Abstract
Hydrogenases are enzymes involved in the bioproduction of hydrogen, a clean alternative energy source whose combustion generates water as the only end product. In this article we identified and characterized a [NiFe] hydrogenase from the marine bacterium Alteromonas macleodii “deep ecotype” with unusual stability toward oxygen and high temperature. The A. macleodii hydrogenase (HynSL) can catalyze both H2 evolution and H2 uptake reactions. HynSL was expressed in A. macleodii under aerobic conditions and reached the maximum activity when the cells entered the late exponential phase. The higher level of hydrogenase activity was accompanied by a greater abundance of the HynSL protein in the late-log or stationary phase. The addition of nickel to the growth medium significantly enhanced the hydrogenase activity. Ni treatment affected the level of the protein, but not the mRNA, indicating that the effect of Ni was exerted at the posttranscriptional level. Hydrogenase activity was distributed ∼30% in the membrane fraction and ∼70% in the cytoplasmic fraction. Thus, HynSL appears to be loosely membrane-bound. Partially purified A. macleodii hydrogenase demonstrated extraordinary stability. It retained 84% of its activity after exposure to 80°C for 2 h. After exposure to air for 45 days at 4°C, it retained nearly 100% of its activity when assayed under anaerobic conditions. Its catalytic activity in the presence of O2 was evaluated by the hydrogen-deuterium (H-D) exchange assay. In 1% O2, 20.4% of its H-D exchange activity was retained. The great stability of HynSL makes it a potential candidate for biotechnological applications.
Biological hydrogen production mediated by hydrogenases or nitrogenases is an attractive solution to generate a renewable energy carrier. Since the process catalyzed by nitrogenases requires ATP, hydrogenases may be more efficient for the large-scale production of H2 as an alternative energy storage molecule. Hydrogenases can catalyze the reversible reduction of protons to molecular H2 according to the equation 2H+ + 2 e− ↔ H2. Depending on the energy demands of the cell, a hydrogenase catalyzes either H2 production to dissipate excess reductant or H2 oxidation to capture the energy in H2 (2, 3, 19). Hydrogenases can be found in a wide variety of microbes, including bacteria, archaea, and unicellular eukaryotes (48). Such microbes may contain one or multiple hydrogenases found in the cytosol, the periplasm, or the cell membrane (47). In addition to its important role in microbial energy metabolism, hydrogenase activity is also involved in other cellular processes, such as methanogenesis, nitrogen fixation, and pathogenesis (47). However, despite its importance to microbial processes, much remains to be understood about the molecular mechanisms for hydrogenase synthesis, assembly, and regulation of gene expression.
Hydrogenases are divided into three distinct groups: [NiFe] hydrogenases, [FeFe] hydrogenases, and [Fe] hydrogenases (44, 48). The [NiFe] hydrogenase represents the largest known group of the hydrogenases (48). Its core enzyme is a heterodimer composed of a large and small subunit and is involved in H2 evolution and uptake reactions in vivo. [NiFe] hydrogenases were crystallized, and the architecture of the catalytic site has been elucidated (50). An intrinsic characteristic of a [NiFe] hydrogenase is the reversible inactivation by O2, while the [FeFe] hydrogenase is irreversibly inactivated by O2 (4). [NiFe] hydrogenases are thus more stable to O2 exposure than [FeFe] hydrogenases.
[NiFe] hydrogenase-mediated processes have been explored for biotechnological applications (32, 46). Photosynthetic and fermentative microbial systems are being developed to use water or carbon sources as feedstock for H2 production (32, 41, 43, 46), in which electrons generated from photosynthesis or oxidation of carbohydrates are driven to the hydrogenases for reduction of protons to H2. [NiFe] hydrogenases could also be applied to in vitro electrochemical apparatuses (23), such as H2 fuel cells, in which the hydrogenases are used as bioelectrocatalysts for proton reduction and H2 oxidation. For a successful biohydrogen production/oxidation system, the [NiFe] hydrogenase needs to be thermostable, tolerant to O2, and catalytically active in O2. Due to the limitations of existing hydrogenases, their application on the industrial scale is not yet successful. Efforts are needed to identify [NiFe] hydrogenases with better stability and catalytic activities. In the present study we identified and characterized a [NiFe] hydrogenase from Alteromonas macleodii and examined its O2 tolerance, thermostability, and catalytic activity.
A. macleodii is a heterotrophic marine bacterium present in surface and deep ocean waters. A. macleodii strain “deep ecotype” (AltDE) was isolated from the deep Mediterranean Sea (27). Whole-genomic sequence analysis shows that this bacterium contains the gene cluster of a putative [NiFe] hydrogenase (HynSL) (21). According to Bergey's Manual of Systematic Bacteriology, AltDE is defined as an aerobic organism, and it is not known to ferment (14). The presence of hydrogenase genes in the genome may indicate the availability of energy-rich H2 that the organism may be adapted to exploit in its native environment (14). It may also reflect a lifestyle of the bacterium in proximity to anaerobic conditions in the ocean depths and near the pelagic brine lakes of the Mediterranean basin (40).
The putative [NiFe] hydrogenase HynSL from AltDE displayed 60% identity to the large subunit and 64% to the small subunit of the stable [NiFe] hydrogenase (HynSL) from the purple sulfur bacterium Thiocapsa roseopersicina (12, 20, 30, 54). In a previous study, we identified an Alteromonas [NiFe] hydrogenase from the Sargasso Sea, which is 99% identical to HynSL in AltDE (30). The expression of its genes cloned from the Sargasso Sea in the foreign host T. roseopersicina generated an active hydrogenase capable of producing H2 (30). The goal of the present study was to determine whether the A. macleodii [NiFe] hydrogenase HynSL is naturally expressed in AltDE, whether it is active, and how it is regulated during the growth cycle. To investigate how environmental factors such as oxygen, nickel, and iron influence the turnover rates of the hydrogenase, we determined activity of HynSL and examined its expression under various growth conditions. We further studied its cellular localization in AltDE cells, examined its thermostability and O2 stability, and measured its catalytic activity in the presence of O2.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains used in the present study are listed in Table 1. A. macleodii was grown aerobically in marine broth or agar (Difco) at 27°C. For anaerobic growth, A. macleodii cells grown aerobically were transferred into flasks containing fresh medium supplemented with 40 mM KNO3 as the electron receptor, sealed with rubber stoppers, and then flushed with argon to remove oxygen. To assess the effect of nickel and iron on the expression of HynSL, A. macleodii cells were cultured in marine broth supplemented with NiCl2 (10, 50, or 100 μM) and/or EDTA-Fe (50 μM). T. roseopersicina was grown anaerobically under illumination in Pfennig's mineral medium or on Pfennig's medium plates as described previously (30). Antibiotics for T. roseopersicina were used at the following concentrations: streptomycin, 5 μg ml−1; gentamicin, 5 μg ml−1; and erythromycin, 50 μg ml−1.
TABLE 1.
Bacterial strains used in this study
| Strain | Genotype and phenotypea | Reference |
|---|---|---|
| A. macleodii “deep ecotype” (AltDE) | Wild type | 27 |
| A. macleodii strain 107 (ATCC 27126) | Wild type | 7 |
| T. roseopersicina GB2131 | hupSLΔ::Gm, hoxHΔ::Em; Gmr Emr | 36 |
Gmr, gentamicin resistance; Emr, erythromycin resistance.
Purification of A. macleodii and T. roseopersicina hydrogenases.
Aerobically grown AltDE cells or anaerobically grown T. roseopersicina cells were resuspended in a sonication buffer containing 20 mM Tris-HCl (pH 7.5), 0.5 mM EDTA, and 1 mM dithiothreitol (DTT). The cell suspensions were sonicated twice (each for 2 min) in an ice-water bath by using a Branson Sonifier 250 ultrasonic cell disruptor as described previously (30). The slurry was centrifuged at 16,000 × g for 20 min, and the supernatant was recovered for column chromatography that was performed at room temperature and under ambient air. DEAE 52-cellulose (Whatman) column was equilibrated with the sonication buffer and used to load the crude cell extracts. After the column was washed with the elution buffer it was eluted with a linear gradient of 0 to 0.6 M NaCl. Eluted fractions were collected and assayed for H2 evolution activities to identify the fractions containing the hydrogenases. Protein concentration was determined as previously described (30).
Preparation of A. macleodii subcellular fractions.
AltDE cells were resuspended in 10 ml of solution A (20 mM Tris-HCl [pH 7.5], 1 M mannitol, 1 mM DTT, 0.5 mM EDTA) supplemented with lysozyme (0.4 mg/ml; Sigma) and then incubated at 37°C for 30 min. The efficiency of cell-wall degradation and spheroplast formation was monitored by microscopy. The spheroplast suspension was centrifuged at 16,000 × g for 10 min, and the supernatant was collected as the periplasmic fraction. The pellet was washed with the solution A and then centrifuged at 16,000 × g for 10 min. Recovered spheroplasts were resuspended in 10 ml of solution B (20 mM Tris-HCl [pH 7.5], 1 mM DTT, 0.5 mM EDTA, 1 mg of RNase/ml, 1 mg of DNase/ml) and incubated at 37°C for 30 min. Cell debris was removed by centrifugation at 10,000 × g for 20 min. The supernatant, containing the cytoplasm and the total-membrane fractions, was further centrifuged at 100,000 × g for 3 h, and the supernatant was harvested as the cytoplasmic fraction. The pellet was washed using solution B, and the membrane fraction was recovered by centrifugation at 100,000 × g for 1 h and then resuspended in 10 ml of solution B. The activities of alkaline phosphatase, malate dehydrogenase, and NADH oxidase in the periplasm, cytoplasm, and membrane fractions were determined, respectively, as previously described (17).
PAGE and Western blotting.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed as described previously (38). Coomassie blue staining of proteins in the gels was carried out by using SimplyBlue SafeStain reagent (Invitrogen). Native PAGE using alkaline polyacrylamide gels (pH 10) was performed as described previously (52). Western blotting was performed with polyclonal rabbit antibodies specific for T. roseopersicina HynL as the primary antibodies.
Hydrogenase activity assays.
In vitro H2 uptake and evolution assays were performed as previously described (30). H2 uptake activity staining (1, 42) was performed after hydrogenase samples were separated by native PAGE. The gel was incubated in 50 mM potassium phosphate buffer (pH 7) supplemented with 2.5 mM oxidized benzyl viologen (BV) in a sealed bottle. After flushing the bottle with H2 for 30 min, it was incubated at 55°C for 5 min. After blue bands appeared, 2,3,5-triphenyl tetrazolium chloride (200 μg/ml) was added to fix the staining. A duplicate gel was run simultaneously for Western blotting to determine the positions of the hydrogenases in the gel.
O2 stability and thermostability assays.
Partially purified A. macleodii hydrogenase was used for O2 stability and thermostability assays. To determine thermostability, the hydrogenase samples were treated for 2 h at 70°C or 80°C. The treated samples, along with the untreated sample (used as a control), were then subjected to in vitro H2 evolution activity assay. O2 stability was examined by determining the H2 evolution activities of hydrogenase samples on the day of purification and after being exposed to air at 4°C for 14 and 45 days, respectively.
H-D exchange assay.
Hydrogen-deuterium (H-D) exchange experiments were performed as previously described (29). Briefly, 0.4 ml each of the partially purified enzyme (∼0.55 mg of protein/ml) was incubated in the presence of 20 mM potassium phosphate buffer (pH 7) and deuterium oxide (D2O; Sigma) in a glass vial sealed with a rubber stopper. After the vessel was flushed out with 10% H2 balanced with argon, various amounts of O2 were added. The reaction mixtures were incubated at 37°C for 2 h, and the concentration of H-D in the headspace of the vials was determined by using an Omnistar quadrupole mass spectrometer (Pfeiffer).
RNA extraction and RT-PCR.
Each RNA sample was extracted from 1 ml of A. macleodii cell culture by using an RNeasy kit (Qiagen) and then treated with a DNA-Free Turbo kit (Ambion) according to the manufacturer's instructions. The RNA samples (500 ng/each) were denatured and run on a 1% agarose gel containing morpholinepropanesulfonic acid buffer and formaldehyde (38). The gel was then stained with ethidium bromide to visualize rRNA that was used as an internal control for RNA quantitation. Reverse transcription-PCR (RT-PCR) was performed as described previously (51) using hynL-specific primers (5′-GTA TGG CAT CGG TGC GTG CT and 5′-CTA GCT GCC CAG ACT CAA CC), hynS-specific primers (5′-GGC AAT GCG AGA AGA CCC TC and 5′-CAG AGC AAC CAA GAC ACG G), or primers at the junction between hynS and hynL (5′-CGT CTT TTG GCG GGA TCC C and 5′-GTA AAA TCA GTT CAA TTC CC). The non-RT control reaction, containing all of the components except the reverse transcriptase, was performed as described above.
RESULTS
Examination of A. macleodii [NiFe] hydrogenase HynSL in AltDE.
Genomic sequencing of AltDE (21) revealed a gene cluster (hynD/hupH/hynS/hynL/hypC/hypA/hypB/hypD/hypF/hypE) coding for a putative [NiFe] hydrogenase and its accessory proteins. This hydrogenase, previously named HyaAB (30), is now renamed HynSL to be consistent with the consensus in the hydrogenase community (48). Two structural genes of the A. macleodii hydrogenase HynSL, renamed hynS (MADE_00346, 999 bp) and hynL (MADE_00347, 1,878 bp), encode a 332-amino-acid small subunit (HynS) and a 625-amino-acid large subunit (HynL), respectively. The hynS/hynL genes of an Alteromonas [NiFe] hydrogenase from the Sargasso sea show 99% identity to their counterpart genes in AltDE and encoded an active hydrogenase in T. roseopersicina (30). Thus, we sought to determine whether the A. macleodii [NiFe] hydrogenase HynSL is naturally expressed in AltDE and whether it is active.
To determine whether AltDE contains an active hydrogenase, we performed H2 evolution and uptake activity assays using crude cell extracts from AltDE cells grown under its normal aerobic growth condition. As expected, no activities were detected in the negative control, A. macleodii strain 107 (Alt107, ATCC 27126), a strain that naturally does not contain any hydrogenase genes (21). Whereas AltDE displayed both H2 evolution (16.4 ± 0.7 nmol H2 mg of protein−1 min−1) and H2 uptake (24.82 ± 2.5 nmol H2 mg of protein−1 min−1) activities at 55°C. These results indicate that AltDE expressed functional hydrogenases. The hydrogenase activity was compared to those at lower temperatures. We observed that the H2 evolution and H2 uptake activity at 55°C was approximately two and five times higher than at 37°C, respectively, suggesting that the enzyme is more active at higher temperatures. We partially purified the A. macleodii hydrogenase from AltDE cells through column chromatography. The hydrogenase activity was eluted as a single peak from the DEAE 52 cellulose column at ∼0.4 M NaCl (Fig. 1 A), which is consistent with the fact that no hydrogenase gene homologs other than hynS and hynL genes were identified in the genome. Although A. macleodii hydrogenase was not purified to homogeneity from this chromatography step, an ∼10-fold increase in purity was reached in the purified fraction (fraction 38), as evidenced by the fact it had a specific activity that was ∼10 times higher (55.45 ± 7.38 nmol of H2 mg−1 min−1 in the purified fraction versus 5.53 ± 0.71 nmol of H2 mg−1 min−1 in original crude extract).
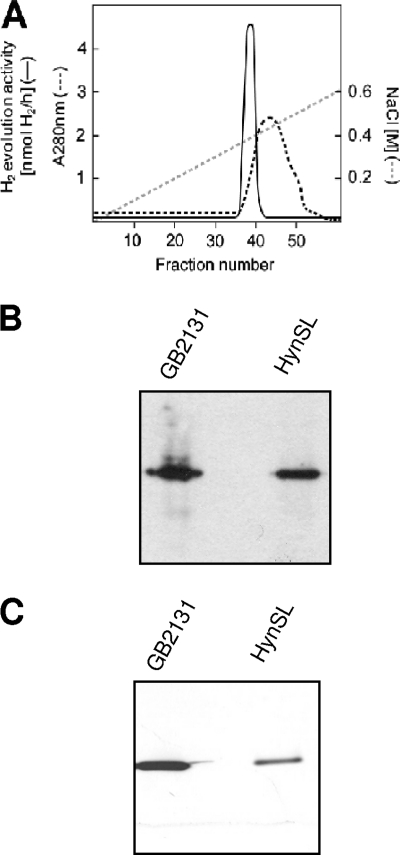
FIG. 1.
Purification and identification of A. macleodii hydrogenase HynSL from AltDE cells. (A) A280 and hydrogenase activity profiles in protein fractions eluted from the DEAE column. The NaCl concentrations (gray dashed line), A280 values (black dashed line), and total hydrogenase activities (solid line) in eluted fractions are indicated separately. (B) H2 uptake activity staining in a native polyacrylamide gel. The gel was incubated in the presence of BV and H2 for the activity staining. (C) Western blotting to detect the purified AltDE hydrogenase in a duplicate native polyacrylamide gel. Anti-TrHynL antibodies were used for immunodetection of A. macleodii HynL. AltDE, crude extract from aerobically grown AltDE cells; HynSL, partially purified A. macleodii hydrogenase (fraction 38 eluted from the DEAE column in panel A); GB2131, a T. roseopersicina strain used as a positive control. Portions of crude extracts (40 μg) and of partially purified A. macleodii HynSL (5 μg) were loaded into each lane for in-gel H2 uptake activity staining and Western blotting assays.
We further evaluated the partially purified A. macleodii hydrogenase by using the H2 uptake activity staining method. The purified hydrogenase sample was separated on two identical native PAGE gels, along with a positive control from T. roseopersicina strain GB2131 (a mutant strain carrying only the T. roseopersicina hydrogenase HynSL). We applied one native PAGE gel for in-gel H2 uptake activity staining and another gel for Western blotting by using antibodies (anti-TrHynL) against the large subunit of T. roseopersicina HynSL, which can specifically recognize the A. macleodii large subunit HynL (30). A single activity band was detected in the positive control strain GB2131 in the native gel stained for H2 uptake activities in the presence of BV and H2 (Fig. 1B). At the position corresponding to this activity band, a specific T. roseopersicina HynSL band (Fig. 1C) was detected on the duplicate native gel subjected to Western blotting with anti-TrHynL, as expected. A single activity band was also detected in the purified AltDE hydrogenase sample in the stained activity gel (Fig. 1B), and its position matched exactly the position of the specific A. macleodii HynSL band detected on the Western blotted gel (Fig. 1C). These results indicate that a hydrogenase from the native PAGE gel had in-gel H2 uptake activity, and it corresponded to A. macleodii hydrogenase HynSL. Overall, our results demonstrate that the A. macleodii hydrogenase HynSL is a functional hydrogenase in AltDE.
Effect of oxygen on hydrogenase expression in AltDE.
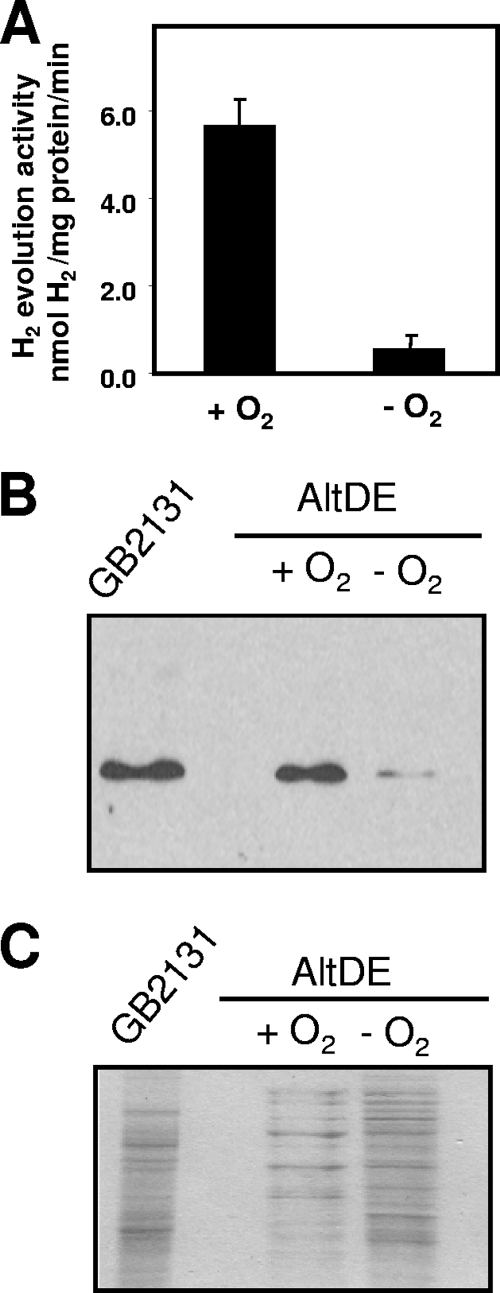
The presence of oxygen has been shown to affect expression of hydrogenase genes, and in most microorganisms hydrogenase expression is limited to microaerobic or anaerobic conditions (49). Therefore, we sought to investigate how oxygen regulates A. macleodii HynSL in AltDE by performing in vitro H2 evolution assays on crude cell extracts from anaerobically and aerobically grown cells. In AltDE cells grown anaerobically in the medium supplemented with KNO3, no hydrogenase activity was detected, whereas in AltDE cells anaerobically incubated in the medium without KNO3, hydrogenase activity was detected, but it was only ∼10% of the activity that we observed in cells incubated under aerobic conditions (Fig. 2 A). We next examined the level of HynSL protein in the protein extracts after aerobic or anaerobic incubation. Western blotting with anti-TrHynL antibodies after native PAGE of crude cell extracts revealed that the differences in activity were also paralleled by protein accumulation (Fig. 2B). Coomassie blue staining of a duplicate gel after SDS-PAGE confirmed that equivalent total protein was loaded to each lane (Fig. 2C). These results illustrate that contrary to most organisms, the accumulation of active A. macleodii HynSL is enhanced in the presence of molecular oxygen.
FIG. 2.
Effect of O2 on A. macleodii hydrogenase abundance and its activity in AltDE. (A) Examination of H2 evolution activity in crude cellular extracts from AltDE cells. The AltDE cells were grown in marine broth medium for 18 h under aerobic conditions (+O2) or incubated under anaerobic conditions (−O2). The bars represent average values ± the standard deviations (SD) of three independent experiments. (B) Immunodetection of HynL in crude cellular extract of AltDE after native PAGE. Crude extracts from the panel A were loaded in the gel, and protein blots were probed with anti-TrHynL antibodies. Crude extract from T. roseopersicina GB2131 was loaded as a positive control. (C) SDS-PAGE gel stained with Coomassie brilliant blue. Portions (40 μg) of total protein were loaded into each lane for Western blotting and Coomassie blue staining.
Effect of metal treatment on hydrogenase expression in AltDE.
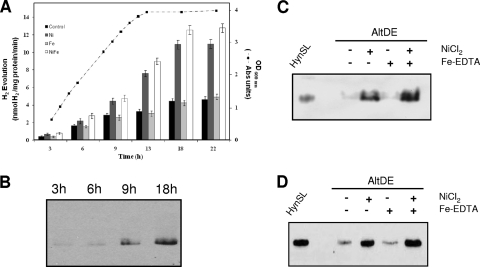
Nickel and iron are essential elements for the assembly and maturation of [NiFe] hydrogenases (10, 11, 53). We first sought to investigate whether adding extra nickel to the growth medium enhances the activity of the A. macleodii hydrogenase HynSL under aerobic growth conditions. Different concentrations (10, 50, and 100 μM) of NiCl2 were tested. Our result shows that the addition of nickel increased the H2 evolution activity in AltDE in direct proportion to the concentration of NiCl2 added to the medium (see Fig. S1 in the supplemental material). However, the addition of NiCl2 did not affect the growth rate of AltDE cells during the 22-h period (see Fig. S2 in the supplemental material). We further examined the effect of nickel on AltDE cells in different growth phases. The AltDE cells, after being treated with or without NiCl2 for 3, 6, 9, 13, 18, and 22 h, were collected, and hydrogenase activities were determined. Based on the results (Fig. 3 A), we have two main observations. First, the addition of NiCl2 (100 μM) increased the H2 evolution activity in AltDE at all of the time points examined (Fig. 3A). Second, in the absence of added NiCl2, the hydrogenase activity increased consistently along the growth curve of AltDE, until cell growth reached the stationary phase (between 13 and 18 h) (Fig. 3A). This increasing hydrogenase activity in untreated cells was paralleled by a strong increase in HynL protein abundance, as evidenced by Western blotting on cells collected at 3-, 6-, 9-, and 18-h time points (Fig. 3B). These results suggest that additional hydrogenase was synthesized or that it was assembled or matured in the late log or stationary phase.
FIG. 3.
Effect of nickel and iron on the activity and expression of A. macleodii HynSL. (A) Examination of A. macleodii hydrogenase activity in AltDE cells aerobically grown in marine broth medium (control), and in the medium supplemented with NiCl2 (Ni-only), Fe-EDTA (Fe-only), or both NiCl2 and Fe-EDTA (Ni+Fe). Growth curve of the AltDE cells in the marine broth medium (control) is presented. H2 evolution activity was assayed using AltDE cells collected at time points as indicated. The bars represent average values ± the SD of three independent experiments. (B) Western blotting to detect expression of HynSL at various time points. Crude extracts from AltDE cells collected at different time points were used for native PAGE. Protein blots were probed with anti-TrHynL antibodies. Lane 1, 3 h (optical density at 600 nm [OD600] = 0.5); lane 2, 6 h (OD600 = 1.12); lane 3, 9 h (OD600 = 2.15); lane 4, 18 h (OD600 = 3.87). (C) H2 uptake activity staining after native PAGE. Protein samples used for the staining were collected from the cells at 18 h. (D) Western blotting of protein samples at 18 h after native PAGE. Anti-TrHynL antibodies were used to detect HynL. Partially purified A. macleodii HynSL was loaded as a control for in-gel H2 uptake activity staining and Western blotting. Portions (40 μg) of total protein were loaded into each lane.
In contrast to the effect of nickel, the addition of Fe-EDTA (50 μM) did not affect the level of hydrogenase activity (Fig. 3A). Additional iron salts, including ferric sulfate, ferric citrate, ferric ammonium citrate, and ferric nitrate were tested to determine whether other forms of iron might be more accessible to the cells and affect hydrogenase activity. No increase in the hydrogenase activity was observed in response to these iron salts (see Fig. S3 in the supplemental material).
To understand the mechanisms involved in the increased A. macleodii HynSL activity in metal-treated cells, we examined HynSL expression at the translational level after the AltDE cells were treated with nickel and iron. The in-gel H2 uptake activity staining coupled with Western blotting was performed by using two identical native PAGE gels. Only cells treated for 18 h were used for examination since the lower in-gel H2 uptake activity at 3-, 6-, and 9-h time points could not be detected due to the limitation of the method. Our result (Fig. 3C) shows that H2 uptake activity was increased after the NiCl2 treatment, whereas the activity stayed the same after the Fe-EDTA treatment, which is consistent with our observation from the H2 evolution activity assay. Western blotting (Fig. 3D) detected higher levels of HynL in cells with Ni-only or Ni+Fe treatments, whereas the Fe-only treatment did not change the level of HynL. These results suggest that the higher levels of hydrogenase activities are due to increased levels of the active A. macleodii HynSL enzyme.
To understand the mechanisms involved in the increased HynSL protein accumulation in Ni-treated cells, we examined hynSL gene transcription. First, RT-PCR amplification of RNA samples with a pair of primers specific for hynS generated a 700-bp specific product (see Fig. S4A in the supplemental material), whereas no products were generated in the no-RT control (see Fig. S4A in the supplemental material), indicating hynS was transcribed. Second, RT-PCR amplification with a pair of primers encompassing the junction region of hynS and hynL generated a 650-bp specific product (see Fig. S4A in the supplemental material), indicating that the hynS and hynL genes were cotranscribed together.
The effect of the Ni and Fe treatment on accumulation of the hynL transcript was examined by RT-PCR in aerobically grown AltDE cells after Ni-only, Fe-only, Ni+Fe, or neither metal (control) treatments. For exponential amplification of hynL transcripts, we determined optimal concentrations of RNA templates in RT-PCRs (see Fig. S4B in the supplemental material). Total RNA was isolated from the cells collected at 3, 6, and 9 h after inoculation. In this experiment the samples obtained at 13, 18, and 22 h were excluded from RNA analysis since the cells had reached the stationary phase, and these samples yielded RNA of poor quality that gave inconsistent results. No variation in the RT-PCR band intensity was observed among the metal-treated samples collected at a single time point (see Fig. S4C in the supplemental material). rRNA visualized in a formaldehyde-denatured agarose gel confirms that similar amounts of RNA sample were added to the RT-PCRs (see Fig. S4C in the supplemental material). Thus, the increase in activity from NiCl2 addition is not likely to be regulated by increased transcription of the hydrogenase genes.
Determination of subcellular localization of A. macleodii HynSL.
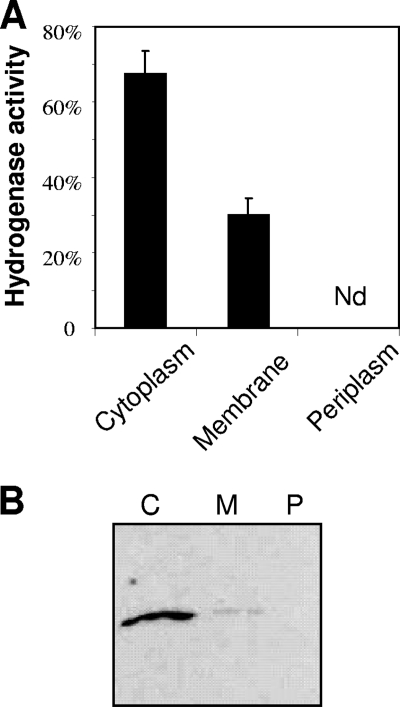
Hydrogenases in microorganisms are targeted to different subcellular locations (47). To determine the subcellular localization of A. macleodii HynSL, we prepared periplasmic, cytoplasmic, and membrane fractions from AltDE cells. To evaluate the purity of these fractions, we determined the activities of their corresponding marker enzymes: alkaline phosphatase was used as the marker for the periplasm fraction, malate dehydrogenase for the cytoplasmic fraction, and NADH oxidase for the membrane fraction. In each case, ∼90% of the total marker enzyme activity was found in its corresponding fraction. The distribution of the A. macleodii hydrogenase activity among these three fractions was examined. The result (Fig. 4 A) shows that the activity was present in cytoplasmic (70%) and membrane (30%) fractions but not in the periplasm. Therefore, the bipartite localization of A. macleodii HynSL between the cytoplasm and the membranes was in a ratio of 7:3. The presence of HynSL in the cytoplasmic and membrane fractions was further confirmed by immunodetection of HynL after native PAGE (Fig. 4B). It was observed that the ratio of the hydrogenase activity between the membrane and cytoplasmic fractions (roughly 1:2) (Fig. 4A) was significantly higher than that of the protein levels between these two fractions (roughly 1:10) (Fig. 4B). This observation suggests that the specific hydrogenase activity in the membrane fraction was significantly higher than in the cytoplasmic fraction.
FIG. 4.
Determination of A. macleodii HynSL localization. (A) Detection of A. macleodii HynSL activity in the cytoplasm, membrane, and periplasm fractions. Bars depict relative activity with respect to the total activity prior to cellular fractioning (100% activity). The data displayed represent average values ± the SD of three independent experiments. Nd, none detected. (B) Immunodetection of A. macleodii HynSL in the cytoplasm, membrane, and periplasm fractions. Three cellular fractions prepared from equal amounts of cells were separated on a native PAGE, and the protein blot was probed with anti-TrHynL antibodies. Lanes: C, the cytoplasmic fraction; P, the periplasmic fraction; M, the membrane fraction. A total of 40 μg of total protein was loaded into each lane for Western blotting.
Examination of thermostability of A. macleodii HynSL.
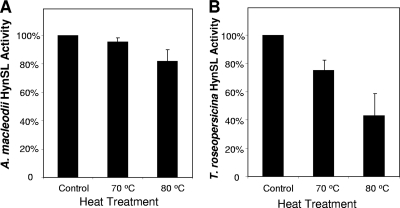
The T. roseopersicina [NiFe] hydrogenase HynSL has extraordinary stability at high temperatures (20). We determined whether A. macleodii hydrogenase HynSL is also thermostable. Aliquots of partially purified A. macleodii HynSL were heated at 70°C or 80°C for 2 h before they were subjected to H2 evolution activity assays. Parallel experiments were also performed on T. roseopersicina HynSL that was partially purified through the DEAE 52 cellulose column. Figure 5 shows the results of thermostability assays, where the activities of the heat-treated hydrogenase samples were compared to the untreated hydrogenase samples. The activity of the untreated samples was used as a standard and normalized to 100%. A. macleodii partially purified HynSL retained 94% of the original activity after the heat treatment at 70°C, whereas, after the 80°C treatment, the enzyme retained 84% of its activity (Fig. 5A). For T. roseopersicina partially purified HynSL 75% of the original activity remained after the treatment at 70°C and less than half (43%) remained following the 80°C treatment (Fig. 5B).
FIG. 5.
Examination of thermostability of A. macleodii HynSL and T. roseopersicina HynSL. (A) Thermostability assay of A. macleodii HynSL. (B) Thermostability assay of T. roseopersicina HynSL. Partially purified HynSL from each organism was incubated under aerobic conditions for 2 h on ice (control), at 70°C, or at 80°C. Aliquots of the three treated samples were used for H2 evolution assays. The activities of the untreated samples (49.30 nmol of H2 mg−1 min−1 for A. macleodii HynSL and 97.85 nmol of H2 mg−1 min−1 for T. roseopersicina HynSL) were used as standards (100% activity). The activities of heat-treated samples were then normalized to control values. The values represent averages ± the SD of three independent experiments.
Examination of O2 stability and catalytic activity of A. macleodii HynSL.
To examine the O2 stability of A. macleodii hydrogenase HynSL, we exposed the partially purified enzyme to air (21% O2) at 4°C over a period of 45 days. The H2 evolution activity of the enzyme was assessed at day 1 (on the day of purification), day 14, and day 45 by performing in vitro H2 evolution activity assays under the anaerobic condition with reduced BV as an artificial electron donor. Our results show that at day 14 and day 45 the A. macleodii hydrogenase retained 100.1% ± 1% and 98.5% ± 0.9% of its activity at day 1, respectively. This result demonstrates that the partially purified HynSL has a remarkably high stability in O2 and that its activity can be fully restored under anaerobic conditions after being exposed to air for such a long period.
We further determined whether A. macleodii HynSL has any catalytic activity in the presence of O2 by using the H-D exchange assay. This assay is based on the intrinsic property of a hydrogenase to catalyze an isotopic exchange reaction between the protons in H2 and the deuterium ions in deuterium oxide (D2O) to yield H-D. Since this reaction does not require any additional electron mediator, it can be used to assess the catalytic properties of the enzyme in O2. The partially purified enzyme was used for H-D exchange activity assays in absence or in the presence of 1%, 3% or 5% O2. Like most bidirectional hydrogenases, A. macleodii HynSL was able to catalyze the H-D exchange reaction when resuspended in D2O in a H2 atmosphere without any O2. When the reaction mixture was exposed to 1% O2 with continuous stirring, the enzyme retained 20.4% ± 1.9% H-D exchange activity, indicating A. macleodii HynSL had catalytic activity in 1% O2. The activity dropped to 5.2% ± 1.4% when the reaction was exposed to 3% O2, and no activity was detected in 5% O2. We performed a parallel study on the T. roseopersicina hydrogenase HynSL, and no H-D exchange activity was detected in three O2 concentrations tested, a finding that is consistent with the previous report (20).
DISCUSSION
This study demonstrates that the [NiFe] hydrogenase HynSL in the marine bacterium A. macleodii AltDE, a homolog of the T. roseopersicina stable [NiFe] hydrogenase HynSL, is naturally expressed and functionally active. A. macleodii HynSL was found in AltDE but not in another A. macleodii strain Alt107, suggesting the enzyme is not conserved among various A. macleodii strains. This lack of conservation is consistent with studies on the genomic sequences in A. macleodii (21). The 4.4-Mb genome of AltDE has 4,102 annotated genes. However, 1,242 of these genes, including all genes in the HynSL gene cluster, were missing in the genome of Alt107. The HynSL gene cluster was discovered in a genomic island of ∼95 kb in the AltDE genome (21), suggesting that the genes of the A. macleodii hydrogenase were likely introduced to A. macleodii through a horizontal gene transfer event. The fact that 65 transposable elements (including one near the gene locus of the A. macleodii hydrogenase) were identified in the AltDE genome (21) further supports this hypothesis.
In addition to the T. roseopersicina hydrogenase HynSL, the A. macleodii hydrogenase HynSL shares strong homology (60 to 69% identity at the protein level) with [NiFe] hydrogenases from Thiobacillus denitrificans ATCC 25259 (Tbd-1375 and -1378), Thioalkalivibrio sp. strain HL-EbGR7 (Tqr7-1266 and -1269), and Allochromatium vinosum DSM 180 (Alividraft-0802 and 08085) (8, 16, 26). The strong homology suggests common origins for these [NiFe] hydrogenases. Among A. macleodii HynSL and its four homologs, Thiobacillus and Thioalkalivibrio hydrogenases are most closely related to each other. Like A. macleodii HynSL, their structural and accessory genes are clustered together. Their gene clusters (hynL/isp2/isp1/hynS/hupH/hynD/hypC/hypA/hypB/hypD/hypF/hypE) share similar gene components and arrangement as the HynSL gene cluster, except that their structural genes are arranged differently. The structural genes hynS and hynL of A. macleodii HynSL are adjacent to each other, while the structural genes hynS and hynL for the Thiobacillus and Thioalkalivibrio hydrogenases are separated by isp1 and isp2 to form the cluster hynL-isp2-isp1-hynS. The protein products of isp1 and isp2 are involved in the in vivo function of the hydrogenase (34); however, no counterparts of isp1 and isp2 are found in AltDE. The other two homologs (16, 35) found in T. roseopersicina and A. vinosum also have the unusual arrangement (hynL-isp2-isp1-hynS) and, unlike A. macleodii HynSL, their structural and accessory genes are scattered throughout the genomes. The 10 proteins encoded by the A. macleodii gene cluster have the overall best matches with their Thiobacillus and Thioalkalivibrio counterparts. These findings suggest that A. macleodii hydrogenase HynSL is more closely related to the Thiobacillus and Thioalkalivibrio hydrogenases than to T. roseopersicina and A. vinosum hydrogenases. The A. macleodii HynSL gene cluster could have been acquired from Thiobacillus denitrificans and Thioalkalivibrio sp. by horizontal gene transfer. However, the G+C content of the HynSL gene locus (46%), which is similar to the overall G+C content of the AltDE genome (44.9%), is substantially lower than those of its homolog hydrogenases (63 to 68%) that are also in the same range as the overall G+C contents of their corresponding bacterial genomes. Therefore, they could have gone through substantial evolution since their genes were first introduced into A. macleodii.
Nickel is an essential component of the catalytic site of [NiFe] hydrogenases (4). Previous reports described the effects of nickel on the hydrogenase expression and activity (5, 18, 31, 37, 45). Our analysis at the functional, protein, and mRNA levels of HynSL in AltDE cells further reveals the mechanisms that govern the effects of nickel. The significantly higher amount of HynSL produced at the posttranscriptional level is likely due to improved translation or more efficient assembly/maturation of the hydrogenase in the presence of nickel. It is also possible that sufficient nickel increases enzyme stability.
Iron is also an essential component of the [NiFe]-catalytic site. However, in contrast to the strong stimulatory effect of nickel on hydrogenase activity, little effect of iron was observed. This could be the result of already high levels of total iron contained in the marine broth media (>400 μM). In this case, iron would not be limiting, and further addition would have no effect on iron metabolism in the cells.
Oxygen is important in the regulation of hydrogenase expression and activity (13, 49). Previous studies reported that the [NiFe] hydrogenases from organisms such as Escherichia coli, Rhizobium japonicum, Bradyrhizobium japonicum, Alcaligenes eutrophus, and T. roseopersicina are upregulated under microaerobic or anaerobic conditions (15, 24, 25, 28, 33). For instance, in E. coli the anaerobic global regulator Fnr (fumarate nitrate reduction) senses low concentrations of O2 and activates the expression of the hypABCDE operon for the biosynthesis of hydrogenases (33). The AltDE genome encodes a homolog of the Fnr protein in E. coli, suggesting AltDE might have a similar regulatory mechanism in response to oxygen limitation. However, we found that both the protein abundance and enzymatic activity of A. macleodii HynSL are downregulated in the absence of oxygen, suggesting that AltDE may possess different cellular machinery for regulating hydrogenase expression.
Protein sequence alignment shows that A. macleodii HynSL belongs to the group 1 hydrogenases, a group of membrane-associated hydrogenases classified by Vignais et al. (48). In conjunction with the sequence alignment, our localization analysis demonstrates that HynSL is loosely membrane-bound. Similar to the cellular localization of the T. roseopersicina stable hydrogenase HynSL (6), A. macleodii HynSL was detected in both soluble and membrane fractions, and the specific activity in the membrane fraction was higher. We speculate that HynSL in the soluble fraction was released from the membrane because the hydrogenase was loosely bound. The finding that A. macleodii HynSL is membrane targeted is consistent with the fact that the small subunit (HynS) of HynSL carries a twin-arginine signal peptide at the N terminus. This signal peptide can target proteins to the membranes and its presence in HynS strongly suggests that A. macleodii HynSL could be targeted to the periplasmic face of the cytoplasmic membrane (9). A gene cluster similar to the Tat operon of E. coli (22, 39) was identified in the genomic sequence of AltDE, supporting that HynSL could be targeted to the membranes through the Tat pathway.
The T. roseopersicina [NiFe] hydrogenase HynSL is remarkably stable at high temperatures and in the presence of O2 and its activities were even higher at 70°C in comparison to lower temperatures (20). The A. macleodii hydrogenase HynSL demonstrated similar enzyme characteristics to T. roseopersicina HynSL, consistent with the existence of strong similarity between these two hydrogenases. Like its homolog, the T. roseopersicina hydrogenase HynSL, the A. macleodii hydrogenase HynSL showed both H2 uptake and H2 evolution activity and the enzyme is also more active at higher temperatures. In the present study, A. macleodii HynSL appeared more thermostable and O2 tolerant than the T. roseopersicina hydrogenase HynSL. However, since hydrogenases used for thermostability and O2 tolerance assays were partially purified, coexisting cellular proteins may possibly provide a stabilizing effect to the hydrogenases. This possible effect makes comparison of the stability between A. macleodii HynSL and T. roseopersicina HynSL complicated. Although the same DEAE column and protocol were used for purification, copurified cellular proteins between two hydrogenase preparations were likely different. Consequently, they could contribute differently to hydrogenase stabilities, if such a stabilizing effect did exist. Therefore, we cannot rule out the possibility that higher stability observed in the A. macleodii hydrogenase preparation was caused by other cellular components in AltDE.
Nevertheless, considering that partially purified A. macleodii HynSL has demonstrated unusual stability at high temperatures and in the presence of O2, this hydrogenase is potentially a good candidate for biotechnological applications. The mechanism behind the stability remains to be determined. Small sequence differences between A. macleodii and T. roseopersicina hydrogenases may cause differences in their structural features associated with stability. Further structure-function analysis should reveal the structural determinants related to stability, which will facilitate designing more stable enzymes that can potentially be used for biotechnological purposes.
Supplementary Material
Acknowledgments
This study was supported by Synthetic Genomics, Inc., and the Hydrogen, Fuel Cells, and Infrastructure Technology Program (DE-FG36-05GO15027) of the U.S. Department of Energy.
We thank Francisco Rodriguez-Valera for kindly providing us with the A. macleodii strain and for sharing unpublished data with us. We also thank Kornel Kovacs for providing us with T. roseopersicina strains. We thank Pin-Ching Maness for helpful suggestions. Finally, we thank Laura Sheahan for kindly editing the manuscript.
Footnotes
Published ahead of print on 21 January 2011.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Ackrell, B. A., R. N. Asato, and H. F. Mower. 1966. Multiple forms of bacterial hydrogenases. J. Bacteriol. 92:828-838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adams, M. W., and A. Kletzin. 1996. Oxidoreductase-type enzymes and redox proteins involved in fermentative metabolisms of hyperthermophilic Archaea. Adv. Protein Chem. 48:101-180. [DOI] [PubMed] [Google Scholar]
- 3.Appel, J., S. Phunpruch, K. Steinmuller, and R. Schulz. 2000. The bidirectional hydrogenase of Synechocystis sp. PCC 6803 works as an electron valve during photosynthesis. Arch. Microbiol. 173:333-338. [DOI] [PubMed] [Google Scholar]
- 4.Armstrong, F. A. 2004. Hydrogenases: active site puzzles and progress. Curr. Opin. Chem. Biol. 8:133-140. [DOI] [PubMed] [Google Scholar]
- 5.Axelsson, R., and P. Lindblad. 2002. Transcriptional regulation of Nostoc hydrogenases: effects of oxygen, hydrogen, and nickel. Appl. Environ. Microbiol. 68:444-447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bagyinka, C., K. L. Kovacs, and E. Rak. 1982. Localization of hydrogenase in Thiocapsa roseopersicina photosynthetic membrane. Biochem. J. 202:255-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baumann, L., P. Baumann, M. Mandel, and R. D. Allen. 1972. Taxonomy of aerobic marine eubacteria. J. Bacteriol. 110:402-429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beller, H. R., et al. 2006. The genome sequence of the obligately chemolithoautotrophic, facultatively anaerobic bacterium Thiobacillus denitrificans. J. Bacteriol. 188:1473-1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berks, B. C., T. Palmer, and F. Sargent. 2003. The Tat protein translocation pathway and its role in microbial physiology. Adv. Microb. Physiol. 47:187-254. [DOI] [PubMed] [Google Scholar]
- 10.Blokesch, M., and A. Bock. 2002. Maturation of [NiFe] hydrogenases in Escherichia coli: the HypC cycle. J. Mol. Biol. 324:287-296. [DOI] [PubMed] [Google Scholar]
- 11.Blokesch, M., A. Magalon, and A. Bock. 2001. Interplay between the specific chaperone-like proteins HybG and HypC in maturation of hydrogenases 1, 2, and 3 from Escherichia coli. J. Bacteriol. 183:2817-2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bogorov, L. V. 1974. The properties of Thiocapsa roseopersicina, strain BBS, isolated from an estuary of the White Sea. Mikrobiologiia 43:326-332. [PubMed] [Google Scholar]
- 13.Bowien, B., and H. G. Schlegel. 1981. Physiology and biochemistry of aerobic hydrogen-oxidizing bacteria. Annu. Rev. Microbiol. 35:405-452. [DOI] [PubMed] [Google Scholar]
- 14.Bowman, J. P., and T. A. McMeekin. 2005. Alteromonadales ord. nov, p. 443-491. In D. J. Brenner, N. R. Krieg, and J. T. Staley (ed.), Bergey's manual of systematic bacteriology, vol. 2. Springer, New York, NY. [Google Scholar]
- 15.Cangelosi, G. A., and M. L. Wheelis. 1984. Regulation by molecular oxygen and organic substrates of hydrogenase synthesis in Alcaligenes eutrophus. J. Bacteriol. 159:138-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dahl, C., et al. 1999. Genes involved in hydrogen and sulfur metabolism in phototrophic sulfur bacteria. FEMS Microbiol. Lett. 180:317-324. [DOI] [PubMed] [Google Scholar]
- 17.de Maagd, R. A., and B. Lugtenberg. 1986. Fractionation of Rhizobium leguminosarum cells into outer membrane, cytoplasmic membrane, periplasmic, and cytoplasmic components. J. Bacteriol. 167:1083-1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Doyle, C. M., and D. J. Arp. 1988. Nickel affects expression of the nickel-containing hydrogenase of Alcaligenes latus. J. Bacteriol. 170:3891-3896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Friedrich, B., and E. Schwartz. 1993. Molecular biology of hydrogen utilization in aerobic chemolithotrophs. Annu. Rev. Microbiol. 47:351-383. [DOI] [PubMed] [Google Scholar]
- 20.Gogotov, I. N., N. A. Zorin, L. T. Serebriakova, and E. N. Kondratieva. 1978. The properties of hydrogenase from Thiocapsa roseopersicina. Biochim. Biophys. Acta 523:335-343. [DOI] [PubMed] [Google Scholar]
- 21.Ivars-Martinez, E., et al. 2008. Comparative genomics of two ecotypes of the marine planktonic copiotroph Alteromonas macleodii suggests alternative lifestyles associated with different kinds of particulate organic matter. ISME J. 2:1194-1212. [DOI] [PubMed] [Google Scholar]
- 22.Jack, R. L., F. Sargent, B. C. Berks, G. Sawers, and T. Palmer. 2001. Constitutive expression of Escherichia coli tat genes indicates an important role for the twin-arginine translocase during aerobic and anaerobic growth. J. Bacteriol. 183:1801-1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Karyakin, A. A., et al. 2005. Hydrogenase electrodes for fuel cells. Biochem. Soc. Trans. 33:73-75. [DOI] [PubMed] [Google Scholar]
- 24.Kim, H., C. Yu, and R. J. Maier. 1991. Common cis-acting region responsible for transcriptional regulation of Bradyrhizobium japonicum hydrogenase by nickel, oxygen, and hydrogen. J. Bacteriol. 173:3993-3999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kovacs, A. T., et al. 2005. An FNR-type regulator controls the anaerobic expression of hyn hydrogenase in Thiocapsa roseopersicina. J. Bacteriol. 187:2618-2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liang, Y., et al. 2009. Increased biological hydrogen production by deletion of hydrogen-uptake system in photosynthetic bacteria. Microbiol. Res. 164:674-679. [DOI] [PubMed] [Google Scholar]
- 27.Lopez-Lopez, A., S. G. Bartual, L. Stal, O. Onyshchenko, and F. Rodriguez-Valera. 2005. Genetic analysis of housekeeping genes reveals a deep-sea ecotype of Alteromonas macleodii in the Mediterranean Sea. Environ. Microbiol. 7:649-659. [DOI] [PubMed] [Google Scholar]
- 28.Maier, R. J., F. J. Hanus, and H. J. Evans. 1979. Regulation of hydrogenase in Rhizobium japonicum. J. Bacteriol. 137:825-829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maness, P. C., S. Smolinski, A. C. Dillon, M. J. Heben, and P. F. Weaver. 2002. Characterization of the oxygen tolerance of a hydrogenase linked to a carbon monoxide oxidation pathway in Rubrivivax gelatinosus. Appl. Environ. Microbiol. 68:2633-2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maroti, G., et al. 2009. Discovery of [NiFe] hydrogenase genes in metagenomic DNA: cloning and heterologous expression in Thiocapsa roseopersicina. Appl. Environ. Microbiol. 75:5821-5830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mattsson, U., and A. Sellstedt. 2002. Nickel affects activity more than expression of hydrogenase protein in Frankia. Curr. Microbiol. 44:88-93. [DOI] [PubMed] [Google Scholar]
- 32.Mertens, R., and A. Liese. 2004. Biotechnological applications of hydrogenases. Curr. Opin. Biotechnol. 15:343-348. [DOI] [PubMed] [Google Scholar]
- 33.Messenger, S. L., and J. Green. 2003. FNR-mediated regulation of hyp expression in Escherichia coli. FEMS Microbiol. Lett. 228:81-86. [DOI] [PubMed] [Google Scholar]
- 34.Palagyi-Meszaros, L. S., et al. 2009. Electron-transfer subunits of the NiFe hydrogenases in Thiocapsa roseopersicina BBS. FEBS J. 276:164-174. [DOI] [PubMed] [Google Scholar]
- 35.Rakhely, G., A. Colbeau, J. Garin, P. M. Vignais, and K. L. Kovacs. 1998. Unusual organization of the genes coding for HydSL, the stable [NiFe] hydrogenase in the photosynthetic bacterium Thiocapsa roseopersicina BBS. J. Bacteriol. 180:1460-1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rakhely, G., et al. 2004. Cyanobacterial-type, heteropentameric, NAD+-reducing NiFe hydrogenase in the purple sulfur photosynthetic bacterium Thiocapsa roseopersicina. Appl. Environ. Microbiol. 70:722-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rowe, J. L., G. L. Starnes, and P. T. Chivers. 2005. Complex transcriptional control links NikABCDE-dependent nickel transport with hydrogenase expression in Escherichia coli. J. Bacteriol. 187:6317-6323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sambrook, J., and D. W. Russell. 2001. Molecular Cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 39.Sargent, F., et al. 1998. Overlapping functions of components of a bacterial Sec-independent protein export pathway. EMBO J. 17:3640-3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sass, A. M., H. Sass, M. J. Coolen, H. Cypionka, and J. Overmann. 2001. Microbial communities in the chemocline of a hypersaline deep-sea basin (Urania Basin, Mediterranean Sea). Appl. Environ. Microbiol. 67:5392-5402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schutz, K., et al. 2004. Cyanobacterial H2 production: a comparative analysis. Planta 218:350-359. [DOI] [PubMed] [Google Scholar]
- 42.Takacs, M., et al. 2008. Formate hydrogenlyase in the hyperthermophilic archaeon, Thermococcus litoralis. BMC Microbiol. 8:88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tamagnini, P., et al. 2002. Hydrogenases and hydrogen metabolism of cyanobacteria. Microbiol. Mol. Biol. Rev. 66:1-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thauer, R. K., et al. 2010. Hydrogenases from methanogenic archaea, nickel, a novel cofactor, and H2 storage. Annu. Rev. Biochem. 79:507-536. [DOI] [PubMed] [Google Scholar]
- 45.Ureta, A. C., J. Imperial, T. Ruiz-Argueso, and J. M. Palacios. 2005. Rhizobium leguminosarum biovar viciae symbiotic hydrogenase activity and processing are limited by the level of nickel in agricultural soils. Appl. Environ. Microbiol. 71:7603-7606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vardar-Schara, G., T. Maeda, and T. Wood. 2007. Metabolically engineered bacteria for producing hydrogen via fermentation. Microbial Biotechnol. 1:107-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vignais, P. M., and B. Billoud. 2007. Occurrence, classification, and biological function of hydrogenases: an overview. Chem. Rev. 107:4206-4272. [DOI] [PubMed] [Google Scholar]
- 48.Vignais, P. M., B. Billoud, and J. Meyer. 2001. Classification and phylogeny of hydrogenases. FEMS Microbiol. Rev. 25:455-501. [DOI] [PubMed] [Google Scholar]
- 49.Vignais, P. M., and A. Colbeau. 2004. Molecular biology of microbial hydrogenases. Curr. Issues Mol. Biol. 6:159-188. [PubMed] [Google Scholar]
- 50.Volbeda, A., et al. 1995. Crystal structure of the nickel-iron hydrogenase from Desulfovibrio gigas. Nature 373:580-587. [DOI] [PubMed] [Google Scholar]
- 51.Weyman, P. D., B. Pratte, and T. Thiel. 2008. Transcription of hupSL in Anabaena variabilis ATCC 29413 is regulated by NtcA and not by hydrogen. Appl. Environ. Microbiol. 74:2103-2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wilquet, V., M. Van de Casteele, D. Gigot, C. Legrain, and N. Glansdorff. 2004. Dihydropteridine reductase as an alternative to dihydrofolate reductase for synthesis of tetrahydrofolate in Thermus thermophilus. J. Bacteriol. 186:351-355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu, L. F., et al. 1989. Nickel deficiency gives rise to the defective hydrogenase phenotype of hydC and fnr mutants in Escherichia coli. Mol. Microbiol. 3:1709-1718. [DOI] [PubMed] [Google Scholar]
- 54.Zorin, N. A., and I. N. Gogotov. 1982. Stability of hydrogenase from the purple sulfur bacteria Thiocapsa roseopersicina. Biokhimiia 47:827-833. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.