl-CoA; or 3-hydroxypropionate, an intermediate in the metabolism of the ubiquitous osmoprotectant dimethylsulfoniopropionate) (130). This makes the pathway best suitable for mixotrophy. It functions in the green non-sulfur phototrophs of the Chloroflexaceae family, which grow preferentially under photoheterotrophic conditions. The only autotrophic representative of this family found so far is Chloroflexus aurantiacus, in which the bi-cycle was discovered. Although its preferable growth mode is photoheterotrophy, it can grow autotrophically, not only in the laboratory but also in situ in hot spring microbial mats (112). The rather limited distribution of the 3-hydroxypropionate bi-cycle suggests that it was a singular invention and evolved in the Chloroflexaceae ancestors. Various widespread marine aerobic phototrophic bacteria (e.g., Erythrobacter sp. strain NAP1) harbor genes of a rudimentary 3-hydroxypropionate bi-cycle required for the conversion of acetyl-CoA to succinyl-CoA (130). Likewise, the heterotrophic Congregibacter litoralis and Nitrococcus mobilis and the photoautotrophic Chloroherpeton thalassium contain the propionyl-CoA synthase gene only. They are probably used for assimilation of acetate, 3-hydroxypropionate, and/or propionate under oligotrophic conditions (130). The 3-hydroxypropinate bi-cycle does not make use of low-potential electron donors like ferredoxin. This is in accordance with the quinone-type of photosynthetic reaction center present in Chloroflexus (18).
An interesting feature of this cycle is the presence of a number of bi- and multifunctional enzymes (Fig. 5): the pathway involving 19 steps requires only 13 enzymes (130). It does not contain oxygen-sensitive steps, and all of its enzymes can function under aerobic conditions (130). Taking into account the high efficiency of the carboxylases of the cycle in comparison to RubisCO, the question arises: why this cycle did not spread over oxygenic phototrophs. Probably, it is not O2 tolerant enough. Although B12-dependent methylmalonyl-CoA mutase can normally be measured and even purified under oxic conditions, the conditions in the cells of oxygenic phototrophs are much more tough: combination of saturated oxygen concentration and light may result in inadequately high rates of inactivation of this radical B12-dependent enzyme and its cofactor, preventing its functioning in these organisms.
ROUTES 5 AND 6: THE 3-HYDROXYPROPIONATE/ 4-HYDROXYBUTYRATE AND DICARBOXYLATE/ 4-HYDROXYBUTYRATE CYCLES
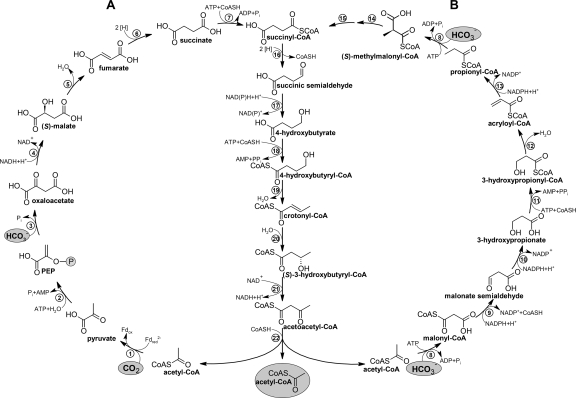
The 3-hydroxypropionate/4-hydroxybutyrate (hydroxypropionate/hydroxybutyrate) (HP/HB) cycle and the dicarboxylate/4-hydroxybutyrate (dicarboxylate/hydroxybutyrate) (DC/HB) cycle are two autotrophic CO2 fixation cycles recently described in Crenarchaeota (13, 46). In both cycles (named together as the 4-hydroxybutyrate cycles), acetyl-CoA and two inorganic carbons are converted to succinyl-CoA, although this is accomplished with different carboxylases (Fig. 6). In the HP/HB cycle, acetyl-CoA/propionyl-CoA carboxylase fixes two molecules of bicarbonate, and in the DC/HB cycle, pyruvate synthase and PEP carboxylase are the two carboxylating enzymes. Yet, the regeneration of acetyl-CoA, the primary CO2 acceptor, from succinyl-CoA proceeds through the same intermediates in both pathways. Succinyl-CoA is reduced to 4-hydroxybutyrate, which is then activated to 4-hydroxybutyryl-CoA. Its dehydratation to crotonyl-CoA is catalyzed by 4-hydroxybutyryl-CoA dehydratase, which is considered a key enzyme of both cycles. Crotonyl-CoA is oxidized to acetoacetyl-CoA and cleaved into two acetyl-CoA molecules, closing the cycle(s) and generating an additional molecule of acetyl-CoA for biosynthesis. While both cycles produce acetyl-CoA, they differ with respect to how they are linked to the central carbon metabolism. The anaerobic DC/HB cycle uses pyruvate synthase to synthesize pyruvate and from there the other central precusor molecules. The aerobic HP/HB cycle requires another half turn of the cycle to make succinyl-CoA, which is oxidatively converted to oxaloacetate, pyruvate, and PEP (31).
FIG. 6.
The 4-hydroxybutyrate cycles of autotrophic CO2 fixation (14). (A) The dicarboxylate/4-hydroxybutyrate cycle functioning in Desulfurococcales and Thermoproteales; (B) the 3-hydroxypropionate/4-hydroxybutyrate cycle functioning in Sulfolobales. Note that succinyl-CoA reductase in Thermoproteales and Sulfolobales uses NADPH (13, 62, 81, 82) and probably reduced ferredoxin (Fdred) in Desulfurococcales (15, 46). Enzymes: 1, pyruvate synthase; 2, pyruvate:water dikinase; 3, PEP carboxylase; 4, malate dehydrogenase; 5, fumarate hydratase; 6, fumarate reductase (natural electron acceptor is not known); 7, succinyl-CoA synthetase; 8, acetyl-CoA/propionyl-CoA carboxylase; 9, malonyl-CoA reductase; 10, malonic semialdehyde reductase; 11, 3-hydroxypropionate-CoA ligase; 12, 3-hydroxypropionyl-CoA dehydratase; 13, acryloyl- CoA reductase; 14, methylmalonyl-CoA epimerase; 15, methylmalonyl-CoA mutase; 16, succinyl-CoA reductase; 17, succinic semialdehyde reductase; 18, 4-hydroxybutyrate-CoA ligase; 19, 4-hydroxybutyryl-CoA dehydratase; 20, crotonyl-CoA hydratase; 21, (S)-3-hydroxybutyryl-CoA dehydrogenase (NAD+); 22, acetoacetyl-CoA β-ketothiolase. Fd, ferredoxin. (Adapted from reference 14.)
Although the 4-hydroxybutyrate cycles share many common enzymes and intermediates, the fundamental difference lies in their sensitivity to oxygen. The enzymes of the HP/HB cycle tolerate oxygen. Even though 4-hydroxybutyryl-CoA dehydratase is a radical enzyme and is inactivated by oxygen in clostridia, it seems to be sufficiently robust to oxygen in Crenarchaeota (15). In contrast, the oxygen sensitivity of some of the enzymes and electron carriers of the DC/HB cycle (i.e., pyruvate synthase and ferredoxin) restricts this cycle to organisms growing under anoxic conditions. Consequently, the HP/HB cycle functions in (micro)aerobic Sulfolobales (13-15), and the DC/HB cycle is present in mostly anaerobic autotrophic representatives of Thermoproteales and Desulfurococcales (14, 46, 81). The presence of genes coding for the characteristic enzymes of the HP/HB cycle in the mesophilic aerobic “marine group I” Archaea (“Thaumarchaeota”) suggests that these abundant marine archaea also use this cycle (13, 41, 117).
Interestingly, Stygiolobus azoricus, a strictly anaerobic representative of Sulfolobales, has turned to an anaerobic lifestyle while keeping its enzyme outfit and using the aerobic HP/HB cycle (15). On the contrary, the anaerobic DC/HB cycle functions in a facultative aerobe of the Desulfurococcales, Pyrolobus fumarii (15). However, its growth is possible only at very low O2 concentrations (up to 0.3% in the gas phase) (17), and the actual conditions in the cytoplasm of actively respiring cells are probably anoxic, especially at the optimal growth temperature of 106°C.
Many Crenarchaeota are (hyper)thermophilic organisms (for example, P. fumarii grows up to 113°C) (17), and their metabolic pathways must cope with the hot environment. Although succinyl-CoA occurring in both cycles can easily be hydrolyzed under thermophilic conditions, its further conversion in the cycle is highly exergonic. Moreover, this conversion is catalyzed by enzymes which are fairly active and exhibit high affinity for succinyl-CoA (13, 62, 82), ensuring its effective conversion into stabile intermediates.
In terms of ATP costs, both cycles are expensive; as usual, the anaerobic one is “cheaper” but uses low-potential electron acceptors (Fig. 6). The synthesis of one pyruvate requires five ATP equivalents in the DC/HB cycle (one pyrophosphate is formed) and nine ATP equivalents in the HP/HB cycle (generating three molecules of pyrophosphate). The synthesis of one triose phosphate (for gluconeogenesis) from pyruvate needs three additional ATP equivalents; however, Archaea usually do not possess large polysaccharide cell walls and therefore require only small amounts of sugars (see, for example, references 39 and 128), leaving pyruvate and acetyl-CoA as the main biosynthetic precursors. Note that pyrophosphate may serve as an energy source, if its hydrolysis is catalyzed by a membrane-bound pyrophosphatase. This would improve the ATP balance of the HP/HB cycle.
As we have seen, other autotrophic pathways are formally more energetically efficient than the 4-hydroxybutyrate cycles: then why have they been this successful during evolution? Possibly, their thermotolerance is an important feature for hyperthermophilic crenarchaea. Moreover, the 4-hydroxybutyrate cycles do not have as high demands for metals and coenzymes as the Wood-Ljungdahl pathway. Furthermore, the postulated functioning of the HP/HB cycle in mesophilic “marine group I” archaea representing ∼20% of all picoplankton cells in the world oceans (58) may indicate that this strategy is highly successful in mesophiles as well. The key to success here is probably the O2 robustness of the cycle; the absence of a wasteful oxygenase side reaction; and its specificity for bicarbonate (not CO2), which is the main inorganic carbon species in slightly alkaline marine water.
Interestingly, the 3-hydroxypropionate part of the HP/HB cycle resembles the first part of the Chloroflexus 3-hydroxypropionate bi-cycle (Fig. 5 and 6). However, the enzymes catalyzing the conversion of malonyl-CoA to propionyl-CoA are not homologous (13, 14, 130), and these pathways evolved independently in archaea and Chloroflexaceae, giving an impressive example of a convergent evolution. This emphasizes that carboxyphosphate, which is used in the biotin-dependent carboxylase reactions, might be an attractive model for carbon fixation during chemoevolution.
COEXISTENCE OF DIFFERENT AUTOTROPHIC PATHWAYS IN ONE SPECIES
An uncultured endosymbiont (gammaproteobacteria) of a deep-sea tube worm possesses the CB cycle and the rTCA cycle and uses them, depending on the energy supply. In a high-energy situation, the symbiont fixes CO2 via the CB cycle, whereas under low-energy conditions, it switches to the energetically more favorable rTCA cycle (70). This allows us to speculate that the conditional usage of different CO2 fixation pathways may be especially advantageous for bacterial symbionts. The regular movement of the host between oxic and anoxic zones results in the periodical (not fluctuational) changes in oxygen and reductant supply and allows the host to combine the energetically efficient anaerobic pathway with the less efficient but aerobic one.
There are further examples of organisms in which the genome analysis suggests the presence of two different autotrophic pathways. Ammonifex degensii (Clostridia) and Ferroglobus placidus (Archaeoglobales, Euryarchaeota) possess genes encoding the key enzymes of the reductive acetyl-CoA pathway; in addition, their genomes harbor genes for the archaeal form III RubisCO as well as for a putative phosphoribulokinase (both genomes are available at http://www.jgi.doe.gov). The possible presence of the CB cycle in these organisms needs to be addressed experimentally. In conclusion, although there is only one example of the coexistence of two autotrophic pathways in one species, this strategy may give an advantage for those organisms living under varying environmental conditions.
REASONS BEHIND DIVERSITY: CONCLUDING REMARKS
According to the “metabolism first” theory, life started in a hydrothermal vent setting in the Hadean ocean with catalytic metal sulfide surfaces or compartments (71, 116), and the reductive acetyl-CoA pathway may have evolved as the first metabolic cycle (14, 34, 84, 123). The further distribution of life drove the first organisms into diverse ecological niches, confronting them with different problems; this led to the appearance of new metabolic strategies. Not only were the conditions different, the organisms diversified as well, thus creating additional factors that influenced their metabolism. Indeed, an autotrophic pathway centered around sugars is probably not beneficial for those organisms, which barely produce them (e.g., autotrophic archaea); the organisms directly reducing ferredoxin during photosynthesis may profit from the usage of this low-potential electron donor for CO2 fixation (e.g., green sulfur bacteria). For the pathways possessing only one or two characteristic enzymes (e.g., the CB cycle or the rTCA cycle), the lateral gene transfer might significantly facilitate their distribution. RubisCO genes are often carried on plasmids (64); even lateral transfer of the more complicated pathways is probably not unprecedented (22).
One of the most important ecological factors in the modern biosphere is the presence and concentration of oxygen. Although the aerobic pathways require generally higher input of energy, the corresponding organisms profit from the higher energetic productivity of aerobic metabolism. Oxygen is an important factor for anaerobes as well, since they often have to cope with fluctuating concentrations of this compound in their environment and therefore need additional protection against oxidative stress.
Life at the thermodynamical limit does not favor the usage of metabolic schemes with high energy demands. For example, anaerobic methane oxidation with sulfate is thermodynamically less favorable than O2-mediated “anaerobic” methane oxidation with nitrite, and autotrophy proceeds in these species either via the reductive acetyl-CoA pathway or the CB cycle, with respect to the energy demands (32, 40). The reversibility of the reductive acetyl-CoA pathway and the rTCA cycle may give additional metabolic plasticity to the organisms possessing these pathways. However, the usage of a more energetically efficient pathway is not necessarily advantageous, since it ensures the higher growth yields but may result in slower growth rates.
(Hyper)thermophiles must cope with the heat instability of some of their metabolites. Such metabolites should be converted quickly and effectively into more stable compounds in order to prevent their breakdown. The triose phosphate glyceraldehyde-3-phosphate is a striking example. This heat-labile compound, probably governing the temperature limits of the CB cycle, is also an intermediate in glycolysis and gluconeogenesis. In (hyper)thermophiles, glycolysis and gluconeogenesis have some deviations from the classical form. In glycolysis, glyceraldehyde-3-phosphate oxidation to 3-phosphoglycerate is irreversible, nonphosphorylative, and highly exergonic (90). In gluconeogenesis, the quick removal of triose phosphates in stabile fructose-6-phosphate is catalyzed by the bifunc- tional (unidirectional) fructose-1,6-bisphosphate aldolase/phosphatase, which is even regarded as one of the determinants of hyperthermophily (87).
How many CO2 fixation pathways exist in nature? Is the autotrophic repertoire exhausted with the six routes described so far? Possibly not. From genome analyses, we know, that some autotrophs do not possess genes of any known autotrophic CO2 fixation pathway (e.g., Pyrobaculum arsenaticum and Ferroplasma acidiphilum) (14). Furthermore, RubisCO deletion mutants of Rhodobacter sphaeroides and Rhodospirillum rubrum were able to grow autotrophically under certain conditions, although the growth rates were extremely low (119). The biochemistry of this phenomenon is unknown (55). Does this hidden pathway participate in CO2 fixation in the wild-type strain, or is this an example of evolution in the laboratory? If the latter is true, it will show how novel pathways could evolve following the mutational loss of the existing features and the experiencing of environmental pressure. With our current knowledge of CO2 fixation pathways, we can now design autotrophic pathways with given properties (9), and even a rational design of novel carboxylases on the scaffold of already known enzymes is possible (30). After more than 70 years of research on autotrophic CO2 fixation, this topic is still an exciting subject for future fundamental and applied research.
Acknowledgments
Our work on autotrophic CO2 fixation was supported by Deutsche Forschungsgemeinschaft and Evonik-Degussa.
I am indebted to Georg Fuchs, Freiburg, for constant support, innumerable discussions and suggestions during the work, and critical reading of the manuscript. I also thank Rudolf K. Thauer, Marburg, for stimulating discussions on autotrophic CO2 fixation and Jan Zarzycki for critical reading of the manuscript.
Footnotes
Published ahead of print on 7 January 2011.
REFERENCES
- 1.Anderson, L. E. 1971. Chloroplast and cytoplasmic enzymes. II. Pea leaf triose phosphate isomerases. Biochim. Biophys. Acta 235:237-244. [DOI] [PubMed] [Google Scholar]
- 2.Aoshima, M. 2007. Novel enzyme reactions related to the citric acid cycle: phylogenetic/functional implications and biotechnological applications. Appl. Microbiol. Biotechnol. 75:249-255. [DOI] [PubMed] [Google Scholar]
- 3.Aoshima, M., and Y. Igarashi. 2008. Nondecarboxylating and decarboxylating isocitrate dehydrogenases: oxalosuccinate reductase as an ancestral form of isocitrate dehydrogenase. J. Bacteriol. 190:2050-2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aoshima, M., M. Ishii, and Y. Igarashi. 2004. A novel enzyme, citryl-CoA synthetase, catalysing the first step of the citrate cleavage reaction in Hydrogenobacter thermophilus TK-6. Mol. Microbiol. 52:751-761. [DOI] [PubMed] [Google Scholar]
- 5.Aoshima, M., M. Ishii, and Y. Igarashi. 2004. A novel enzyme, citryl-CoA lyase, catalysing the second step of the citrate cleavage reaction in Hydrogenobacter thermophilus TK-6. Mol. Microbiol. 52:763-770. [DOI] [PubMed] [Google Scholar]
- 6.Aoshima, M., M. Ishii, and Y. Igarashi. 2006. A novel oxalosuccinate-forming enzyme involved in the reductive carboxylation of 2-oxoglutarate in Hydrogenobacter thermophilus TK-6. Mol. Microbiol. 62:748-759. [DOI] [PubMed] [Google Scholar]
- 7.Badger, M. R., and E. J. Bek. 2008. Multiple Rubisco forms in proteobacteria: their functional significance in relation to CO2 acqusition by the CBB cycle. J. Exp. Bot. 59:1525-1541. [DOI] [PubMed] [Google Scholar]
- 8.Badger, M. R., and G. D. Price. 2003. CO2 concentrating mechanisms in cyanobacteria: molecular components, their diversity and evolution. J. Exp. Bot. 54:609-622. [DOI] [PubMed] [Google Scholar]
- 9.Bar-Even, A., E. Noor, N. E. Lewis, and R. Milo. 2010. Design and analysis of synthetic carbon fixation pathways. Proc. Natl. Acad. Sci. U. S. A. 107:8889-8894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beatty, J. T., et al. 2005. An obligately photosynthetic bacterial anaerobe from a deep-sea hydrothermal vent. Proc. Natl. Acad. Sci. U. S. A. 102:9306-9310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beh, M., G. Strauss, R. Huber, K. O. Stetter, and G. Fuchs. 1993. Enzymes of the reductive citric acid cycle in the autotrophic eubacterium Aquifex pyrophilus and in the archaebacterium Thermoproteus neutrophilus. Arch. Microbiol. 160:306-311. [Google Scholar]
- 12.Berg, I. A., O. I. Keppen, E. N. Krasil'nikova, N. V. Ugol'kova, and R. N. Ivanovsky. 2005. Carbon metabolism of filamentous anoxygenic phototrophic bacteria of the family Oscillochloridaceae. Microbiology 74:258-264. (Translated from Mikrobiologiia.) [PubMed] [Google Scholar]
- 13.Berg, I. A., D. Kockelkorn, W. Buckel, and G. Fuchs. 2007. A 3-hydroxypropionate/4-hydroxybutyrate autotrophic carbon dioxide assimilation pathway in Archaea. Science 318:1782-1786. [DOI] [PubMed] [Google Scholar]
- 14.Berg, I. A., et al. 2010. Autotrophic carbon fixation in Archaea. Nat. Rev. Microbiol. 8:447-460. [DOI] [PubMed] [Google Scholar]
- 15.Berg, I. A., W. H. Ramos-Vera, A. Petri, H. Huber, and G. Fuchs. 2010. Study of the distribution of autotrophic CO2 fixation cycles in Crenarchaeota. Microbiology 156:256-269. [DOI] [PubMed] [Google Scholar]
- 16.Biegel, E., and V. Müller. 2010. Bacterial Na+-translocating ferredoxin:NAD+ oxidoreductase. Proc. Natl. Acad. Sci. U. S. A. 107:18138-18142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Blöchl, E., et al. 1997. Pyrolobus fumarii, gen. and sp. nov., represents a novel group of archaea, extending the upper temperature limit for life to 113 degrees C. Extremophiles 1:14-21. [DOI] [PubMed] [Google Scholar]
- 18.Bryant, D. A., and N.-U. Frigaard. 2006. Prokaryotic photosynthesis and phototrophy illuminated. Trends Microbiol. 14:488-496. [DOI] [PubMed] [Google Scholar]
- 19.Caldwell, P. E., M. R. MacLean, and P. R. Norris. 2007. Ribulose bisphosphate carboxylase activity and a Calvin cycle gene cluster in Sulfobacillus species. Microbiology 153:2231-2240. [DOI] [PubMed] [Google Scholar]
- 20.Campbell, B. J., J. L. Stein, and S. C. Cary. 2003. Evidence of chemolithoautotrophy in the bacterial community associated with Alvinella pompejana, a hydrothermal vent polychaete. Appl. Environ. Microbiol. 69:5070-5078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cegelski, L., and J. Schaefer. 2006. NMR determination of photorespiration in intact leaves using in vivo 13CO2 labeling. J. Magn. Reson. 178:1-10. [DOI] [PubMed] [Google Scholar]
- 22.Chistoserdova, L., J. A. Vorholt, R. K. Thauer, and M. E. Lidstrom. 1998. C1 transfer enzymes and coenzymes linking methylotrophic bacteria and methanogenic Archaea. Science 281:99-102. [DOI] [PubMed] [Google Scholar]
- 23.Cleland, W. W., T. J. Andrews, S. Gutteridge, F. C. Hartman, and G. H. Lorimer. 1998. Mechanism of Rubisco—the carbamate as general base. Chem. Rev. 98:549-561. [DOI] [PubMed] [Google Scholar]
- 24.Dawson, R. M. C., D. C. Elliott, W. H. Elliott, and K. M. Jones. 1986. Data for biochemical research. Clarendon Press, Oxford, United Kingdom.
- 25.Drake, H. L., A. S. Gößner, and S. L. Daniel. 2008. Old acetogens, new light. Ann. N. Y. Acad. Sci. 1125:100-128. [DOI] [PubMed] [Google Scholar]
- 26.Eden, G., and G. Fuchs. 1983. Autotrophic CO2 fixation in Acetobacterium woodii. II. Demonstration of enzymes involved. Arch. Microbiol. 135:68-73. [Google Scholar]
- 27.Eisenhut, M., et al. 2008. The photorespiratory glycolate metabolism is essential for cyanobacteria and might have been conveyed endosymbiontically to plants. Proc. Natl. Acad. Sci. U. S. A. 105:17199-17204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ekiel, I., G. D. Sprott, and G. B. Patel. 1985. Acetate and CO2 assimilation in Methanothrix concilii. J. Bacteriol. 162:905-908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ellis, R. J. 1979. The most abundant protein on Earth. Trends Biochem. Sci. 4:241-244. [Google Scholar]
- 30.Erb, T. J., V. Brecht, G. Fuchs, M. Müller, and B. E. Alber. 2009. Carboxylation mechanism and stereochemistry of crotonyl-CoA carboxylase/reductase, a carboxylating enoyl-thioester reductase. Proc. Natl. Acad. Sci. U. S. A. 106:8871-8876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Estelmann, S., et al. 2011. Labeling and enzyme studies of the central carbon metabolism in Metallosphaera sedula. J. Bacteriol. [Epub ahead of print.] doi: 10.1128/JB.01155-10. [DOI] [PMC free article] [PubMed]
- 32.Ettwig, K. F., et al. 2010. Nitrite-driven anaerobic methane oxidation by oxygenic bacteria. Nature 464:543-548. [DOI] [PubMed] [Google Scholar]
- 33.Evans, M. C. W., B. B. Buchanan, and D. I. Arnon. 1966. A new ferredoxin-dependent carbon reduction cycle in a photosynthetic bacterium. Proc. Natl. Acad. Sci. U. S. A. 55:928-934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fuchs, G. 1989. Alternative pathways of autotrophic carbon dioxide fixation in autotrophic bacteria, p. 365-382. In H. G. Schlegel (ed.), Biology of autotrophic bacteria. Science Tech, Madison, WI.
- 35.Fuchs, G., and E. Stupperich. 1978. Evidence for an incomplete reductive carboxylic acid cycle in Methanobacterium thermoautotrophicum. Arch. Microbiol. 118:121-125. [DOI] [PubMed] [Google Scholar]
- 36.Fuchs, G., E. Stupperich, and G. Eden. 1980. Autotrophic CO2 fixation in Chlorobium limicola. Evidence for the operation of a reductive tricarboxylic acid cycle in growing cells. Arch. Microbiol. 128:64-71. [Google Scholar]
- 37.Geelhoed, J. S., R. Kleerebezem, D. Y. Sorokin, A. J. Stams, amd M. C. van Loosdrecht. 2010. Reduced inorganic sulfur oxidation supports autotrophic and mixotrophic growth of Magnetospirillum strain J10 and Magnetospirillum gryphiswaldense. Environ. Microbiol. 12:1031-1040. [DOI] [PubMed] [Google Scholar]
- 38.Goltsman, D. S., et al. 2009. Community genomic and proteomic analyses of chemoautotrophic iron-oxidizing “Leptospirillum rubarum” (group II) and “Leptospirillum ferrodiazotrophum” (group III) bacteria in acid mine drainage biofilms. Appl. Environ. Microbiol. 75:4599-4615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gonzalez, O., et al. 2008. Reconstruction, modeling & analysis of Halobacterium salinarum R-1 metabolism. Mol. Biosyst. 4:148-159. [DOI] [PubMed] [Google Scholar]
- 40.Hallam, S. J., et al. 2004. Reverse methanogenesis: testing the hypothesis with environmental genomics. Science 305:1457-1462. [DOI] [PubMed] [Google Scholar]
- 41.Hallam, S. J., et al. 2006. Pathways of carbon assimilation and ammonia oxidation suggested by environmental genomic analyses of marine Crenarchaeota. PLoS Biol. 4:e95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hattori, S., A. S. Galushko, Y. Kamagata, and B. Schink. 2005. Operation of the CO dehydrogenase/acetyl coenzyme A pathway in both acetate oxidation and acetate formation by the syntrophically acetate-oxidizing bacterium Thermacetogenium phaeum. J. Bacteriol. 187:3471-3476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hernandez, J. M., S. H. Baker, S. C. Lorbach, J. M. Shively, and F. R. Tabita. 1996. Deduced amino acid sequence, functional expression, and unique enzymatic properties of the form I and form II ribulose bisphosphate carboxylase/oxygenase from the chemoautotrophic bacterium Thiobacillus denitrificans. J. Bacteriol. 178:347-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Herter, S., G. Fuchs, A. Bacher, and W. Eisenreich. 2002. A bicyclic autotrophic CO2 fixation pathway in Chloroflexus aurantiacus. J. Biol. Chem. 277:20277-20283. [DOI] [PubMed] [Google Scholar]
- 45.Holo, H. 1989. Chloroflexus aurantiacus secretes 3-hydroxypropionate, a possible intermediate in the assimilation of CO2 and acetate. Arch. Microbiol. 151:252-256. [Google Scholar]
- 46.Huber, H., et al. 2008. A dicarboxylate/4-hydroxybutyrate autotrophic carbon assimilation cycle in the hyperthermophilic archaeum Ignicoccus hospitalis. Proc. Natl. Acad. Sci. U. S. A. 105:7851-7856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hügler, M., and S. M. Sievert. 2011. Beyond the Calvin cycle: autotrophic carbon fixation in the ocean. Annu. Rev. Mar. Sci. 3:10.1-10.29. [DOI] [PubMed] [Google Scholar]
- 48.Hügler, M., C. O. Wisen, G. Fuchs, C. D. Taylor, and S. M. Sievert. 2005. Evidence for autotrophic CO2 fixation via the reductive citric acid cycle by members of the epsilon subdivision of proteobacteria. J. Bacteriol. 187:3020-3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hügler, M., H. Huber, S. J. Molyneaux, C. Vetriani, and S. M. Sievert. 2007. Autotrophic CO2 fixation via reductive citric acid cycle in different lineages within the phylum Aquificae: evidence for two ways of citrate cleavage. Environ. Microbiol. 9:81-92. [DOI] [PubMed] [Google Scholar]
- 50.Imanaka, H., T. Fukui, H. Atomi, and T. Imanaka. 2002. Gene cloning and characterization of fructose-1,6-bisphosphate aldolase from the hyperthermophilic archaeon Thermococcus kodakaraensis KOD1. J. Biosci. Bioeng. 94:237-243. [DOI] [PubMed] [Google Scholar]
- 51.Ivanovsky, R. N., et al. 1999. Evidence for the presence of the reductive pentose phosphate cycle in a filamentous anoxygenic photosynthetic bacterium, Oscillochloris trichoides strain DG-6. Microbiology 145:1743-1748. [DOI] [PubMed] [Google Scholar]
- 52.Ivanovsky, R. N., N. V. Sintsov, and E. N. Kondratieva. 1980. ATP-linked citrate lyase activity in the green sulfur bacterium Chlorobium limicola forma thiosulfatophilum. Arch. Microbiol. 128:239-241. [Google Scholar]
- 53.Jordan, D. B., and W. L. Orgen. 1981. Species variation in the specificity of ribulose bisphosphate carboxylase/oxygenase. Nature 291:513-515. [Google Scholar]
- 54.Joshi, G. S., et al. 2009. Differential accumulation of form I RubisCO in Rhodopseudomonas palustris CGA010 under photoheterotrophic growth conditions with reduced carbon sources. J. Bacteriol. 191:4243-4250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Joshi, H. M., and F. R. Tabita. 2000. Induction of carbon monoxide dehydrogenase to facilitate redox balancing in a ribulose bisphosphate carboxylase/oxygenase-deficient mutant strain of Rhodospirillum rubrum. Arch. Microbiol. 173:193-199. [DOI] [PubMed] [Google Scholar]
- 56.Kai, Y., et al. 1999. Three-dimensional structure of phosphoenolpyruvate carboxylase: a proposed mechanism for allosteric inhibition. Proc. Natl. Acad. Sci. U. S. A. 96:823-828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kanao, T., M. Kawamura, T. Fukui, H. Atomi, T. Imanaka. 2002. Characterization of isocitrate dehydrogenase from the green sulfur bacterium Chlorobium limicola. A carbon dioxide-fixing enzyme in the reductive tricarboxylic acid cycle. Eur. J. Biochem. 269:1926-1931. [DOI] [PubMed] [Google Scholar]
- 58.Karner, M. B., E. F. DeLong, and D. M. Karl. 2001. Archaeal dominance in the mesopelagic zone of the Pacific Ocean. Nature 409:507-510. [DOI] [PubMed] [Google Scholar]
- 59.Keeley, J. E., and P. W. Rundell. 2003. Evolution of CAM and C4 carbon-concentrating mechanisms. Int. J. Plant Sci. 164:S55-S77. [Google Scholar]
- 60.Kerfeld, C. A., et al. 2005. Protein structures forming the shell of primitive bacterial organelles. Science 309:936-938. [DOI] [PubMed] [Google Scholar]
- 61.Kerfeld, C. A., S. Heinhorst, and G. C. Cannon. 2010. Bacterial microcompartments. Annu. Rev. Microbiol. 64:391-408. [DOI] [PubMed] [Google Scholar]
- 62.Kockelkorn, D., and G. Fuchs. 2009. Malonic semialdehyde reductase, succinic semialdehyde reductase, and succinyl-coenzyme A reductase from Metallosphaera sedula: enzymes of the autotrophic 3-hydroxypropionate/ 4-hydroxybutyrate cycle in Sulfolobales. J. Bacteriol. 191:6352-6362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kreel, N. E., and F. R. Tabita. 2007. Substitutions at methionine 295 of Archaeoglobus fulgidus ribulose-1,5-bisphosphate carboxylase/oxygenase affect oxygen binding and CO2/O2 specificity. J. Biol. Chem. 282:1341-1351. [DOI] [PubMed] [Google Scholar]
- 64.Kusian, B., and B. Bowien. 1997. Organization and regulation of cbb CO2 assimilation genes in autotrophic bacteria. FEMS Microbiol. Rev. 21:135-155. [DOI] [PubMed] [Google Scholar]
- 65.Lee, J. H., et al. 2009. Expression and regulation of ribulose 1,5-bisphosphate carboxylase/oxygenase genes in Mycobacterium sp. strain JC1 DSM 3803. J. Microbiol. 47:297-307. [DOI] [PubMed] [Google Scholar]
- 66.Levicán, G., J. A. Ugalde, N. Ehrenfeld, A. Maass, and P. Parada. 2008. Comparative genomic analysis of carbon and nitrogen assimilation mechanisms in three indigenous bioleaching bacteria: predictions and validations. BMC Genomics 9:581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ljungdahl, L. G. 1986. The autotrophic pathway of acetate synthesis in acetogenic bacteria. Annu. Rev. Microbiol. 40:415-450. [DOI] [PubMed] [Google Scholar]
- 68.Lorimer, G. H., and T. J. Andrews. 1973. Plant photorespiration—an inevitable consequence of the existence of atmospheric oxygen. Nature 243:359-360. [Google Scholar]
- 69.Lücker, S., et al. 2010. A Nitrospira metagenome illuminates the physiology and evolution of globally important nitrite-oxidizing bacteria. Proc. Natl. Acad. Sci. U. S. A. 107:13479-13484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Markert, S., et al. 2007. Physiological proteomics of the uncultured endosymbiont of Riftia pachyptila. Science 315:247-250. [DOI] [PubMed] [Google Scholar]
- 71.Martin, W., J. Baross, D. Kelley, and M. J. Russell. 2008. Hydrothermal vents and the origin of life. Nat. Rev. Microbiol. 6:805-814. [DOI] [PubMed] [Google Scholar]
- 72.Miura, A., M. Kameya, H. Arai, M. Ishii, and Y. Igarashi. 2008. A soluble NADH-dependent fumarate reductase in the reductive citric acid cycle of Hydrogenobacter thermophilus TK-6. J. Bacteriol. 190:7170-7177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nakagawa, S., and K. Takai. 2008. Deep-sea vent chemoautotrophs: diversity, biochemistry and ecological significance. FEMS Microbiol. Ecol. 65:1-14. [DOI] [PubMed] [Google Scholar]
- 74.Ormerod, J. 2003. ‘Every dogma has its day’: a personal look at carbon metabolism in photosynthetic bacteria. Photosynth. Res. 76:135-143. [DOI] [PubMed] [Google Scholar]
- 75.Phillips, S. A., and P. J. Thornalley. 1993. The formation of methylglyoxal from triosephosphates. Investigation using a specific assay for methylglyoxal. Eur. J. Biochem. 212:101-105. [DOI] [PubMed] [Google Scholar]
- 76.Price, G. D., and M. R. Badger. 1989. Expression of human carbonic anhydrase in the cyanobacterium Synechococcus PCC7942 creates a high CO2-requiring phenotype: evidence for a central role for carboxysomes in the CO2 concentrating mechanism. Plant Physiol. 91:505-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Price, G. D., M. R. Badger, F. J. Woodger, and B. M. Long. 2008. Advances in understanding the cyanobacterial CO2-concentrating-mechanism (CCM): functional components, Ci transporters, diversity, genetic regulation and prospects for engineering into plants. J. Exp. Bot. 59:1441-1461. [DOI] [PubMed] [Google Scholar]
- 78.Ragsdale, S. W. 2008. Enzymology of the Wood-Ljungdahl pathway of acetogenesis. Ann. N. Y. Acad. Sci. 1125:129-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ragsdale, S. W., and M. Kumar. 1996. Nickel-containing carbon monoxide dehydrogenase/acetyl-CoA synthase. Chem. Rev. 96:2515-2539. [DOI] [PubMed] [Google Scholar]
- 80.Ragsdale, S. W., and E. Pierce. 2008. Acetogenesis and the Wood-Ljungdahl pathway of CO2 fixation. Biochim. Biophys. Acta 1784:1873-1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ramos-Vera, W. H., I. A. Berg, and G. Fuchs. 2009. Autotrophic carbon dioxide assimilation in Thermoproteales revisited. J. Bacteriol. 191:4286-4297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ramos-Vera, W. H., M. Weiss, E. Strittmatter, D. Kockelkorn, and G. Fuchs. 2011. Identification of missing genes and enzymes of autotrophic carbon fixation in Crenarchaeota. J. Bacteriol. [Epub ahead of print.] doi: 10.1128/JB.01156-10. [DOI] [PMC free article] [PubMed]
- 83.Roberts, J. R., W.-P. Lu, and S. W. Ragsdale. 1992. Acetyl-coenzyme A synthesis from methyltetrahydrofolate, CO, and coenzyme A by enzymes purified from Clostridium thermoaceticum: attainment of in vivo rates and identification of rate-limiting steps. J. Bacteriol. 174:4667-4676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Russell, M. J., and W. Martin. 2004. The rocky roots of the acetyl-CoA pathway. Trends Biochem. Sci. 29:358-363. [DOI] [PubMed] [Google Scholar]
- 85.Sarles, L. S., and F. R. Tabita. 1983. Derepression of the synthesis of D-ribulose 1,5-bisphosphate carboxylase/oxygenase from Rhodospirillum rubrum. J. Bacteriol. 153:458-464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sato, T., H. Atomi, and T. Imanaka. 2007. Archaeal type III RuBisCOs function in a pathway for AMP metabolism. Science 315:1003-1006. [DOI] [PubMed] [Google Scholar]
- 87.Say, R., and G. Fuchs. 2010. Fructose 1,6-bisphosphate aldolase/phosphatase may be an ancestral gluconeogenic enzyme. Nature 464:1077-1081. [DOI] [PubMed] [Google Scholar]
- 88.Schauder, R., A. Preuß, M. Jetten, and G. Fuchs. 1989. Oxidative and reductive acetyl-CoA/carbon monoxide dehydrogenase pathway in Desulfobacterium autotrophicum. 2. Demonstration of the enzymes of the pathway and comparison of CO dehydrogenase. Arch. Microbiol. 151:84-89. [Google Scholar]
- 89.Schauder, R., F. Widdel, and G. Fuchs. 1987. Carbon assimilation pathways in sulfate-reducing bacteria. II. Enzymes of a reductive citric acid cycle in the autotrophic Desulfobacter hydrogenophilus. Arch. Microbiol. 148:218-225. [Google Scholar]
- 90.Schönheit, P. 2008. Glycolysis in hyperthermophiles, p. 99-112. In F. Robb, G. Antranikian, D. Grogan, and A. Driessen (ed.), Thermophiles. biology and technology at high temperature. CRC Press, Boca Raton, FL.
- 91.Schübbe, S., et al. 2009. Complete genome sequence of the chemolithoautotrophic marine magnetotactic coccus strain MC-1. Appl. Environ. Microbiol. 75:4835-4852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Shiba, H., T. Kawasumi, Y. Igarashi, T. Kodama, and Y. Minoda. 1985. The CO2 assimilation via the reductive tricarboxylic acid cycle in an obligately autotrophic, aerobic hydrogen-oxidizing bacterium, Hydrogenobacter thermophilus. Arch. Microbiol. 141:198-203. [Google Scholar]
- 93.Shima, S., et al. 2001. Characterization of a heme-dependent catalase from Methanobrevibacter arboriphilus. Appl. Environ. Microbiol. 67:3041-3045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Shively, J. M., F. Ball, D. H. Brown, and R. E. Saunders. 1973. Functional organelles in prokaryotes: polyhedral inclusions (carboxysomes) of Thiobacillus neapolitanus. Science 182:584-586. [DOI] [PubMed] [Google Scholar]
- 95.Shively, J. M., G. van Keulen, and W. G. Meijer. 1998. Something from almost nothing: carbon dioxide fixation in chemoautotrophs. Annu. Rev. Microbiol. 52:191-230. [DOI] [PubMed] [Google Scholar]
- 96.Spreitzer, R. J. 2003. Role of the small subunit in ribulose-1,5-bisphosphate carboxylase/oxygenase. Arch. Biochem. Biophys. 414:141-149. [DOI] [PubMed] [Google Scholar]
- 97.Strauss, G., and G. Fuchs. 1993. Enzymes of a novel autotrophic CO2 fixation pathway in the phototrophic bacterium Chloroflexus aurantiacus, the 3-hydroxypropionate cycle. Eur. J. Biochem. 215:633-643. [DOI] [PubMed] [Google Scholar]
- 98.Strous, M., et al. 2006. Deciphering the evolution and metabolism of an anammox bacterium from a community genome. Nature 440:790-794. [DOI] [PubMed] [Google Scholar]
- 99.Stupperich, E., and B. Kräutler. 1988. Pseudo vitamin B12 or 5-hydroxybenzimidazolyl-cobamide are the corrinoids found in methanogenic bacteria. Arch. Microbiol. 149:268-271. [Google Scholar]
- 100.Tabita, F. R. 1999. Microbial ribulose 1,5-bisphosphate carboxylase/oxygenase: a different perspective. Photosynth. Res. 60:1-28. [Google Scholar]
- 101.Tabita, F. R., et al. 2007. Function, structure, and evolution of the RubisCO-like proteins and their RubisCO homologs. Microbiol. Mol. Biol. Rev. 71:576-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Tabita, F. R., T. E. Hanson, S. Satagopan, B. H. Witte, and N. E. Kreel. 2008. Phylogenetic and evolutionary relationships of RubisCO and the RubisCO-like proteins and the functional lessons provided by diverse molecular forms. Philos. Trans. R. Soc. Lond. B Biol. Sci. 363:2629-2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Tabita, F. R., S. Satagopan, T. E. Hanson, N. E. Kreel, and S. S. Scott. 2008. Distinct form I, II, III, and IV Rubisco proteins from the three kingdoms of life provide clues about Rubisco evolution and structure/function relationships. J. Exp. Bot. 59:1515-1524. [DOI] [PubMed] [Google Scholar]
- 104.Takai, K., et al. 2005. Enzymatic and genetic characterization of carbon and energy metabolism by deep-sea hydrothermal chemolithoautotrophic isolates of Epsilonproteobacteria. Appl. Environ. Microbiol. 71:7310-7320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Takai, K., et al. 2008. Cell proliferation at 122 degrees C and isotopically heavy CH4 production by a hyperthermophilic methanogen under high-pressure cultivation. Proc. Natl. Acad. Sci. U. S. A. 105:10949-10954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Tcherkez, G. G., G. D. Farquhar, and T. J. Andrews. 2006. Despite slow catalysis and confused substrate specificity, all ribulose bisphosphate carboxylases may be nearly perfectly optimized. Proc. Natl. Acad. Sci. U. S. A. 103:7246-7251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Thauer, R. K. 2008. Biologische Methanbildung: Eine erneuerbare Energiequelle von Bedeutung?, p. 119-137. In P. Gruss and F. Schüth (ed.), Die Zukunft der Energie. C. H. Beck, Munich, Germany.
- 108.Thauer, R. K., A. K. Kaster, H. Seedorf, W. Buckel, and R. Hedderich. 2008. Methanogenic archaea: ecologically relevant differences in energy conservation. Nat. Rev. Microbiol. 6:579-591. [DOI] [PubMed] [Google Scholar]
- 109.Thauer, R. K., D. Möller-Zinkhan, and A. M. Spormann. 1989. Biochemistry of acetate catabolism in anaerobic chemotrophic bacteria. Annu. Rev. Microbiol. 43:43-67. [DOI] [PubMed] [Google Scholar]
- 110.Tourova, T. P., E. M. Spiridonova, I. A. Berg, B. B. Kuznetsov, and D. Y. Sorokin. 2006. Occurrence, phylogeny and evolution of ribulose-1,5bisphosphate carboxylase/oxygenase genes in obligately chemolithoautotrophic sulfur-oxidizing bacteria of the genera Thiomicrospira and Thioalkalimicrobium. Microbiology 152:2159-2169. [DOI] [PubMed] [Google Scholar]
- 111.Tourova, T. P., and E. M. Spiridonova. 2009. Phylogeny and evolution of the ribulose 1,5-bisphosphate carboxylase/oxygenase genes in prokaryotes. Mol. Biol. (Moscow) 43:713-728. [PubMed] [Google Scholar]
- 112.van der Meer, M. T., S. Schouten, J. W. de Leeuw, and D. M. Ward. 2000. Autotrophy of green non-sulphur bacteria in hot spring microbial mats: biological explanations for isotopically heavy organic carbon in the geological record. Environ. Microbiol. 2:428-435. [DOI] [PubMed] [Google Scholar]
- 113.Van Gemerden, H., and J. Mas. 1995. Ecology of phototrophic sulfur bacteria, p. 49-85. In R. E. Blankenship, M. T. Madigan, and C. E. Bauer (ed.), Anoxygenic photosynthetic bacteria. Kluwer Academic Publishers, Dordrecht, Netherlands.
- 114.Vorholt, J. A., D. Hafenbradl, K. O. Stetter, and R. K. Thauer. 1997. Pathways of autotrophic CO2 fixation and of dissimilatory nitrate reduction to N2O in Ferroglobus placidus. Arch. Microbiol. 167:19-23. [DOI] [PubMed] [Google Scholar]
- 115.Vorholt, J. A., J. Kunow, K. O. Stetter, and R. K. Thauer. 1995. Enzymes and coenzymes of the carbon monooxide dehydrogenase pathway for autotrophic CO2 fixation in Archaeoglobus lithotrophicus and the lack of carbon monoxide dehydrogenase in the heterotrophic A. profundus. Arch. Microbiol. 163:112-118. [Google Scholar]
- 116.Wächtershäuser, G. 2007. On the chemistry and evolution of the pioneer organism. Chem. Biodivers. 4:584-602. [DOI] [PubMed] [Google Scholar]
- 117.Walker, C. B., et al. 2010. Nitrosopumilus maritimus genome reveals unique mechanisms for nitrification and autotrophy in globally distributed marine crenarchaea Proc. Natl. Acad. Sci. U. S. A. 107:8818-8823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Wang, X., D. L. Falcone, and F. R. Tabita. 1993. Reductive pentose phosphate-independent CO2 fixation in Rhodobacter sphaeroides and evidence that ribulose bisphosphate carboxylase/oxygenase activity serves to maintain the redox balance of the cell. J. Bacteriol. 175:3372-3379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Wang, X., H. V. Modak, and F. R. Tabita. 1993. Photolithoautotrophic growth and control of CO2 fixation in Rhodobacter sphaeroides and Rhodospirillum rubrum in the absence of ribulose bisphosphate carboxylase-oxygenase. J. Bacteriol. 175:7109-7114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Weimer, P. J., and J. G. Zeikus. 1979. Acetate assimilation pathway in Methanosarcina barkeri. J. Bacteriol. 137:332-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Wildman, S. G. 2002. Along the trail from fraction I protein to Rubisco (ribulose bisphosphate carboxylase-oxygenase). Photosynth. Res. 73:243-250. [DOI] [PubMed] [Google Scholar]
- 122.Williams, T. J., C. L. Zhang, J. H. Scott, and D. A. Bazylinski. 2006. Evidence for autotrophy via the reverse tricarboxylic acid cycle in the marine magnetotactic coccus strain MC-1. Appl. Environ. Microbiol. 72:1322-1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Wood, H. G. 1991. Life with CO or CO2 and H2 as a source of carbon and energy. FASEB J. 5:156-163. [DOI] [PubMed] [Google Scholar]
- 124.Yamamoto, M., H. Arai, M. Ishii, and Y. Igarashi. 2006. Role of two 2-oxoglutarate:ferredoxin oxidoreductases in Hydrogenobacter thermophilus under aerobic and anaerobic condition. FEMS Microbiol. Lett. 263:189-193. [DOI] [PubMed] [Google Scholar]
- 125.Yamamoto, M., T. Ikeda, H. Arai, M. Ishii, and Y. Igarashi. 2010. Carboxylation reaction catalyzed by 2-oxoglutarate:ferredoxin oxidoreductases from Hydrogenobacter thermophilus. Extremophiles 14:79-85. [DOI] [PubMed] [Google Scholar]
- 126.Yeates, T. O., C. A. Kerfeld, S. Heinhorst, G. C. Cannon, and J. M. Shively. 2008. Protein-based organelles in bacteria: carboxysomes and related microcompartments. Nat. Rev. Microbiol. 6:681-691. [DOI] [PubMed] [Google Scholar]
- 127.Yoshizawa, Y., K. Toyoda, H. Arai, M. Ishii, and Y. Igarashi. 2004. CO2-responsive expression and gene organization of three ribulose-1,5-bisphosphate carboxylase/oxygenase enzymes and carboxysomes in Hydrogenovibrio marinus strain MH-110. J. Bacteriol. 186:5685-5691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Yu, J.-P., J. Ladapo, and W. B. Whitman. 1994. Pathway of glycogen metabolism in Methanococcus maripaludis. J. Bacteriol. 176:325-332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Zakharchuk, L. M., et al. 2003. Activity of the enzymes of carbon metabolism in Sulfobacillus sibiricus under various conditions of cultivation. Microbiology 72:553-557. (Translated from Mikrobiologiia.) [PubMed] [Google Scholar]
- 130.Zarzycki, J., V. Brecht, M. Müller, and G. Fuchs. 2009. Identifying the missing steps of the autotrophic 3-hydroxypropionate CO2 fixation cycle in Chloroflexus aurantiacus. Proc. Natl. Acad. Sci. U. S. A. 106:21317-21322. [DOI] [PMC free article] [PubMed] [Google Scholar]