Abstract
Five thousand mutants of Herbaspirillum seropedicae SmR1 carrying random insertions of transposon pTnMod-OGmKmlacZ were screened for differential expression of LacZ in the presence of naringenin. Among the 16 mutants whose expression was regulated by naringenin were genes predicted to be involved in the synthesis of exopolysaccharides, lipopolysaccharides, and auxin. These loci are probably involved in establishing interactions with host plants.
The betaproteobacterium Herbaspirillum seropedicae is an endophytic diazotroph that forms nitrogen-fixing associations with maize (Zea mays), rice (Oryza sativa), sorghum (Sorghum bicolor), and sugar cane (Saccharum officiarum), as well as such diverse plants as bananas (Musa spp.) and pineapple (Ananas comosus) (1, 5, 27). As such, H. seropedicae is a potential nitrogen biofertilizer that also produces phytohormones that may stimulate the growth of plants (4). Studies demonstrated that the inoculation of rice with H. seropedicae promoted a yield increase equivalent to treatment with 40 kg N/ha (2). Coinoculation of micropropagated sugar cane with Gluconacetobacter diazotrophicus and Herbaspirillum sp. resulted in an increase of the total plant biomass (25).
Exactly how H. seropedicae colonizes Gramineae and other plants is not known, and there is even less information available on the role of plant metabolites in the regulation of bacterial invasion and colonization of the inner tissues. That such associations involve molecular communication between the host plant and bacteria, resulting in modified patterns of gene expression, is clear from studies of other rhizospheric bacteria (15). In legume-Rhizobium interactions, flavonoids released by plant roots induce sets of genes involved in nodulation. As a result, lipochitooligosaccharides are excreted and symbiotic forms of exopolysaccharides (EPSs), lipopolysaccharides (LPSs), and glucans synthesized, all of which modulate the nodulation process (8, 15, 26). Previous studies showed that naringenin (50 μmol/liter) stimulated the root colonization of Arabidopsis thaliana by H. seropedicae (16) and the intercellular colonization of wheat roots by Azorhizobium caulinodans (33).
To identify bacterial genes whose expression is under the control of the flavonoid naringenin, the chromosome of H. seropedicae strain SmR1 (23) was randomly mutagenized using a lacZ-Km-Gm cassette carried by the plasmid pTnMod-OGmKmlacZ, and strains resistant to both kanamycin and gentamicin were selected (30). Twelve thousand mutant strains were obtained, and 5,000 screened for differential expression of the promoterless lacZ reporter gene in the presence of naringenin (50 μmol/liter) using NFbHP-malate-agar medium (19) containing X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) (30 μg/ml). One hundred ninety-six mutants bearing mutated genes potentially controlled by naringenin were preselected, and their β-galactosidase activities determined (7, 24). Of these, 16 isolates carried mutated genes that reacted differently to the presence of naringenin: 4 of the mutants carried genes that were upregulated, and 12 carried genes that were downregulated by naringenin (Table 1). The mutated genes of these strains were identified by sequencing as described previously (30).
TABLE 1.
Identification of genes from H. seropedicae that are regulated by the flavonoid naringenin
| Mutant straina | Accession no. of the mutated locus | Deduced gene productb | Relative naringenin regulation (avg expression ± SD) shown by: |
|
|---|---|---|---|---|
| β-Gal activityc | RT-PCRd | |||
| MHS01 | YP_003775401.1 | EPS biosynthesis protein (EpsG) | Downregulated (0.31 ± 0.05) | Downregulated (0.07 ± 0.045) |
| MHS02 | YP_003775614 | Probable O-antigen acetylase | Downregulated (0.34 ± 0.09) | Downregulated (0.23 ± 0.11) |
| MHS03 | YP_003773926.1 | Conserved hypothetical protein | Downregulated (0.50 ± 0.01) | Downregulated (0.52 ± 0.082) |
| MHS04 | YP_003774406 | Probable acyltransferase protein | Downregulated (0.44 ± 0.07) | Downregulated (0.43 ± 0.047) |
| MHS05 | YP_003773531.1 | Probable muropeptide permease of the major facilitator superfamily protein (AmpG) | Downregulated (0.33 ± 0.05) | Downregulated (0.12 ± 0.043) |
| MHS06 | YP_003778011.1 | Probable acyl CoA:acetate/3-ketoacid CoA transferase, beta subunit protein | Upregulated (1.65 ± 0.21) | Upregulated (1.23 ± 0.012) |
| MHS07 | YP_003774121.1 | Probable tRNA (5-methylaminomethyl-2-thiouridylate)-methyltransferase protein (TrmU) | Downregulated (0.66 ± 0.11) | Downregulated (0.24 ± 0.063) |
| MHS08 | YP_003776865.1 | Hypothetical protein | Upregulated (1.66 ± 0.07) | Upregulated (1.13 ± 0.09) |
| MHS09 | YP_003775366.1 | Probable 5-aminolevulinic acid synthase protein | Upregulated (2.04 ± 0.03) | Downregulated (0.93 ± 0.046) |
| MHS10 | YP_003776866.1 | Hypothetical protein | Downregulated (0.18 ± 0.001) | Downregulated (0.71 ± 0.01) |
| MHS11 | YP_003778130.1 | Conserved hypothetical protein | Upregulated (1.22 ± 0.015) | Downregulated (0.81 ± 0.056) |
| MHS12 | YP_003775661.1 | Probable NAD-dependent aldehyde dehydrogenase protein | Downregulated (0.27 ± 0.03) | Downregulated (0.258 ± 0.075) |
| MHS13 | YP_003777134.1 | Hypothetical protein | Downregulated (0.54 ± 0.12) | Downregulated (0.21 ± 0.038) |
| MHS14 | YP_003777655.1 | Probable indole pyruvate ferredoxin oxidoreductase, alpha and beta subunits protein | Downregulated (0.042 ± 0.02) | Downregulated (0.15 ± 0.015) |
| MHS15 | YP_003777590.1 | Glucosyltransferase hypothetical protein | Downregulated (0.54 ± 0.12) | Downregulated (0.23 ± 0.04) |
| MHS16 | YP_003775234.1 | Conserved hypothetical protein | Downregulated (0.12 ± 0.04) | Downregulated (0.17 ± 0.053) |
Sixteen insertion mutants of H. seropedicae SmR1 carry genes whose expression is modulated by naringenin.
BLASTn and SMART comparisons of the genes with publicly available databases (accessed on 28 July 2010) are shown.
Expression levels were measured as β-galactosidase (β-Gal)-specific activities of the parental strain and lacZ fusion mutants, and relative expression is reported as the ratio of β-galactosidase activity in the presence of 50 μmol/liter naringenin to β-galactosidase activity in the absence of naringenin.
Relative expression was determined by the 2−ΔΔCT (threshold cycle) method (22); the relative expression of the epsB gene located in the same operon as epsG was 0.082 ± 0.01. The results represent three replicate experiments. The RT-PCR experiments were performed using Power SYBR green master mix (Applied Biosystems, Carlsbad, CA), and reactions were run on a Step One Plus real-time PCR system (Applied Biosystems).
The fraction of the genome covered by the set of insertion mutants was calculated according to the method of Jacobs et al. (17). The total length of the H. seropedicae genome is 5,513,887 bp, containing 4,737 open reading frames (ORFs) with an average length of 1,029 bp. Since a total of 5,000 mutants were tested and assuming that half of these (2,500) would have the lacZ gene inserted in the mutated gene transcription orientation, approximately 38% of the genome was covered by the mutant collection screened (17).
Among the mutants in genes downregulated (2- to 3-fold) by naringenin, four were affected in genes probably involved in the synthesis of cell wall components. Strain MHS01 was mutated in epsG, which is involved in exopolysaccharide biosynthesis; MHS02 in a gene for a probable O-antigen acetylase; MHS05 in ampG, which codes for a muropeptide permease of the major facilitator superfamily; and MHS15 in a gene coding for a probable glucosyl transferase bearing low similarity to GumH, a glucosyl transferase from Xylella fastidiosa that is involved in exopolysaccharide biosynthesis. Interestingly, strain MHS03 was mutated in a gene coding for a conserved protein with no function assigned but located in an operon together with two genes coding for putative outer membrane proteins. One of these is a probable outer membrane porin of the OmpA family, suggesting that the mutated gene may also play a role in outer membrane structure. LPSs consist of an O-antigen, a lipid A, and core oligosaccharides and are present as a monolayer on the outer membrane of Gram-negative bacterial cells. Strain MHS02 carries a mutation that may affect the acetylation of the O-antigen part of the LPS of H. seropedicae. Similar changes to the O-antigen of rhizobia result in altered symbiotic phenotypes (9, 21).
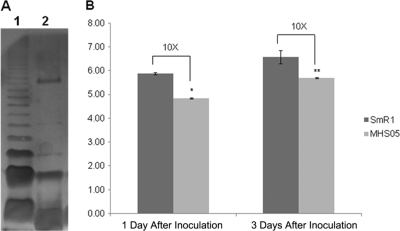
Changes in bacterial outer surface structures, such as LPS and EPS, often result in increased sensitivity to cationic peptide antibiotics, detergents, and various stresses and affect symbiotic development (9, 10, 18, 20, 21, 31). Accordingly, we challenged the collection of mutants with increasing concentrations of sodium dodecyl sulfate (SDS). In contrast to the parental strain, isolates with mutations in genes related to the synthesis of EPS (MHS01) and muropeptide permease (MHS05) were unable to grow in the presence of 0.5% (wt/vol) SDS (data not shown). Since mutants affected in LPS synthesis had an altered sensitivity to SDS, we analyzed the LPS profiles of isolates MHS01 and MHS05. LPSs were extracted using the phenol procedure (13) and separated on denaturing polyacrylamide gels (20). Only mutation of the muropeptide permease gene caused significant changes to the LPS profile (Fig. 1A). Although the band pattern is similar to that of the parental strain, the amount of LPS recovered per cell of strain MSH05 is clearly reduced, suggesting that LPS may not be qualitatively different but is present in a lower concentration in the ampG strain's outer membrane. The AmpG protein is located in the membrane of Gram-negative bacteria and is necessary for cell wall peptidoglycan recycling (11). The peptidoglycan is both a highly complex and essential macromolecule of bacterial cell walls (6). The results suggest that the lack of AmpG in strain MSH05 leads to an anomaly in the peptidoglycan layer affecting the synthesis or targeting of LPS in H. seropedicae.
FIG. 1.
LPS profiles and colonization of the roots of Zea mays by the parental H. seropedicae strain SmR1 and ampG mutant MHS05. (A) Sodium deoxycholate-PAGE electrophoresis patterns of LPS isolated from the parental strain (lane 1) and the mutant derivative (lane 2) grown in NFbHPN-malate medium. (B) Colonization of Z. mays roots. Results are shown as the average numbers of bacteria·g−1 fresh root ± standard deviations. The values above the columns represent the difference in colonization between parental and mutant strains. Asterisks indicate statistically (Student t test) significant differences between the parental strain and the ampG mutant in colonization of maize roots. *, P < 0.005; **, P < 0.01.
To check whether any of the mutations influenced the colonization of maize (Zea mays), the protocol of Balsanelli et al. (3) was used to test bacterial cell adhesion to root surface and internal colonization. No difference was observed between the parental strain (SmR1) and mutants in growth and root surface attachment (data not shown). On the other hand, the endophytic population of MHS05 was reduced by 10-fold compared to that of the parental strain both 1 and 3 days after inoculation, suggesting that the modification in LPS in this strain is important for bacterial establishment in plant tissues (Fig. 1B). Previously, Balsanelli et al. (3) showed that strains unable to synthesize LPS were severely impaired in maize colonization. These authors also showed that the rmlB gene, whose product is involved in LPS biosynthesis, was upregulated by naringenin (4-fold), in agreement with our results showing that naringenin can modulate genes involved in the biosynthesis and modification of LPS. All other strains had the same pattern of colonization as the parental strain (data not shown).
Genes coding for a putative methyltransferase (MHS07), a probable acyltransferase (MHS04), an aldehyde dehydrogenase (MHS12), and indole pyruvate ferredoxin oxidoreductase (MHS14) were also downregulated by naringenin. Since the mutated indole pyruvate ferredoxin oxidoreductase in strain MHS14 may be involved in indole acetic acid (IAA) biosynthesis, the results suggest that naringenin may modulate indole acetic acid synthesis in H. seropedicae. The relationship of plant flavonoid and auxin production was also observed in Azospirillum brasilense, where mutation of ipdC of the IAA biosynthetic pathway caused a reduction of exudation of flavonoids by maize roots (14), and flavonoids induce IAA production in Rhizobium NGR234 (32). In the latter organism, the genes y4wE (class 2 amino transferase) or yw4F (monooxygenase) are regulated by flavonoids through NodD.
Four strains were mutated in genes upregulated by naringenin. These include those coding for a putative acyl coenzyme A (CoA):acetate/3-ketoacid CoA transferase (MHS06), a hypothetical protein (MHS08), an aminotransferase similar to 5-aminolevulinic acid synthase (MHS09), and a MarR transcription regulator that may be involved in multidrug resistance (MHS11). The deduced product of the gene mutated in strain MHS08 is similar to a protein of Vibrio cholera of the type II secretory pathway. This system is used by many Gram-negative bacteria, mainly to secrete extracellular enzymes that are associated with the degradation of host tissue (28, 29). The gene coding for the aminotransferase (MHS09) is located in an operon together with a gene coding for a permease of the MFS family, which is possibly related to the tetracenomycin C resistance protein. Cho et al. (12) identified a similar multidrug resistance permease gene of Escherichia coli that is upregulated by an infusion containing polyphenols. In strain MHS16, the gene mutated is located in an operon together with another acetyltransferase, an organization that is conserved in Burkholderia vietnamiensis. Thus, altogether, at least three different genes coding for acyltransferases seem to have their expression affected by naringenin.
Four other mutations were in hypothetical genes of H. seropedicae whose products had no significant matches in all databases (MHS03, MHS10, MHS13, and MHS16). The genes mutated in strains MHS03 and MHS10 probably code for membrane proteins.
Real-time PCR analysis was used to validate the data obtained with lacZ fusions. To do this, total RNA was isolated from cultures grown in the presence and absence of naringenin (50 μmol/liter) using the Trizol procedure (Sigma Corp., St. Louis, MO). Two micrograms of RNA was used to synthesize cDNA with a high-capacity cDNA reverse transcription kit (Applied Biosystems, Carlsbad, CA). The 16S rRNA gene was used as the internal control. Primers for the gene epsB located in the same operon as epsG were also used, to confirm the coregulation of genes related to EPS synthesis.
The RT-PCR analyses confirmed regulation of the mutated genes by naringenin except for strains MHS08, MHS09, and MHS11 (Table 1). In these three strains, the β-galactosidase activity indicated upregulation by naringenin, whereas RT-PCR indicated no regulation (MHS08 and MHS09) or even slight downregulation (MHS11), raising the possibility of posttranscriptional regulation of these genes. Finally, the epsB and epsG genes responded similarly to naringenin, suggesting that the eps operon is regulated by the flavonoid.
Our results show that H. seropedicae responds to the plant flavonoid naringenin by modifying the pattern of gene expression. The products of four genes with altered expression seem to be involved in the synthesis of the outer membrane of the cell wall. Changes in the cell surface are documented in other bacteria that interact with plants (e.g., rhizobia) (9, 20, 31) and probably play a role in the interaction between H. seropedicae and its host plants. Furthermore, the data indicate that naringenin may also regulate genes involved in phytohormone production in H. seropedicae.
Acknowledgments
This work was supported by the Brazilian agencies CAPES and INCT-FBN/CNPq.
We thank Roseli Prado and Julieta Pie for technical assistance.
Footnotes
Published ahead of print on 21 January 2011.
REFERENCES
- 1.Baldani, J. I., V. L. D. Baldani, L. Seldin, and J. Döbereiner. 1986. Characterization of Herbaspirillum seropedicae gen. nov., sp. nov., a root-associated nitrogen-fixing bacterium. Int. J. Syst. Bacteriol. 33:167-172. [Google Scholar]
- 2.Baldani, J. I., and V. L. Baldani. 2005. History on the biological nitrogen fixation research in graminaceous plants: special emphasis on the Brazilian experience. An. Acad. Bras. Cienc. 77:549-579. [DOI] [PubMed] [Google Scholar]
- 3.Balsanelli, E., et al. 2010. Herbaspirillum seropedicae rfbB and rfbC genes are required for maize colonization. Environ. Microbiol. 12:2233-2244. [DOI] [PubMed] [Google Scholar]
- 4.Bastián, F., et al. 1998. Production of indole-3-acetic acid and gibberellins A1 and A3 by Acetobacter diazotrophicus and Herbaspirillum seropedicae in chemically-defined culture media. Plant Growth Regul. 24:7-11. [Google Scholar]
- 5.Boddey, R. M., et al. 1995. Biological nitrogen fixation associated with sugar cane and rice: contributions and prospects for improvement. Plant Soil 74:195-209. [Google Scholar]
- 6.Boneca, I. G. 2005. The role of peptidoglycan in pathogenesis. Curr. Opin. Microbiol. 8:46-53. [DOI] [PubMed] [Google Scholar]
- 7.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 8.Broughton, W. J., S. Jabbouri, and X. Perret. 2000. Keys to symbiotic harmony. J. Bacteriol. 182:5641-5652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Broughton, W. J., et al. 2006. Flavonoid-inducible modifications to rhamnan O antigens are necessary for Rhizobium sp. strain NGR234-legume symbiosis. J. Bacteriol. 188:3654-3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Campbell, G. R. O., B. L. Reuhs, and G. C. Walker. 2002. Chronic intra-cellular infection of alfalfa nodules by Sinorhizobium meliloti requires correct lipopolysaccharide core. Proc. Natl. Acad. Sci. U. S. A. 99:3928-3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chahboune, A., M. Decaffmeyer, R. Brasseur, and B. Joris. 2005. Membrane topology of the Escherichia coli AmpG permease required for recycling of cell wall anhydromuropeptides and AmpC beta-lactamase induction. Antimicrob. Agents Chemother. 49:1145-1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cho, Y. S., N. L. Schiller, H. Y. Kahng, and K. H. Oh. 2007. Cellular responses and proteomic analysis of Escherichia coli exposed to green tea polyphenols. Curr. Microbiol. 55:501-506. [DOI] [PubMed] [Google Scholar]
- 13.Currie, C. G., and I. R. Poxton. 1999. The lipopolysaccharide core type of Escherichia coli O157:H7 and other non-O157 verotoxin-producing E. coli. FEMS Immunol. Med. Microbiol. 24:57-62. [DOI] [PubMed] [Google Scholar]
- 14.Dardanelli, M. S., et al. 2008. Effect of Azospirillum brasilense coinoculated with Rhizobium on Phaseolus vulgaris flavonoids and Nod factor production under salt stress. Soil Biol. Biochem. 40:2713-2721. [Google Scholar]
- 15.Deakin, W. J., and W. J. Broughton. 2009. Symbiotic use of pathogenic strategies: rhizobial protein secretion systems. Nat. Rev. Microbiol. 7:312-320. [DOI] [PubMed] [Google Scholar]
- 16.Gough, C., et al. 1997. Specific flavonoids promote intercellular root colonization of Arabidopsis thaliana by Azorhizobium caulinodans ORS571. Mol. Plant Microbe Interact. 10:560-570. [DOI] [PubMed] [Google Scholar]
- 17.Jacobs, M. A., et al. 2003. Comprehensive transposon mutant library of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U. S. A. 100:14339-14344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jofré, E., A. Lagares, and G. Mori. 2004. Disruption of dTDP-rhamnose biosynthesis modifies lipopolysaccharide core, exopolysaccharide production and root colonization in Azospirillum brasilense. FEMS Microbiol. Lett. 231:267-275. [DOI] [PubMed] [Google Scholar]
- 19.Klassen, G., F. O. Pedrosa, E. M. Souza, S. Funayama, and L. U. Rigo. 1997. Effect of nitrogen compounds on nitrogenase activity in Herbaspirillum seropedicae SMR1. Can. J. Microbiol. 43:887-891. [Google Scholar]
- 20.Le Quéré, A. J., et al. 2006. Structural characterization of a K-antigen capsular polysaccharide essential for normal symbiotic infection in Rhizobium sp. NGR234. Deletion of the rkpMNO locus prevents synthesis of 5,7-diacetamido-3,5,7,9-tetradeoxy-non-2-ulosonic acid. J. Biol. Chem. 281:28981-28992. [DOI] [PubMed] [Google Scholar]
- 21.Lerouge, I., and J. Vanderleyden. 2002. O-antigen structural variation: mechanisms and possible roles in animal/plant-microbe interactions. FEMS Microbiol. Rev. 26:17-47. [DOI] [PubMed] [Google Scholar]
- 22.Livak, K. J., and T. D. Schmittgen. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods 25:402-408. [DOI] [PubMed] [Google Scholar]
- 23.Machado, I. M., M. G. Yates, H. B. Machado, E. M. Souza, and F. O. Pedrosa. 1996. Cloning and sequencing of the nitrogenase structural genes nifHDK of Herbaspirillum seropedicae. Braz. J. Med. Biol. Res. 29:1599-1602. [PubMed] [Google Scholar]
- 24.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 25.Pedraza, R. O. 2008. Recent advances in nitrogen-fixing acetic acid bacteria. Int. J. Food Microbiol. 125:25-35. [DOI] [PubMed] [Google Scholar]
- 26.Perret, X., C. Staehelin, and W. J. Broughton. 2000. Molecular basis of symbiotic promiscuity. Microbiol. Mol. Biol. Rev. 64:180-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pimentel, J. P., et al. 1991. Dinitrogen fixation and infection of grass leaves by Pseudomonas rubrisubalbicans and Herbaspirillum seropedicae. Plant Soil 137:61-65. [Google Scholar]
- 28.Pugsley, A. P. 1993. The complete general secretory pathway in gram-negative bacteria. Microbiol. Rev. 57:50-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sandkvist, M. 2001. Biology of type II secretion. Mol. Microbiol. 40:271-283. [DOI] [PubMed] [Google Scholar]
- 30.Schwab, S., et al. 2007. Identification of NH4+-regulated genes of Herbaspirillum seropedicae by random insertional mutagenesis. Arch. Microbiol. 187:379-386. [DOI] [PubMed] [Google Scholar]
- 31.Staehelin, C., et al. 2006. Exo-oligosaccharides of Rhizobium sp. NGR234 are required for symbiosis with various legumes. J. Bacteriol. 188:6168-6178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Theunis, M., H. Kobayashi, W. J. Broughton, and E. Prinsen. 2004. Flavonoids, NodD1, NodD2 and nod-box NB15 modulate expression of the y4wEFG locus that is required for indole-3-acetic acid synthesis in Rhizobium sp. NGR234. Mol. Plant Microbe Interact. 17:1153-1161. [DOI] [PubMed] [Google Scholar]
- 33.Webster, G., et al. 1998. The flavonoid naringenin stimulates the intercellular colonization of wheat roots by Azorhizobium caulinodans. Plant Cell Environ. 21:373-383. [Google Scholar]