Abstract
129I is of major concern because of its mobility in the environment, excessive inventory, toxicity (it accumulates in the thyroid), and long half-life (∼16 million years). The aim of this study was to determine if bacteria from a 129I-contaminated oxic aquifer at the F area of the U.S. Department of Energy's Savannah River Site, SC, could accumulate iodide at environmentally relevant concentrations (0.1 μM I−). Iodide accumulation capability was found in 3 out of 136 aerobic bacterial strains isolated from the F area that were closely related to Streptomyces/Kitasatospora spp., Bacillus mycoides, and Ralstonia/Cupriavidus spp. Two previously described iodide-accumulating marine strains, a Flexibacter aggregans strain and an Arenibacter troitsensis strain, accumulated 2 to 50% total iodide (0.1 μM), whereas the F-area strains accumulated just 0.2 to 2.0%. Iodide accumulation by FA-30 was stimulated by the addition of H2O2, was not inhibited by chloride ions (27 mM), did not exhibit substrate saturation kinetics with regard to I− concentration (up to 10 μM I−), and increased at pH values of <6. Overall, the data indicate that I− accumulation likely results from electrophilic substitution of cellular organic molecules. This study demonstrates that readily culturable, aerobic bacteria of the F-area aquifer do not accumulate significant amounts of iodide; however, this mechanism may contribute to the long-term fate and transport of 129I and to the biogeochemical cycling of iodine over geologic time.
129I is a major by-product of nuclear fission that is of concern because of its mobility in the environment, excessive inventory, long half-life (∼16 million years), and potential toxicity due to bioaccumulation through the food chain and bioconcentration in the thyroid gland (16, 17, 22). Currently, 146 Ci of 129I is inventoried in soils at two U.S. Department of Energy (DOE) sites, the Hanford Site and the Savannah River Site (SRS), where it has been identified as a key risk driver in contaminated soils and groundwater (21, 24). Furthermore, the global inventory of 129I will increase significantly if just a fraction of the expected “Nuclear Renaissance” is realized (e.g., between China and India alone, 50 to 60 new nuclear reactors are expected to come online by the year 2020) (16, 50). Thus, it is critical to understand the environmental behavior of 129I in order to rigorously assess storage and disposal options for current and future stockpiles of 129I.
In general, little information is available about the chemical properties and mobility of iodine in subsurface aquifers, particularly its tendency to form organo-iodine or its mobility as organo-iodine. However, our own measurements of groundwater from several of the F-area aquifer bore holes found that organo-iodine could account for up to 25% of total iodine (43, 52). The various isotopes of iodine can be strongly bound to macromolecular organic matter, which can significantly decrease or increase its transport, bioavailability, and transfer to humans, depending on the molecular weight and physicochemical properties of the resulting iodine-organic matter species (15, 19, 41-43, 49).
Microbial activity has been linked to the production of organo-iodine and sorption of iodine to soil, and a small but growing body of literature has implicated microbial oxidases, perhydrolases, and particularly peroxidases in the halogenation of soil organic matter (4, 10, 20, 23, 27, 32-34, 37, 38). Yet, details concerning the mechanisms and bacterial species or groups involved are lacking. The most-recent advances in research concerning microbial-iodine interactions have been contributed by Amachi et al. (1-8, 18). In a series of papers, Amachi's research group has isolated (i) iodide-accumulating bacteria (IAB) from marine sediments that concentrate iodide by a factor of 6 × 103 (6), (ii) iodide-oxidizing bacteria from seawater and natural gas brine water that transform iodide into I2 and volatile organo-iodine species (8), and (iii) iodide-methylating bacteria from a variety of soil and seawater samples (4, 5). For the marine IAB, Amachi et al. (2) proposed a model whereby extracellular H2O2, generated by glucose oxidase, oxidizes iodide to I2 or hypoiodous acid (HIO) via an unidentified haloperoxidase. HIO is then transported across the cell membrane via a facilitated diffusion-type mechanism. Once inside the cell, HIO either is reduced to iodide or forms organo-iodine.
Notably, iodide-oxidizing and iodine-accumulating strains were readily obtained from marine or brine waters but not from surface soils (22 samples from a rice paddy, upland field, and forest soils) (6, 7). The only study to date examining the potential for microorganisms from subsurface aquifers to associate with iodine was performed using enrichments from anoxic natural gas formation waters that contain exceedingly high concentrations of iodine (∼120 mg liter−1) and a set of 16 anaerobic bacterial strains obtained from culture collections that included sulfate and iron reducers, denitrifiers, and methanogens (3). The results indicated very limited adsorption or accumulation of iodine by anaerobic microorganisms.
Overall, the research conducted thus far demonstrates that microbial activity is an important factor in iodine biogeochemical cycling (particularly in enhancing iodine binding to high-molecular-weight organic matter) and leads us to hypothesize that select soil and sediment bacteria could be capable of influencing the chemical behavior of iodide (the most common form of 129I found in groundwater) via accumulation, volatilization, and oxidation under aerobic conditions. In particular, we wish to develop an understanding of the role that microorganisms play in the formation of organo-iodine in the subsurface and how they influence iodine mobility. As an initial step toward this overarching goal, the aim of this study was to determine if naturally occurring bacteria from a 129I-contaminated oxic aquifer at the Savannah River Site, SC, could accumulate iodide at environmentally relevant concentrations.
MATERIALS AND METHODS
Isolation of bacteria.
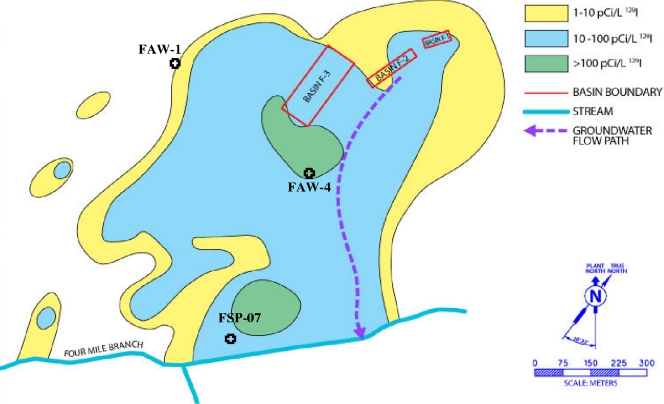
Sediment samples were collected from three locations and multiple depths within the F-area plume zone (10 to 100 pCi/liter 129I) at the Savannah River Site, SC (Fig. 1). Sediments from the first two locations, FAW-4 and FAW-1, were collected from a sandy/clay aquifer 13.1 to 20.7 m and 24.1 to 25.9 m below the surface, respectively. The other sampling location, FSP-07, was an organic-rich wetland zone (0.15 to 1.2 m below the surface). Two approaches, sonication or pyrophosphate based, were used to isolate microbes directly from each of the subsurface soils. For the sonication method, soil samples (1 g) were suspended in 100 ml sterile distilled water (dH2O) and stirred for 15 min. A portion (1 ml) of this soil slurry was sonicated for 15 s to generate a 10−2 dilution of the original soil slurry. For the pyrophosphate method, soil samples (1 g) were suspended in 100 ml sterile phosphate-buffered saline (pH 7.0) containing 25 mM pyrophosphate and stirred for 30 min and then allowed to settle for 30 min to generate a 10−2 dilution of the original soil slurry.
FIG. 1.
Map of the F area at the Savannah River Site with 129I concentrations in groundwater. Mixed radioactive wastes originating from three storage basins (red rectangles) have seeped into the groundwater and have, over time, created a groundwater plume that flows toward Four Mile Branch Creek, a tributary to the Savannah River. Sediment samples for this study were collected at FAW-4 (13.1 to 20.7 m below the surface) and FAW-1 (20.7 to 25.9 m below the surface) in a sandy/clay aquifer and at FSP-07 (0.15 to 1.2 m below the surface) in an organic-rich wetland zone.
Subsequent dilutions (10−3 to 10−6) were prepared from the sonication- and pyrophosphate-generated soil slurries in both R2A (1/10 strength; Difco) and DNB (Oxoid) medium. A portion (100 μl) of each dilution (10−2 to 10−6) series prepared in R2A was spread onto R2A (1/10 strength) plates solidified with either noble agar (Difco) or gellum gum (Sigma Aldrich), with or without cycloheximide (100 μg ml−1; Sigma Aldrich). A portion (100 μl) of each dilution (10−2 to 10−6) series prepared in DNB was spread onto DNB agar plates, with or without cycloheximide (100 μg ml−1). Plates were incubated at room temperature in the dark and monitored for growth over a period of 1 week to 3 months. As colonies with unique morphologies appeared on the plates, they were transferred to fresh R2A (1/10 strength) or DNB plates. In this manner, 325 distinct morphotypes were isolated and stored (agar plates at 4°C and glycerol stocks at −80°C) for further analysis.
Enrichment cultures containing 1 mM iodide were also prepared with sediments collected from the aquifer and wetland zones as means to isolate IAB. Enrichments were generated by suspending sediments (0.05 g) in 50-ml conical tubes containing (i) 20 ml dH2O, (ii) 20 ml 10% nutrient broth (Oxoid), or (iii) 20 ml 10% nutrient broth plus 50 μg ml−1 cycloheximide. Iodide was added (as KI; Sigma) to give a final concentration of 1 mM. This resulted in a total of 24 enrichment cultures (8 sediment samples, with 3 treatments for each sediment sample). The enrichments were incubated in the dark at room temperature and agitated by tube inversion once each day. At 2, 9, and 22 weeks, 50 μl from each tube was transferred to sterile tubes containing 5 ml R2A (1/10 strength) to generate a 10−2 dilution. Dilutions of 10−4 and 10−6 were then prepared in R2A (1/10 strength), and 25 μl of each dilution was spread on both 25% DNB-IS (IS consists of 1.2 g liter−1 KI and 1g liter−1 soluble starch) and 25% R2A-IS agar plates. In this manner, 29 distinct morphotypes were isolated and stored (agar plates at 4°C and glycerol stocks at −80°C) for further analysis.
16S rRNA gene sequencing and phylogenetic analysis.
To determine the taxonomic identity of the isolates, a portion of the 16S rRNA gene was amplified and sequenced. Crude bacterial lysate was collected by placing a small amount of isolated colonies into 10 μl Tris-EDTA (TE) buffer (pH 7.5) with sterile pipette tips. Amplification was performed using 1 μl of the crude lysate as a template in 50-μl reaction mixtures containing 5 μl 10× buffer, MgCl2 (2.5 mM), the 27f primer (5′-GAGTTTGATCMTGGCTCAG-3′) (10 pmol), the 1492r primer (5′-GGTTACCTTGTTACGACTT-3′) (10 pmol), deoxynucleoside triphosphates (200 μM each), and 1.25 U of HotStar Taq DNA polymerase (Qiagen). Reactions were amplified in a PTC-100 thermocycler (MJ Research, Inc.) as follows: 95°C for 10 min and 32 cycles of 95°C for 1 min, 45°C for 45 s, and 72°C for 1 min, followed by a 7-min elongation step. PCRs were visualized on a 1.2% agarose gel to ensure amplification of a single product of the expected size. The 16S rRNA gene amplicon was purified with a MinElute PCR purification kit (Qiagen), and the concentration of the purified product was adjusted to ∼15 ng μl−1 in dH2O using a Nanodrop 1000 spectrophotometer (Thermo Scientific). Sequencing of the purified PCR products was performed by the Georgia Sequencing Facility at the University of Georgia, Athens, GA.
Phylogenetic similarity of 16S rRNA sequences was first determined with BLAST searches of the GenBank (http://www.ncbi.nlm.nih.gov/genbank/index.html) and the RDP classifier (13), followed by maximum-likelihood reconstruction of phylogenetic trees with ARB (48).
Culture conditions.
Two marine iodide-accumulating bacterial (IAB) strains, Flexibacter aggregans NBRC15975 and Arenibacter troitsensis JCM11736, were purchased from the NITE Biological Research Center, Chibin, Japan, and used as reference strains for the iodide accumulation assay. The two reference strains and all microbial isolates from the F area were grown on marine agar 2216 (Difco) and 1/4-strength R2A agar medium (EMD), respectively.
Iodide accumulation screening.
Two methods using 125I− as a tracer were combined to determine the iodide-accumulating ability from F-area microbes. The first used autoradiography, similar to the procedure of Amachi et al. (7). A mixture of I−, 0.016 μM stable I− (as NaI; Sigma), and 0.80 kBq ml−1 125I− was applied evenly to R2A agar plates. Microbial isolates from the F area were inoculated onto the plates using sterile toothpicks and incubated for 3 days in the dark at room temperature (∼27°C). A small portion of microbial colonies, ∼0.25 mm in diameter, was transferred with sterile toothpicks into 7-ml glass vials containing 4 ml of scintillation cocktail (Ecolume). After the colonies were vortexed for 1 min, the radioactivity of accumulated 125I was determined by a liquid scintillation counter (Beckman Coulter LS6500) for 10 min as a preliminary screen. Colonies testing positive for 125I accumulation were then transferred from the agar plates via a traditional plate lift technique onto a nitrocellulose membrane filter (82-mm diameter; Whatman Optitran BA-S 85). As a control for the background, liquid on the agar surface was also transferred onto the nitrocellulose membrane. The membranes were dried at room temperature for 30 min and then exposed to a maximum-sensitivity autoradiography film (Kodak BioMax) at −20°C for 14 days in the dark. Film was processed with developer, a deionized-water stop bath, and fixer according the manufacturer's instructions (Kodak).
For the second method for assessment of iodide accumulation, microbial isolates from the F area and the two reference marine IAB strains (the F. aggregans and A. troitsensis strains) were cultivated in 50-ml conical tubes containing 5 ml of nutrient broth and marine broth, respectively. Stable I− was added to the cultures at a concentration (0.1 μM) that reflected the ambient concentration of 127I− (stable iodine) in the F area. 125I− (1.33 kBq liter−1) was added to the cultures as the tracer. Incubations were performed at room temperature in the dark with continuous shaking (150 rpm). After a 24-h incubation period, a subsample (100 μl) was collected and was immediately transferred into a scintillation vial. Radioactivity of 125I was determined by liquid scintillation counting for 10 min and referred to as “125I activity in the microbial culture.” To investigate the effect of hydrogen peroxide on cellular iodide accumulation, H2O2 (final concentration, 5 mM) was added to a portion (4 ml) of the cell-iodide mixture after the initial 24-h incubation period, and the incubation was continued for an additional 4 to 24 h. Portions of the cell suspension (900 μl) were collected immediately after the initial 24 h of incubation (before H2O2 addition) or 4 or 24 h after H2O2 addition. The collected cell suspension was centrifuged at 16,000 × g for 20 min at 4°C. The supernatant was discarded, and the cell pellet was washed 3 times and suspended in 900 μl of fresh medium (without added stable I− and 125I−). To determine “cell-associated 125I,” a portion (100 μl) of the washed cell suspension was measured by scintillation counting for 10 min (count errors were consistently below 20%). The iodide-accumulating ability (percent accumulation) of microbial cells was calculated as the ratio of “cell-associated 125I” versus “125I activity in the microbial culture.” Cell densities in aliquots were measured at 600 nm with a spectrophotometer (Turner SP8001). Correlations of bacterial dry weight with optical density at 600 nm (OD600) for each target bacterium were obtained during different periods of exponential growth through linear regression, resulting in an r2 value higher than 0.98 for each strain.
Effect of chloride on iodide-accumulating abilities.
Bacteria were grown in two types of aqueous minimal media, M9 and Cl−-deficient M9 (M9X). M9 contained 47.75 mM Na2HPO4, 22.04 mM KH2PO4, 8.56 mM NaCl, 18.70 mM NH4Cl, 2 mM MgSO4, 0.1 mM CaCl2, 22.22 mM glucose, and 0.5% yeast extract. M9X was prepared by replacing NaCl, NH4Cl, and CaCl2 with the same concentrations of Na2SO4, (NH4)2SO4, and Ca(OH)2, respectively. Cells grown in M9 and M9X media were exposed to 2 different concentrations of stable I− (0.1 and 10 μM) while maintaining the same concentration of 125I− (1.33 kBq liter−1). Incubations were performed in the dark at room temperature with continuous shaking at 150 rpm for 24 h. H2O2 (final concentration, 5 mM) was then added to the microbial culture. After an additional 24-h incubation, aliquots were collected for the determination of cellular 125I activity and cell density as described above.
Effect of pH and sodium azide on iodide-accumulating abilities.
To evaluate iodide accumulation as a function of pH, IAB from the F area were grown in the M9 medium containing 0.5% yeast extract at room temperature in the dark with shaking (150 rpm). When the optical density (600 nm) of the cultures reached ∼0.8, cells were harvested by centrifugation (3,500 × g at 20°C for 15 min) and washed 2 times with pH-adjusted minimal medium (pH 4 to 9). The minimal medium contained 8.6 mM NaCl, 18.7 mM NH4Cl, 2 mM MgSO4, 0.1 mM CaCl2, 22.2 mM glucose, and 50 mM KH2PO4. Acetate buffer, phosphate buffer, and borate buffer were used to adjust the pH from 4 to 5, from 6 to 8, and to 9, respectively. To avoid bursting or shrinking of cells by transferring them from M9 to pH-adjusted medium, the ionic strength of pH-adjusted medium was maintained at 177 mM, which was the same ionic strength as in the M9 medium. Cell pellets were suspended in pH-adjusted minimal medium containing stable I− (0.1 μM), 125I− (1.33 kBq liter−1) and H2O2 (5 mM).
In order to investigate the role of heme-containing enzymes or other active cell processes on iodide accumulation, a parallel assay was performed with the addition of sodium azide (NaN3). NaN3 (10 mM) was added to the cell suspension to inhibit enzymatic activities before addition of I− and H2O2. Cell suspensions were then incubated at room temperature in the dark for 24 h, after which aliquots were collected for the determination of 125I activity and cell density as described above.
Iodide desorption.
A procedure similar to that of MacLean et al. (30), with slight modifications, was performed to determine if iodine desorption could occur after accumulation. After iodide accumulation at pH 4 to 9 (described above), cells were collected via centrifugation (16,000 × g for 20 min at 4°C), suspended in M9 medium (pH 7), and incubated with end-over-end shaking (150 rpm) for 2 h. Aliquots were then collected for the determination of 125I activity and cell density as described above.
Statistical analyses.
Analyses of variance (ANOVA) were performed to evaluate the significant differences in iodide-accumulating abilities as a function of iodide concentrations, as well as the presence versus absence of chloride ions. Tukey's post hoc tests were used to compare means of values for iodide-accumulating abilities from significant (P < 0.05) ANOVA test results. The statistical analyses were performed using SPSS 16.0 (SPSS Institute, Inc.).
RESULTS
Isolation and phylogenetic analysis of F-area soil bacteria.
A total of 325 aerobic microbes were directly isolated from subsurface sediments of the F area at the Savannah River Site (Fig. 1). Of these isolates, 32% were cultured from sandy/clay sediments of an aquifer (sites FAW-1 and FAW-4, 13.1 to 25.9 m below the surface) and 29% and 39% were cultured from the dark, organic rich sediments of the seep zone (site FSP-07), 0.15 to 1 m and 1 to 1.2 m below the surface, respectively. Analysis of the 16S rRNA gene revealed that the isolates were members of four phyla within the domain Bacteria. The majority of isolates (55%) were classified as Proteobacteria, followed by Firmicutes (20%), Actinobacteria (17%), and Bacteroidetes (8%). No obvious trends relating the phylogeny of the isolates to their environmental source (seep zone versus sand/clay aquifer or depth of the sediment) or the isolation method employed (pyrophosphate versus sonication or gellan gum versus noble agar) were observed (data not shown).
A yellow coloration was noted on the tubes of 2/24 enrichment cultures (8 sediment sources, with 3 enrichment conditions for each culture; see Materials and Methods) after 22 weeks of incubation in the dark. A strong I2 smell was also detected after the caps of the discolored conical tubes were opened, indicating the transformation of I− to I2 (8). The two enrichment cultures that exhibited yellow coloration were derived from the seep zone (FSP-07) sediments collected 0.15 to 1 and 1 to 1.2 m below the surface, and both were incubated in dH2O with 1 mM I−. Isolation efforts from the 24 enrichment cultures yielded 29 distinct morphotypes. Analysis of 16S rRNA gene sequences revealed that these 29 strains were members of the same 4 phyla, as the isolates recovered directly from sediments. The majority of isolates from the enrichment cultures (76%) were classified as Proteobacteria, followed by Actinobacteria (14%), Bacteroidetes (7%), and Firmicutes (3%).
Screening for iodide-accumulating strains.
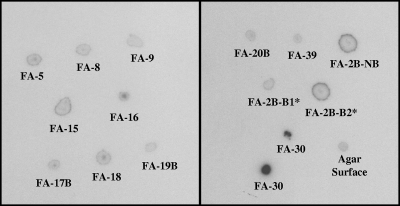
Forty-two strains, representing the phylogenetic diversity of isolates from F-area soils and enrichment cultures, were screened for their ability to accumulate 125I− during growth on R2A agar. Cell material from 14 strains exhibited detectable radioactivity by liquid scintillation counting following growth in the presence of 125I−. Cell mass from colonies of these 14 isolates and liquid from the surface of the 125I− R2A plates was transferred to a nitrocellulose membrane, which was exposed to film for 14 days. An obvious solid black circle was observed where the film had been exposed to cell material from strain FA-30, indicating that this strain accumulated iodide relatively strongly (Fig. 2). Autoradiographic impressions from four of the isolates, FA-5, FA-16, FA-17B, and FA-18, showed a slightly darker region in the center of each image, whereas impressions from FA-15, FA-2B-NB, and FA-2B-B* showed a dark halo around the outer border of each of their respective images (Fig. 2). These distinct, strain-specific autoradiographic patterns were repeatable, suggesting that the strains accumulate or interact with 125I− differently.
FIG. 2.
Autoradiographic image of cell material from 14 F-area isolates (FA-30 was blotted in duplicate) that had been grown on R2A plates containing 125I−. The “Agar Surface” dot shows the background radioactivity from the surface of an R2A plate containing 125I−.
A second method, in which bacteria were grown in liquid medium containing 125I−, was used to assess iodide accumulation among 139 of the F-area isolates (the 42 strains initially screened by autoradiographic analysis of colony material and 97 additional strains) as well as the two I−-accumulating marine bacterial strains, the F. aggregans strain and the A. troitsensis strain. With the use of this method, nine of the F-area strains exhibited the potential to accumulate iodide. However, upon reexamination using the same procedure with 3 additional washes, only one F-area strain (FA-30) and the marine strains consistently exhibited appreciable 125I-accumulating abilities (Table 1) .
TABLE 1.
Iodide accumulation by F-area bacterial isolates and two known IAB isolates
| Bacterial isolate(s) | Without H2O2 |
With H2O2 added at 4 h |
With H2O2 added at 24 h |
|||
|---|---|---|---|---|---|---|
| Cell activity (cpm)a | Accumulation (%)b | Cell activity (Bq/ml) | Accumulation (%) | Cell activity (Bq/ml) | Accumulation (%) | |
| F. aggregans NBRC15975c | 726 | 2.410 | 637 | 2.096 | 2,097 | 7.246 |
| A. troitsensis JCM11736c | 10,170 | 25.482 | 10,365 | 25.609 | 17,635 | 41.489 |
| FA-30 | 105 | 0.383 | 120 | 0.428 | 315 | 1.100 |
| FA-2C-B* | 33 | 0.157 | 61 | 0.296 | 121 | 0.593 |
| FA-191 | 27 | 0.099 | 40 | 0.154 | 67 | 0.251 |
| Other strainsd | 31 (±18) | 0.143 (±0.087) | 26 (±13) | 0.125 (±0.066) | 28 (±17) | 0.134 (±0.082) |
Activity associated with cells washed 3 times following incubation of cells in the presence of 125I for 24, 28, or 56 h. H2O2 was added after 24 h for the 28- and 56-h incubations.
(Number of cpm 125I in cell pellet after incubation/number of cpm 125I in supernatant after incubation) × 100.
Marine strains previously identified as IAB (6).
Average values ± SD for the 133 strains that did not consistently accumulate iodide.
It has been reported that H2O2 plays a role in iodide accumulation and oxidation by marine bacteria (6, 18); thus, iodide accumulation was assessed for the same 139 strains and 2 marine IAB strains in liquid cultures as described above, except that H2O2 was added to the cultures either 4 or 24 h prior to harvesting of the cells. H2O2 addition resulted in greater iodide accumulation for FA-30 and the two marine strains and enabled the identification of two additional iodide-accumulating strains, FA-2C-B* and FA-191 (Table 1). In each of these strains, iodide accumulation was greater in cells that had been exposed to H2O2 for a longer period of time (24 versus 4 h). However, the three IAB strains isolated from the F area, FA-30, FA-2C-B*, and FA-191, exhibited an iodide-specific accumulation that was 2 orders of magnitude less than that observed for the marine IAB strains, the F. aggregans and A. troitsensis strains (Table 1). Among the F-area isolates, the only strain that exhibited an iodide accumulation phenotype with both the plate and the liquid culture assays was FA-30 (among the F-area isolates, FA-30 also exhibited the strongest 125I− accumulation phenotype with the use of both assays).
Phylogenic analysis of the partial 16S rRNA genes (750 to 900 bp) from FA-30, FA-2C-B*, and FA-191 revealed that these strains were members of the Actinobacteria, Firmicutes, and Proteobacteria phyla, respectively. FA-30 was most closely related to various Streptomyces and Kitasatospora species (16S rRNA gene similarity, ∼94%), FA-2C-B* was closely related to Bacillus mycoides L2S8 (EU221418; 98% similarity), and FA-191 was most closely related to various Ralstonia and Cupriavidus species (97 to 98% similarity). FA-191 and FA-30 were cultured from seep zone (FSP-07) sediments, 0.15 to 1 and 1 to 1.2 m below the surface, respectively, whereas strain FA-2C-B* was isolated from one of the I− enrichment cultures. No IAB were found among the isolates obtained from the deeper, oligotrophic sandy/clay aquifer material.
Impact of chloride on iodide accumulation.
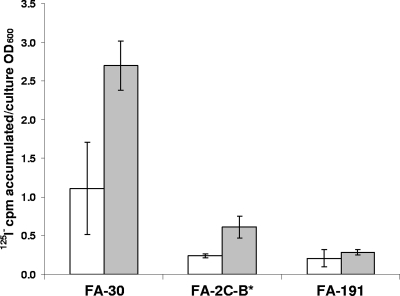
To evaluate whether Cl− present in M9 medium could inhibit iodide accumulation by the F-area IAB, cells of FA-30, FA-2C-B*, and FA-191 were incubated in medium with and without added chloride (27 mM total Cl− concentration, added as 8.56 mM NaCl, 18.70 mM NH4Cl, and 0.1 mM CaCl2). Strains FA-30 and FA-2C-B* accumulated more iodide in the presence of chloride (P > 0.05), whereas chloride did not have a significant effect on I− accumulation in strain FA-191 (Fig. 3).
FIG. 3.
Iodide accumulation by FA-30, FA-2C-B*, and FA-191 in M9 medium containing 27 mM chloride ions (gray bars) versus that in medium without chloride salts (white bars). The experiment was performed using 0.1 μM iodide. Bars represent average amounts of iodide accumulated per cell culture biomass (OD600), and error bars show standard deviations (SD) (n = 3).
Impact of pH and sodium azide on iodide accumulation.
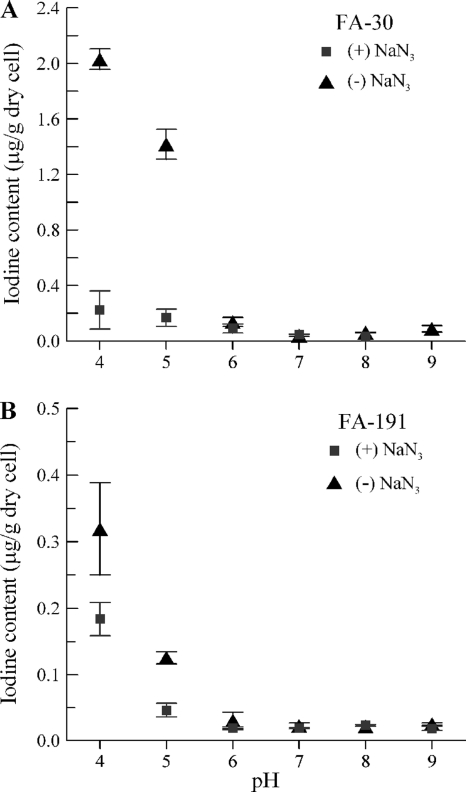
The iodine-accumulating ability of strains FA-30, FA-2C-B*, and FA-191 in liquid cultures grown in pH-adjusted medium (pH 4 to 9) containing iodide (0.1 μM) was evaluated. Between pH 4 and pH 6, there were significant decreases in cellular iodine content in strains FA-30 and FA-191 as the pH increased (Fig. 4). Above pH 6, the cellular iodine content was low, <0.1 μg/g dry cell weight, and either gradually decreased with increasing pH (FA-30) or remained at a constant low level with increasing pH (FA-191). A relationship between pH and iodide accumulation was not observed for FA-2C-B* (data not shown).
FIG. 4.
Correlation between pH and iodide accumulation by FA-30 (A) and FA-191 (B) in the presence (squares) or absence (triangles) of sodium azide. Symbols represent average amounts of iodide accumulated per cell culture biomass (μg g dry cell weight−1), and error bars show standard deviations (n = 3).
Strains FA-30 and FA-191 were tested in a parallel pH assay where sodium azide (NaN3; 10 mM) was included. Both strains exhibited lower I− accumulation when incubated with NaN3, particularly below pH 6.0 (Fig. 4). At pH 4.0, cells of FA-191 and FA-30 accumulated 40% and 90% less I−, respectively, in the presence of NaN3. At pH levels of >6.0, I− accumulation was essentially unaffected by NaN3 (it should be noted, however, that the cellular iodine content of cells incubated in medium with a pH value of >6.0 was barely above background levels).
Desorption of iodine from IAB.
Desorption experiments were conducted with strains FA-30 and FA-191 to evaluate the nature of the interaction between iodide and the cells. Desorption of iodine as a function of pH was not observed in cells of FA-30 or FA-191 (only ±10% variation in the cellular iodine content of the cells before and after the desorption step) (data not shown).
DISCUSSION
Mobility of radioactive iodine in the subsurface environment is affected by the iodine's chemical speciation and interactions with soil constituents, including minerals, organic matter, and microorganisms. Based on thermodynamic principles, the main iodine species in SRS F-area groundwater and sediments should be iodide, which is thought to have the highest subsurface mobility, i.e., to be least sorbed or taken up by sediments, compared to iodate and organo-iodine (33, 40, 43). However, organo-iodine has been found to contribute a significant fraction (up to 25%) of total iodine in groundwater from the F area (43, 52). The extent to which iodide binds to or is incorporated within bacterial cells in oxic subsurface aquifers has not previously been investigated; thus, we tested the iodide-accumulating ability of 139 phylogenetically distinct bacterial isolates from F-area sediments.
Three aerobic bacterial strains from the F area, FA-30, FA-2C-B*, and FA-191, were shown to accumulate iodide. Iodide accumulation by these three strains was significantly different from the background value observed for 136 other strains evaluated (consistently 2 to 10 times higher), and in the case of FA-30, this result was validated by two different approaches, (i) autoradiography of cell material grown on 125I− agar plates and (ii) liquid scintillation analysis of washed cell material that had been grown in liquid medium containing 125I−. Compared to that of IAB isolated from marine sources, however, the iodide-accumulating capacity of the F-area strains was quite small. Amachi et al. (6) reported accumulation of 80 to 90% of total iodide by various marine strains, including F. aggregans NBRC15975 and A. troitsensis JCM11736, when these strains were incubated in the presence of 0.1 μM I−. Under our experimental conditions (i.e., much more rigorous cell washing and higher pH [7.0 versus 6.0] than those used by Amachi et al.), F. aggregans and A. troitsensis accumulated 2 to 50% total iodide (0.1 μM), whereas F-area strains FA-30, FA-2C-B*, and FA-191 accumulated 0.2 to 1.5% of the total iodide (0.1 μM).
Another difference between the IAB isolated from F-area sediments and the IAB previously identified from marine sediments (6) is that the latter microorganisms were classified exclusively within the Flavobacteriaceae family of the Bacteroidetes, whereas the F-area IAB represented three phyla, Actinobacteria (FA-30, most closely related to Streptomyces and Kitasatospora spp.), Firmicutes (FA-2C-B*, a putative Bacillus sp.), and Betaproteobacteria (FA-191, closely related to Ralstonia/Cupriavidus spp.). As these three phyla are often dominant members of terrestrial soil microbial communities (they comprise >90% of the isolates obtained from F-area sediments) whereas Bacteroidetes spp. are more common in marine environments, it appears that iodine accumulation is not restricted to a distinct phylogenetic lineage(s) but rather is manifested in select bacterial taxa adapted to their respective environments.
There are two mechanisms that provide the most parsimonious explanation for iodide accumulation by bacteria. One is electrostatic adsorption of I− by positively charged functional groups, such as amines from proteins and peptides, present on the surface of the cell. For example, MacLean et al. (30) used Bacillus subtilis to model iodide-bacterium adsorption through electrostatic interaction and found that it readily adsorbed iodide at pH values of <4 and that iodide could then be quickly desorbed (∼2 h) by raising the pH to 7.0. In that study, an aqueous solution of diluted HNO3 was used as the experimental wash solution, precluding other halide ions from competing with iodide for electrostatic interaction with bacterial cells. In the present study, the three terrestrial IAB strains were incubated and maintained in a M9 minimal medium solution that contained ∼27 mM chloride ions, which is ∼2.7 × 105 times higher than the iodide concentration (0.1 μM) present in the experimental assays. Iodide accumulation by FA-30, FA-2C-B*, and FA-191 was not inhibited by chloride ions (27 mM) (Fig. 3). Indeed, at low iodide concentrations (0.1 μM), I− accumulation by FA-30 and FA-2C-B* was greater in M9 medium than in chloride-deficient M9 medium (we note that there was a difference in ionic strength between the standard M9 medium and chloride-deficient M9 medium used in this experiment). Furthermore, pH-dependent desorption tests failed to reveal reversibility of the interaction between iodide and cells of FA-30 and FA-191. These results indicate that I− accumulation by the F-area terrestrial strains was not due to electrostatic surface adsorption when pH levels were ≥4.0.
The second possible mechanism for adsorption between iodide and cellular constituents is electrophilic substitution resulting in iodination of organic molecules. Strong electrophiles such as HOI/I2/I3− produced by biotic or abiotic processes from I− could attack aromatic rings or other organic moieties of the cell and replace -H with -I to form a stable organo-iodine bond (12, 31, 44). Given the high concentration of organic matter in a bacterial culture, it is reasonable to expect that the oxidized I species, i.e., HOI/I2/I3−, could react with it. Several observations signify that the iodide accumulation phenotype exhibited by F-area strains FA-30, FA-2C-B*, and FA-191 likely proceeds via this mechanism. First, the iodide-accumulating abilities from FA-30, FA-2C-B*, FA-191 were all facilitated by the addition of H2O2. Since H2O2 is a strong oxidant, I− is readily oxidized to I2/I3− without enzymatic catalysis, especially under acidic conditions. However, at the near-neutral pH values (pH 6 to 7) that were used during the screening portion of our study, H2O2 consistently stimulated iodide accumulation in just 5 of 141 strains tested (including the two marine strains). Thus, it is unlikely that H2O2, itself acting as an oxidizing agent, transformed I− into a highly reactive species capable of binding nonspecifically to cell material (though at lower pH values [<4 to 5], this mechanism could be more relevant). Alternatively, haloperoxidases, which are found in animals, plants (including algae), fungi, and bacteria, utilize H2O2 as a cosubstrate and are considered the primary enzyme system responsible for nonspecific halogenation of organic substrates in nature (11, 45, 46). NaN3 significantly inhibited I− accumulation in the F-area strains, implicating the involvement of an enzymatic driven process, such as an active transport system or heme haloperoxidases, which are NaN3 sensitive (45). The F-area strains also exhibited increased I− accumulation with decreasing pH, characteristic of many haloperoxidases, which exhibit optimal activity under acidic conditions (14, 36, 38). Although our data support electrophilic substitution or internalization rather than electrostatic adsorption to the cell surface, the precise nature and location of bacterially bound I− in these terrestrial strains and the accumulation mechanism remain to be determined.
Background concentrations of stable I in F-area groundwater range from 10 to 100 nM, and plume concentrations of 129I are typically ∼60 pCi/liter (2.4 nM) at “hot spots” but can reach levels of >900 pCi/liter in the organic-rich seep zone. Our results demonstrate that the majority (98%) of aerobic bacteria isolated from F-area sediments do not accumulate iodide (<0.2% accumulation) at ambient I− concentrations (0.1 μM), and the three IAB strains that were identified accumulate less than 2% I− under environmental conditions (aerobic; pH 4 to 9; 0.1 μM total I−) associated with most of the F-area plume (at the center of the plume, pH values as low as 3.2 have been documented, where electrostatic adsorption of I− by bacterial cells as demonstrated by MacLean et al. [30] could play a role in I− transport). Our experiments were conducted with dense cell cultures (≥1 × 109 cells ml−1), whereas cell numbers in groundwater from the sandy/clay aquifer of the F area are lower than 1 × 104 cells ml−1 (data not shown). At these cell concentrations, cellular accumulation of I− would be exceedingly low. Furthermore, each of the IAB strains identified in this study was isolated from the seep zone sediments, not the sandy/clay aquifer material. These results indicate that IAB are most likely not responsible for the high fraction of organo-iodine (up to 25% of total iodine) that has been measured in groundwater of the F-area subsurface aquifer above the seep zone (43, 51). However, our ongoing experiments with iodine-oxidizing bacteria from F-area soils thus far indicate that this pathway for organo-iodine formation is more significant.
Our multifaceted, carefully controlled approach allowed us to definitively identify an IAB phenotype that was 1 to 2 orders of magnitude less, in terms of specific iodine accumulation activity, than that previously established for bacteria from very different environments (i.e., brines). This is important for several reasons. 129I has an extremely long half-life (∼16 million years), and its production is increasing each year. The DOE and other entities are tasked with modeling the long-term (centuries to thousands of years) fate and transport of 129I. Over decades or centuries, I− accumulated by bacterial cells and covalently attached to cellular constituents could conceivably make its way to the organo-iodine pool through cell lysis and possible incorporation into more-refractory organic soil material (e.g., humic or fulvic acids). Even when bacteria, whose biomass typically accounts for 1% or less of sedimentary organic matter (such as the F-area seep zone sediments) (25), incorporate less than 2% of iodine into their cells, this process could contribute appreciably to the organo-iodine pool over the long term. Similar mechanisms have been proposed to explain chloride retention in forest soils and peat bogs over decades to centuries (10). Carefully controlled, long-term column studies are needed to examine the extent that IAB, such as those identified in this study, affect 129I speciation and mobility in F-area seep zone sediments. Finally, uncultivated bacterial species yet to be discovered from the F area or fungi may be capable of much higher levels of iodide accumulation (9). We are currently examining that possibility through a microautoradiography-fluorescence in situ hybridization (MAR-FISH) approach (29).
Acknowledgments
We thank Melanie Dunn for providing assistance in the pH effect assays.
This work was funded by the U.S. Department of Energy's Subsurface Biogeochemical Research Program within the Office of Science (DE-FG02-08ER64567, modification no. 002) and partially supported by Welch Grant BD0046.
Footnotes
Published ahead of print on 28 January 2011.
REFERENCES
- 1.Amachi, S. 2008. Microbial contribution to global iodine cycling: volatilization, accumulation, reduction, oxidation, and sorption of iodine. Microbes Environ. 23:269-276. [DOI] [PubMed] [Google Scholar]
- 2.Amachi, S., K. Kimura, Y. Muramatsu, H. Shinoyama, and T. Fujii. 2007. Hydrogen peroxide-dependent uptake of iodine by marine Flavobacteriaceae bacterium strain C-21. Appl. Environ. Microbiol. 73:7536-7541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amachi, S., K. Minami, I. Miyasaka, and S. Funkanaga. 2010. Ability of anaerobic microorganisms to associate with iodine: 125I tracer experiments using laboratory strains and enriched microbial communities from subsurface formation water. Chemosphere 79:349-353. [DOI] [PubMed] [Google Scholar]
- 4.Amachi, S., et al. 2003. Microbial participation in iodine volatilization from soils. Environ. Sci. Technol. 37:3885-3890. [DOI] [PubMed] [Google Scholar]
- 5.Amachi, S., Y. Kamagata, T. Kanagawa, and Y. Muramatsu. 2001. Bacteria mediate methylation of iodine in marine and terrestrial environments. Appl. Environ. Microbiol. 67:2718-2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Amachi, S., Y. Mishima, H. Shinoyama, Y. Muramatsu, and T. Fujii. 2005. Active transport and accumulation of iodide by newly isolated marine bacteria. Appl. Environ. Microbiol. 71:741-745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Amachi, S., Y. Muramatsu, H. Shinoyama, and T. Fujii. 2005. Application of autoradiography and a radiotracer method for the isolation of iodine-accumulating bacteria. J. Radioanal. Nucl. Chem. 266:229-233. [Google Scholar]
- 8.Amachi, S., et al. 2005. Isolation of iodide-oxidizing bacteria from iodide-rich natural gas brines and seawaters. Microb. Ecol. 49:547-557. [DOI] [PubMed] [Google Scholar]
- 9.Ban-nai, T., Y. Muramatsu, and S. Amachi. 2006. Rate of iodine volatilization and accumulation by filamentous fungi through laboratory cultures. Chemosphere 65:2216-2222. [DOI] [PubMed] [Google Scholar]
- 10.Bastviken, D., et al. 2007. Chloride retention in forest soil by microbial uptake and by natural chlorination of organic matter. Geochim. Cosmochim. Acta 71:3182-3192. [Google Scholar]
- 11.Butler, A., and M. Sandy. 2009. Mechanistic considerations of halogenating enzymes. Nature 460:848-854. [DOI] [PubMed] [Google Scholar]
- 12.Christiansen, J. V., and L. Carlsen. 1991. Iodinated humic acids, p. 467-474. In B. Allard, H. Boren, and A. Grimvall (ed.), Humic substances in the aquatic and terrestrial environment, vol. 33. Springer-Verlag Berlin, Berlin, Germany. [Google Scholar]
- 13.Cole, J. R., et al. 2009. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 37:D141-D145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Colin, C., et al. 2003. The brown algal kelp Laminaria digitata reatures distinct bromoperoxidase and iodoperoxidase activities. J. Biol. Chem. 278:23545-23552. [DOI] [PubMed] [Google Scholar]
- 15.Cook, P. L. M., P. D. Carpenter, and E. C. V. Butler. 2000. Speciation of dissolved iodine in the waters of a humic-rich estuary. Mar. Chem. 69:179-192. [Google Scholar]
- 16.Denham, M., D. Kaplan, and C. Yeager. 2009. Groundwater radioiodine: prevalence, biogeochemistry, and potential remedial approaches. SRNL-STI-2009-00463; TRN: US200921%%175. Savannah River National Laboratory, Aiken, SC.
- 17.Fuge, R. 2005. Soils and iodine deficiency, p. 417-433. In O. Selinus et al. (ed.), Essentials of medical geology. Elsevier, Amsterdam, Netherlands.
- 18.Fuse, H., H. Inoue, K. Murakami, O. Takimura, and Y. Yamaoka. 2003. Production of free and organic iodine by Roseovarius spp. FEMS Microbiol. Lett. 229:189-194. [DOI] [PubMed] [Google Scholar]
- 19.Gilfedder, B. S., M. Petri, and H. Biester. 2009. Iodine speciation and cycling in fresh waters: a case study from a humic rich headwater lake (Mummelsee). J. Limnol. 68:396-408. [Google Scholar]
- 20.Heumann, K. G., et al. 2000. Aging of dissolved halogenated humic substances and the microbiological influence on this process. Acta Hydrochim. Hydrobiol. 28:193-201. [Google Scholar]
- 21.Hiergesell, R. A., et al. 2008. Inventory of residual radioactive material at the projected Savannah River Site end state. SRNL-STI-2008-00380, Rev. 0. Savannah River National Laboratory, Aiken, SC.
- 22.Hou, X. L., et al. 2009. A review on speciation of iodine-129 in the environmental and biological samples. Anal. Chim. Acta 632:181-196. [DOI] [PubMed] [Google Scholar]
- 23.Johanson, K. J. 2000. Iodine in soil. SKB technical reports, no. TR-00-21. Swedish Nuclear Fuel and Waste Management Company, Stockholm, Sweden.
- 24.Kincaid, C. T., et al. 2006. Inventory data package for Hanford assessments. PNNL-15829, Rev. 0. Pacific Northwest National Laboratory, Richland, WA.
- 25.Kindler, R., A. Miltner, H.-H. Richnow, and M. Kästner. 2006. Fate of gram-negative bacterial biomass in soil-mineralization and contribution to SOM. Soil Biol. Biochem. 38:2860-2870. [Google Scholar]
- 26.Reference deleted.
- 27.Küpper, F. C., et al. 1998. Iodine uptake in Laminariales involves extracellular, haloperoxidase-mediated oxidation of iodide. Planta 207:163-171. [Google Scholar]
- 28.Reference deleted.
- 29.Li, H. P., et al. 2010. The potential role of microbes on iodine-129 mobility in groundwater relevant to long-term stewardship of DOE sites, abstr. LBNL-43E-2010, p. 199. Abstr. Subsurf. Biogeochem. Res. Program Princ. Invest. Meet., Washington, DC, 28 to 31 March 2010.
- 30.MacLean, L. C. W., R. E. Martinez, and D. A. Fowle. 2004. Experimental studies of bacteria—iodide adsorption interactions. Chem. Geol. 212:229-238. [Google Scholar]
- 31.Moulin, V., P. Reiller, B. Amekraz, and C. Moulin. 2001. Direct characterization of iodine covalently bound to fulvic acids by electrospray mass spectrometry. Rapid Commun. Mass. Spectrom. 15:2488-2496. [DOI] [PubMed] [Google Scholar]
- 32.Muramatsu, Y., S. Yoshida, S. Uchida, and A. Hasebe. 1996. Iodine desorption from rice paddy soil. Water Air Soil Pollut. 86:359-371. [Google Scholar]
- 33.Muramatsu, Y., S. Yoshida, U. Fehn, S. Amachi, and Y. Ohmomo. 2004. Studies with natural and anthropogenic iodine isotopes: iodine distribution and cycling in the global environment. J. Environ. Radioact. 74:221-232. [DOI] [PubMed] [Google Scholar]
- 34.Ortiz-Bermúdez, P., C. H. Kolby, E. Srebotnik, and K. E. Hammel. 2007. Chlorination of lignin by ubiquitous fungi has a likely role in global organochlorine production. Proc. Natl. Acad. Sci. U. S. A. 104:3895-3900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reference deleted.
- 36.Pommier, J., L. Sokoloff, and J. Nunez. 1973. Enzymatic iodination of protein: kinetics of iodine formation and protein iodination catalyzed by horse-radish peroxidase. Eur. J. Biochem. 38:497-506. [DOI] [PubMed] [Google Scholar]
- 37.Radlinger, G., and K. G. Heumann. 2000. Transformation of iodide in natural and wastewater systems by fixation on humic substances. Environ. Sci. Technol. 34:3932-3936. [Google Scholar]
- 38.Renganathan, V., K. Miki, and M. H. Gold. 1987. Haloperoxidase reactions catalyzed by lignin peroxidase, an extracellular enzyme from the basidiomycete Phanerochaete chrysosporium. Biochemistry 26:5127-5132. [Google Scholar]
- 39.Reference deleted.
- 40.Santschi, P. H., and K. A. Schwehr. 2004. I-129/I-127 as a new environmental tracer or geochronometer for biogeochemical or hydrodynamic processes in the hydrosphere and geosphere: the central role of organo-iodine. Sci. Total Environ. 321:257-271. [DOI] [PubMed] [Google Scholar]
- 41.Schwehr, K. A., and P. H. Santschi. 2003. Sensitive determination of iodine species, including organo-iodine, for freshwater and seawater samples using high performance liquid chromatography and spectrophotometric detection. Anal. Chim. Acta 482:59-71. [Google Scholar]
- 42.Schwehr, K. A., P. H. Santschi, and D. Elmore. 2005. The dissolved organic iodine species of the isotopic ratio of I-129/I-127: a novel tool for tracing terrestrial organic carbon in the estuarine surface waters of Galveston Bay, Texas. Limnol. Oceanogr. Methods 3:326-337. [Google Scholar]
- 43.Schwehr, K. A., P. H. Santschi, D. I. Kaplan, C. M. Yeager, and R. Brinkmeyer. 2009. Organo-iodine formation in soils and aquifer sediments at ambient concentrations. Environ. Sci. Technol. 43:7258-7264. [DOI] [PubMed] [Google Scholar]
- 44.Steinberg, S. M., et al. 2008. Immobilization of fission iodine by reaction with insoluble natural organic matter. J. Radioanal. Nucl. Chem. 277:175-183. [Google Scholar]
- 45.Van Pée, K.-H. 1996. Biosynthesis of halogenated metabolites by bacteria. Annu. Rev. Microbiol. 50:375-399. [DOI] [PubMed] [Google Scholar]
- 46.Wagner, C., M. El Omari, and G. M. Koìnig. 2009. Biohalogenation: nature's way to synthesize halogenated metabolites. J. Nat. Prod. 72:540-553. [DOI] [PubMed] [Google Scholar]
- 47.Reference deleted.
- 48.Wolfgang, L., et al. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wong, G. T. F., and X. H. Cheng. 1998. Dissolved organic iodine in marine waters: Determination, occurrence and analytical implications. Mar. Chem. 59:271-281. [Google Scholar]
- 50.World Nuclear Association. 2009. India, China & NPT. World Nuclear Association, London, United Kingdom. http://www.world-nuclear.org/info/inf80.html.
- 51.Zhang, S., et al. 2010. Determination of 127I and 129I speciation in environmental waters using a novel gas chromatography-mass spectrometry method. Environ. Sci. Technol. 44:9042-9048. [DOI] [PubMed] [Google Scholar]