Abstract
Shiga toxin (Stx)-producing Escherichia coli (STEC) strains are a diverse group of food-borne pathogens with various levels of virulence for humans. In this study, we describe the use of a combination of multiple real-time PCR assays for the screening of 400 raw-milk cheeses for the five main pathogenic STEC serotypes (O26:H11, O103:H2, O111:H8, O145:H28, and O157:H7). The prevalences of samples positive for stx, intimin-encoding gene (eae), and at least one of the five O group genetic markers were 29.8%, 37.3%, and 55.3%, respectively. The H2, H7, H8, H11, and H28 fliC alleles were highly prevalent and could not be used as reliable targets for screening. Combinations of stx, eae variants, and O genetic markers, which are typical of the five targeted STEC serotypes, were detected by real-time PCR in 6.5% of the cheeses (26 samples) and included stx-wzxO26-eae-β1 (4.8%; 19 samples), stx-wzxO103-eae-ɛ (1.3%; five samples), stx-ihp1O145-eae-γ1 (0.8%; three samples), and stx-rfbEO157-eae-γ1 (0.3%; one sample). Twenty-eight immunomagnetic separation (IMS) assays performed on samples positive for these combinations allowed the recovery of seven eaeβ1-positive STEC O26:H11 isolates, whereas no STEC O103:H2, O145:H28, or O157:H7 strains could be isolated. Three stx-negative and eaeβ1-positive E. coli O26:[H11] strains were also isolated from cheeses by IMS. Colony hybridization allowed us to recover STEC from stx-positive samples for 15 out of 45 assays performed, highlighting the difficulties encountered in STEC isolation from dairy products. The STEC O26:H11 isolates shared the same virulence genetic profile as enterohemorrhagic E. coli (EHEC) O26:H11, i.e., they carried the virulence-associated genes EHEC-hlyA, katP, and espP, as well as genomic O islands 71 and 122. Except for one strain, they all contained the stx1 variant only, which was reported to be less frequently associated with human cases than stx2. Pulsed-field gel electrophoresis (PFGE) analysis showed that they displayed high genetic diversity; none of them had patterns identical to those of human O26:H11 strains investigated here.
Enterohemorrhagic Escherichia coli (EHEC) strains are a subset of Shiga toxin (Stx)-producing E. coli (STEC) strains that are isolated from human patients and are responsible for severe clinical symptoms, such as hemorrhagic colitis (HC) and the potentially lethal hemolytic uremic syndrome (HUS) (28, 29). Although most outbreaks of HC and HUS have been attributed to serotype O157:H7/H−, an increasing number of human infections are caused by other serotypes, such as O26:H11/H−, O103:H2, O111:H8/H−, and O145:H28/H− (13, 20, 28). O157:H7/H− and these four serotypes were classified into seropathotypes A and B, respectively, which occur most frequently in human diseases and outbreaks and in patients with severe symptoms (30).
Shiga toxins, the main virulence factors contributing to pathogenicity, consist of two major types, Stx1 and Stx2, each including several variants (47). In addition to Stx, typical EHEC strains carry on their chromosomes the locus for enterocyte effacement (LEE), a large pathogenicity island that is shared with enteropathogenic E. coli (EPEC). The LEE is responsible for attaching and effacing (A/E) lesions on enterocytes (34); it encodes several virulence factors, including the outer membrane adhesin intimin and its translocated receptor Tir, as well as components of a type III secretion machinery and its effector proteins. Intimin is involved in the tight attachment of bacteria to the enterocytes. It is encoded by the eae gene, 18 types and 9 subtypes of which have been described on the basis of its variable 3′ region (25). The EHEC serotypes O157:H7 and O145:H28 are known to be associated with the eae-γ1 subtype, whereas EHEC O26:H11, O103:H2, and O111:H8 harbor the eae-β1, eae-ɛ, and eae-γ2/θ subtypes, respectively (7, 11, 37, 51). Additional candidate pathogenicity islands, or “O islands” (OI), that may contribute to virulence have been identified. They include OI-71 and OI-122, which contain a number of type III non-LEE-encoded effector (nle) genes whose presence correlates with outbreak and HUS potential (17). Finally, EHEC O157:H7 contains a large plasmid with putative virulence genes encoding the EHEC-hemolysin (EHEC-hlyA), a serine protease (espP), and a catalase peroxidase (katP). However, strains lacking one or more of these genes have been involved in HUS, and high variability in the gene composition of this large plasmid has also been reported in various STEC serotypes (14).
Domestic ruminants have long been identified as a major reservoir for STEC (29). Although STEC transmission to humans is frequently associated with consumption of raw or undercooked meat, raw milk and dairy products have also been implicated in human disease (4). Detection and isolation of STEC in foodstuffs by traditional culture methods is rather laborious and time-consuming and is complicated by the lack of common biochemical characteristics distinguishing most STEC from other E. coli strains. Development of rapid methods for the detection of the most pathogenic STEC strains is essential to ensure the safety of food products. A sequential approach based on real-time PCR assays specific for stx, wzxO26, wzxO103, wbdIO111, ihp1O145, and rfbEO157 genetic markers has been described for the detection of STEC O26, O103, O111, O145, and O157 in foods (3, 40). A similar strategy that includes an additional eae amplification step has been proposed by working group 6 of the European Committee for Standardization (CEN) TC275 for a technical specification (currently under International Organization for Standardization [ISO] evaluation) regarding the detection of these five STEC serogroups in foods (21).
Recently, we developed real-time PCR assays for detection of the eae variants and fliC alleles associated with the five most common EHEC serotypes (32). The aim of the present study was to investigate whether these newly developed assays could further refine the diagnostic result obtained from PCR analysis of foods. To this end, the prevalence of stx, O group genetic markers, eae variants, and fliC alleles typical of EHEC O26:H11, O103:H2, O111:H8, O145:H28, and O157:H7 was investigated in 400 raw-milk cheeses. STEC and E. coli strains belonging to the five targeted serotypes were then isolated for confirmation of the PCR results, and their virulence traits were characterized. As the term EHEC includes a clinical connotation (28, 29), the stx-positive E. coli isolates recovered from food are designated STEC (not EHEC) here, irrespective of the serotype.
MATERIALS AND METHODS
Food samples, enrichment, and DNA extraction.
Soft cheeses and smear semihard uncooked cheeses produced from raw cow's milk (n = 265) and unpasteurized goat's milk cheeses (n = 135) were collected over a 5-month period (March to August 2009) in various French retail stores. They were immediately analyzed or held at 4°C before use. Twenty-five grams of cheese was subjected to enrichment, and bacterial DNAs were extracted as described elsewhere (32). DNA extracts (100 μl) were stored at 4°C before PCR analysis.
Screening of cheeses for EHEC-associated genetic markers.
Two-microliter aliquots of DNA extracts were subjected to the following real-time PCR assays: an internally controlled PCR for the detection of the stx gene (2), a simplex PCR for the detection of eae (35), a quadruplex PCR for the detection of four eae variants (eae-β1, eae-γ1, eae-ɛ, and eae-γ2/θ), and simplex and multiplex PCR assays for the detection of five fliC alleles (fliCH2, fliCH7, fliCH8, fliCH11, and fliCH28) (32). A multiplex real-time PCR was used for the detection of the O group markers as described by Perelle et al. (40) with the following modifications: the O103-specific primers and probe targeted wzxO103 (39), and the primers and probes specific for rfbEO157 and wbdIO111 were not incorporated in the reaction mixture and were used in two separate simplex PCR assays. All primers and probes specific for stx, eae, and the five O genetic markers were those specified in the CEN technical specification project (21). All the PCRs were performed on a LightCycler 480 instrument (Roche Diagnostics). The cycle threshold (CT) value was defined as the PCR cycle at which the fluorescent signal exceeded the background level. The CT was determined automatically by the Lightcycler 480 software by the second derivative maximum method.
Isolation of STEC strains from naturally contaminated cheeses and characterization of their virulence gene profiles.
Isolation of STEC strains was performed within 1 day after PCR analysis of cheeses. Colony hybridization was carried out as described previously (22). As previous analyses of food products showed that no STEC strains could be isolated by colony hybridization from enriched broths with high stx CT values corresponding to low STEC concentrations (2), only cheese samples with stx CT values of <30 were subjected to this technique. For isolation of STEC O26, O103, O111, O145, and O157, immunomagnetic separation (IMS) (Dynabeads, Invitrogen, Cergy Pontoise, France) was performed manually as recommended by the manufacturer. IMS-concentrated bacteria were plated onto TBX agar (Bio-Rad Laboratories, Marnes-la-Coquette, France), cefixime-tellurite-sorbitol-MacConkey agar (CT-SMAC), and a recently described selective differential medium for E. coli O26, O103, O111, and O145 (41). Suspected colonies were confirmed as E. coli by using an API 20E test (bioMérieux). Strains were further characterized for their O:H antigens by agglutination assays (36, 46) and by real-time PCR as described above. Conventional PCR was used for detection of the virulence markers katP (15); espP (14); OI-122-associated pagC, ent, and efa1 (30); and OI-71-associated nleA (17). Real-time PCR was used for detection of stx, eae, and eae variants as described above. Subtypes of stx were identified by PCR (F. Scheutz, L. D. Teel, L. Beutin, D. Piérard, G. Buvens, H. Karch, A. Mellmann, A. Caprioli, R. Tozzoli, A. D. O'Brien, A. R. Melton-Celsa, S. Persson, and N. A. Strockbine, unpublished data), according to the subtyping nomenclature established at the 7th International Symposium on Shiga Toxin (Verocytotoxin)-Producing Escherichia coli Infections (Buenos Aires, 10 to 13 May 2009). The presence of EHEC-hlyA, saa, bfpA, and EPEC adherence factor (EAF) plasmid was determined by DNA dot blot hybridization as previously described (45). The hemolysis phenotype and Shiga toxin production were assessed on red blood cell agar (8) and with a Vero cell assay (36), respectively. The alpha-hemolytic phenotype was further confirmed by PCR detection of the α-hlyA gene as described previously (7).
PFGE.
Pulsed-field gel electrophoresis (PFGE) was performed as described by Ribot et al. (43). DNAs were digested with XbaI and BlnI enzymes (Roche Diagnostics), and the restriction fragments were resolved at 14°C on 1% Seakem gold agarose gels (Cambrex, Emerainville, France) on a Chef-DR-III system (Bio-Rad Laboratories, Germany). Pulse times were ramped with 2 s and 2.2 s at the beginning and 64 s and 63.8 s at the end for separation of DNA fragments digested with XbaI and BlnI, respectively. XbaI-digested Salmonella enterica serovar Braenderup H9812 DNA was used as a molecular size standard. After being stained with ethidium bromide (10 μg ml−1), gels were visualized on GelDocEQ (Bio-Rad Laboratories), and the PFGE profiles were analyzed using Bionumerics software (Applied Math, Sint-Martens-Latem, Belgium). The PFGE typing scheme included seven human O26:H11 isolates (CB6307, ED-21, VTH7, EH284, EH324, 5917/97, and 6061/96) and 11 O26:H11 strains recovered at the French National Reference Laboratory for E. coli (VetAgro Sup) from various types of samples, i.e., cattle feces collected in two French dairy farms in 2009 (FFL1-1, FFL2-6, FFL3-12, FV2-33, FV3-11, FV4-14, and FV5-36); cheeses analyzed in 2005 (FR14-18), 2008 (51-2) and 2009 (F15-313); and ewe's milk analyzed in 2007 (L23A).
RESULTS
Monitoring of EHEC-associated genetic markers in raw-milk cheeses.
A total of 400 raw-milk cheeses were screened for the presence of genetic markers associated with EHEC O26:H11, O103:H2, O111:H8, O145:H28, and O157:H7 (Table 1). The stx gene was detected in 29.8% of the samples. More than half (55.3%) of the samples contained at least one of the five EHEC O group genetic markers; the most frequent O group marker was ihp1O145 (45.8%), followed by wzxO103 (23.8%), wzxO26 (11.3%), and rfbEO157 (5.8%). Only one sample (0.3%) contained wbdIO111. Many of the samples (87.3%) contained at least one of the five EHEC fliC alleles (Table 1), and the prevalences of the fliC alleles individually ranged from 38.5% (fliCH28) to 65.5% (fliCH8). The eae gene was present in 37.3% of the samples. At least one of the four variants eae-β1, eae-γ1, eae-ɛ, and eae-γ2/θ was detected in 32.3% of the samples; the most frequent variants were eae-β1 (21.8%) and eae-γ2/θ (15.5%), followed by eae-ɛ (2.8%) and eae-γ1 (1.3%) (Table 1). The concomitant presence of several eae variants was observed for 31 (7.75%) samples. The presence of both stx and eae was observed for 68 (17%) samples, and the simultaneous presence of stx, eae, and at least one of the five EHEC O group markers was detected in 61 (15.3%) samples (Table 1). Twenty-six (6.5%) samples were positive for both stx and at least one of the four main EHEC associations of markers wzxO26-eae-β1, wzxO103-eae-ɛ, ihp1O145-eae-γ1, and rfbEO157-eae-γ1 (Table 1). The most frequent combination identified was stx-wzxO26-eae-β1 (19 samples; 4.8%), followed by stx-wzxO103-eae-ɛ (5 samples; 1.3%), stx-ihp1O145-eae-γ1 (3 samples; 0.8%), and stx-rfbEO157-eae-γ1 (one sample; 0.3%). The prevalences of these four combinations remained the same if a fliC allele was included in each of the combinations (Table 1). None of the samples were positive for the combination stx-wbdIO111-eae-γ2/θ.
TABLE 1.
Detection of EHEC-associated genetic markers in 400 raw-milk cheese samples
| Genetic markers targeted by real-time PCR (either alone or in combination with each other) | Positive samples/400 tested |
|
|---|---|---|
| No. | % | |
| Individual markers | ||
| stx | 119 | 29.8 |
| eaea | 149 | 37.3 |
| eae-β1 | 87 | 21.8 |
| eae-γ1 | 5 | 1.3 |
| eae-ɛ | 11 | 2.8 |
| eae-γ2/θ | 62 | 15.5 |
| eae-β1, eae-γ1, eae-ɛ, eae-γ2/θb | 129c | 32.3 |
| wzxO26 | 45 | 11.3 |
| wzxO103 | 95 | 23.8 |
| wbdIO111 | 1 | 0.3 |
| ihp1O145 | 183 | 45.8 |
| rfbEO157 | 20 | 5 |
| wzxO26, wzxO103, wbdIO111, ihp1O145, rfbEO157d | 221 | 55.3 |
| fliCH2 | 244 | 61.0 |
| fliCH7 | 234 | 58.5 |
| fliCH8 | 262 | 65.5 |
| fliCH11 | 197 | 49.3 |
| fliCH28 | 154 | 38.5 |
| fliCH2, fliCH7, fliCH8, fliCH11, fliCH28e | 349 | 87.3 |
| Combinations of markers | ||
| stx-eae | 68 | 17 |
| stx-wzxO26-eae | 22 | 5.5 |
| stx-wzxO26-eae-β1 | 19 | 4.8 |
| stx-wzxO26-eae-β1-fliCH11 | 19 | 4.8 |
| stx-wzxO103-eae | 31 | 7.8 |
| stx-wzxO103-eae-ɛ | 5 | 1.3 |
| stx-wzxO103-eae-ɛ-fliCH2 | 5 | 1.3 |
| stx-wbdIO111-eae | 0 | 0 |
| stx-wbdIO111-eae-γ2/θ | 0 | 0 |
| stx-ihp1O145-eae | 51 | 12.8 |
| stx-ihp1O145-eae-γ1 | 3 | 0.8 |
| stx-ihp1O145-eae-γ1-fliCH28 | 3 | 0.8 |
| stx-rfbEO157-eae | 4 | 1 |
| stx-rfbEO157-eae-γ1 | 1 | 0.3 |
| stx-rfbEO157-eae-γ1-fliCH7 | 1 | 0.3 |
| stx-(wzxO26, wzxO103, wbdIO111, ihp1O145, rfbEO157)f | 89 | 22.3 |
| stx-(wzxO26, wzxO103, wbdIO111, ihp1O145, rfbEO157)-eaeg | 61 | 15.3 |
| stx-(wzxO26, wzxO103, wbdIO111, ihp1O145, rfbEO157)-(eae-β1, eae-γ1, eae-ɛ, eae-γ2/θ)h | 54 | 13.5 |
| stx-wzxO26-eae-β1, stx-wzxO103-eae-ɛ, stx-wbdIO111-eae-γ2/θ, stx-ihp1O145-eae-γ1, stx-rfbEO157-eae-γ1i | 26j | 6.5 |
Detection of the eae gene with universal primers/probe.
Detection of at least one of the four eae variants eae-β1, eae-γ1, eae-ɛ, and eae-γ2/θ.
Twenty-two samples contained eae-β1 and eae-γ2/θ; two samples contained eae-β1 and eae-ɛ; two samples contained eae-ɛ and eae-γ2/θ; one sample contained eae-β1 and eae-γ1; two samples contained eae-β1, eae-γ2/θ, and eae-ɛ; and two samples contained eae-β1, eae-γ2/θ, and eae-γ1.
Detection of at least one of the five serogroup genetic markers wzxO26, wzxO103, wbdIO111, ihp1O145, and rfbEO157.
Detection of at least one of the five fliC alleles specific for H2, H7, H8, H11, and H28 flagellar antigens.
Detection of stx and at least one of the five serogroup genetic markers.
Detection of stx, at least one of the five serogroup genetic markers and eae.
Detection of stx, at least one of the five serogroup genetic markers, and at least one of the four eae variants.
Detection of at least one of the five stx-serogroup-eae variant marker combinations.
Two samples contained two combinations of markers each (i.e., stx-wzxO26-eae-β1 plus stx-ihp1O145-eae-γ1 and stx-ihp1O145-eae-γ1 plus stx-rfbEO157-eae-γ1).
Isolation of STEC strains from raw-milk cheeses and characterization of their virulence profiles.
All 26 samples that tested positive by real-time PCR for stx-wzxO26-eae-β1, stx-wzxO103-eae-ɛ, stx-ihp1O145-eae-γ1, and/or stx-rfbEO157-eae-γ1 were subjected to IMS assays. As two samples contained two combinations each (Table 1), a total of 28 IMS assays were performed. Despite frequent nonspecific binding of bacteria to the IMS particles (data not shown), 10 E. coli strains were isolated out of 19 IMS O26 assays; they corresponded to seven STEC O26:H11/[H11] (the brackets represent strains whose serotype could be determined only by real-time PCR and not by conventional serotyping) strains and to stx-negative E. coli O26:[H11] (306.D and 315.2) and O26:H32 (355.2) (Table 2 ). No E. coli O103, O145, or O157 strains could be isolated from the 5 IMS O103, 3 IMS O145, and 1 IMS O157 assays performed, respectively. Eleven additional IMS assays (7 O26, 1 O103, 2 O145, and 1 O157) performed on cheese samples that were negative for stx but positive for serogroup/eae variant associations allowed us to isolate two stx-negative E. coli strains belonging to O26:[H11] (04.2) and O103:H25 (167.2) serotypes (Table 2).
TABLE 2.
Characterization of the 28 E. coli strains isolated from raw-milk cheeses
| Strain | Isolationa | Serotypeb | Presence off: |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| stx (subtypec) | eae (type) | Hemo- lysise | EHEC-hlyA | katP | espP | nleA | pagC | ent | efa1 | saa | EAF | bfpA | |||
| STEC | |||||||||||||||
| 33.1 | C | O2:H27 | + (stx2a) | − | − | + | − | − | − | − | − | − | − | − | − |
| 162.5 | C | O2:H27 | + (stx2a) | − | − | + | − | − | − | − | − | − | − | − | − |
| 246.3 | C | O2:H27 | + (stx2a) | − | − | + | − | − | − | − | − | − | − | − | − |
| 104.5 | C | O8:H19 | + (stx2a + stx2d) | − | E-hly | + | − | − | − | − | − | − | − | − | − |
| 303.1 | C | O8:H19 | + (stx2ND) | − | − | − | − | − | − | − | − | − | − | − | − |
| 170.2 | I, C | O26:H11 | + (stx1a) | + (β1) | E-hly | + | + | + | + | − | + | + | − | − | − |
| 238.2 | I | O26:H11 | + (stx1a) | + (β1) | E-hly | + | + | + | + | − | + | + | − | − | − |
| 129.2 | I | O26:[H11] | + (stx1a) | + (β1) | E-hly | + | + | + | + | − | + | + | − | − | − |
| 245.3 | I | O26:[H11] | + (stx1a) | + (β1) | E-hly | + | + | + | + | − | + | + | − | − | − |
| 260.3 | I | O26:[H11] | + (stx1a) | + (β1) | E-hly | + | + | + | + | − | + | + | − | − | − |
| 283.4 | I | O26:[H11] | + (stx1a) | + (β1) | E-hly | + | + | + | + | − | + | + | − | − | − |
| 277.2 | I | O26:[H11] | + (stx1a + stx2a) | + (β1) | E-hly | + | + | − | + | − | + | + | − | − | − |
| 161.2 | C | O91:H10 | + (stx2d) | − | − | − | − | − | − | − | − | − | − | − | − |
| 282.2 | C | O128ab:[H2] | + (stx1c) | − | E-hly | + | − | − | − | − | − | − | − | − | − |
| 124.1 | C | O166:[H28]d | + (stx2b + stx2c) | − | − | − | − | − | − | − | − | − | − | − | − |
| 91.1 | C | O174:H2 | + (stx1a) | − | E-hly | + | − | + | − | + | − | − | + | − | − |
| 299.3 | C | O175:H16 | + (stx2a) | − | E-hly | + | − | − | − | − | − | − | + | − | − |
| 311.2 | C | O175:H16 | + (stx2d) | − | − | − | − | − | − | − | − | − | − | − | − |
| 381.1 | C | O178:H19 | + (stx2c) | − | − | − | − | − | − | + | − | − | − | − | − |
| 225.4 | C | ONT:H10 | + (stx1c) | − | − | − | − | − | − | − | − | − | − | + | − |
| 47.1 | C | Or:H10 | + (stx1c) | − | − | − | − | − | − | − | − | − | − | + | − |
| 180.E | C | Or:H10 | + (stx1c) | − | − | − | − | − | − | − | − | − | − | + | − |
| Non-STEC | |||||||||||||||
| 04.2 | I | O26:[H11] | − | + (β1) | α-hly | − | − | − | + | − | + | + | − | − | − |
| 315.2 | I | O26:[H11] | − | + (β1) | α-hly | − | − | − | + | − | + | + | − | − | − |
| 306.D | I | [O26]:[H11] | − | + (β1) | α-hly | − | − | − | + | − | + | + | − | − | − |
| 355.2 | I | O26:H32 | − | − | − | − | − | − | − | − | − | − | − | − | − |
| 167.2 | I | O103:H25 | − | + (γ2/θ) | E-hly | + | − | − | − | + | + | + | − | − | − |
| 275.1 | C | O133:H29d | − | − | − | − | − | − | − | − | − | − | − | − | − |
C, colony hybridization; I, immunomagnetic separation.
Or, O rough. O and H types in brackets represent strains whose serotype could be determined by real-time PCR only and not by conventional serotyping.
stx subtypes refer to prototype organisms (GenBank accession numbers in parentheses) as follows: stx1a, O157-EDL933 (M19473); stx1c, O174-DG131-3 (Z36901); stx2a, O157-EDL933 (X07865); stx2b, O118-EH250 (AF043627); stx2c, O157-E32511 (M59432); stx2d, O73-C165-02 (DQ059012); ND, not determined.
These strains tested positive for ihp1O145 by real-time PCR.
E-hly and α-hly, EHEC hemolytic and alpha-hemolytic phenotypes on red blood cell agar, respectively.
+, present; −, absent.
Colony hybridization was applied to 45 stx-positive samples with low stx CT values (see Materials and Methods), including 8 out of the 26 samples containing stx-serogroup-eae variant marker combinations. This allowed the recovery of one STEC O26:H11 isolate that showed the same characteristics as the STEC O26:H11 isolate obtained by IMS from the same sample (Table 2), and both isolates were therefore considered the same strain. Fifteen additional STEC strains were recovered by colony hybridization from stx-positive samples that were not investigated by IMS. They belonged to nine different serotypes, i.e., O2:H27 (n = 3), O nontypeable (ONT)/O rough:H10 (n = 3), O8:H19 (n = 2), O175:H16 (n = 2), O91:H10, O128ab:[H2], O166:[H28], O174:H2, and O178:H19 (one strain each) (Table 2). One E. coli O133:H29 strain was also isolated, but it was not confirmed as stx positive (275.1 [Table 2]). It was recovered from an stx-eae-ihp1O145-positive cheese sample and tested positive for ihp1O145, as was also the case for the STEC O166:[H28] strain 124.1 (Table 2).
All the isolates were characterized for the presence of the genes stx1, stx2, eae, EHEC-hlyA, katP, espP, saa, nleA, pagC, ent, and efa1, as well as for the presence of the EAF plasmid. Twenty-two E. coli strains possessed at least one stx gene (Table 2) and were cytotoxic to Vero cells (data not shown). All the STEC O26:[H11] strains shared the same virulence gene profile (stx1a eae-β1 EHEC-hlyA katP espP nleA ent efa1), except for one strain (277.2) that carried an additional stx2a gene and was espP negative (Table 2). The three E. coli O26:[H11] isolates 04.2, 306.D, and 315.2 carried the same virulence genes as STEC O26:H11 except that they lacked an stx gene and the plasmid-associated genes EHEC-hlyA, katP, and espP (Table 2). They also differed from STEC O26:H11 by the production of an α-hemolysin phenotype on red blood cell agar (Table 2). Among the non-O26 STEC strains isolated, none belonged to the four other main EHEC serotypes targeted here, and none contained the eae gene (Table 2). No strains contained the bundle-forming pilus bfpA gene, and only three STEC ONT/O rough:H10 (stx1c-positive) strains carried the EAF plasmid (Table 2). Two STEC O174:H2 (91.1) and O175:H16 (299.3) isolates contained the EHEC-hlyA gene and the autoagglutinating adhesin-encoding gene saa found in many eae-negative STEC strains (27); the former also carried the espP and pagC genes (Table 2). The stx-negative E. coli O103:H25 isolate (167.2) possessed the eae-γ2/θ variant and the three OI-122-encoded virulence markers tested (Table 2).
Typing of E. coli O26 strains by PFGE.
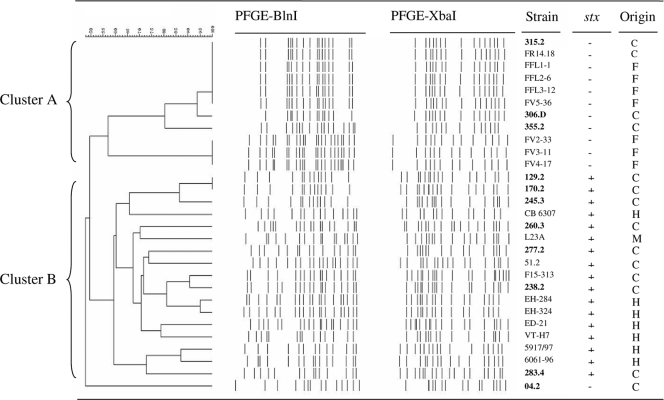
To examine the relatedness among the E. coli O26 isolates recovered from cheeses, their XbaI and BlnI restriction patterns were analyzed by PFGE. This typing scheme was also applied to 18 additional O26:H11 strains from various origins (see Materials and Methods). PFGE produced 13 to 22 fragments ranging in size from ca. 40 to 700 kb. A dendrogram analysis of PFGE patterns showed high heterogeneity among the 29 E. coli O26 strains examined, with 21 distinct XbaI-BlnI patterns obtained (Fig. 1). Two clusters designated A (11 strains; >56% similarity) and B (17 strains; >58% similarity) were identified (Fig. 1). Cluster A comprised four PFGE patterns. The first pattern was common to the α-hemolysin-producing stx-negative O26:H11 strain 315-2 recovered here and to one cheese (FR14-18) and four cattle (FFL1-1, FFL2-6, FFL3-12, and FV5-36) stx-negative O26:H11 isolates recovered elsewhere. This pattern showed >90% similarity to that of the α-hemolysin-producing stx-negative O26:H11 strain 306D. A third and a fourth pattern were found in the O26:H32 strain (355.2) recovered here and in three cattle O26:H11 strains (FV2-33, FV3-11, and FV4-17), respectively. Cluster B included 7 and 10 stx-positive O26:H11 strains of human and cheese origins, respectively. Two cheese isolates (129.2 and 170.2) shared a single PFGE pattern, and two human isolates (EH284 and EH324) were 95% similar. All the other strains had more distantly related patterns. Finally, the α-hemolysin-producing and stx-negative O26:H11 isolate 04.2 showed a PFGE pattern with less than 50% similarity to the others and thus clustered separately.
FIG. 1.
Pulsed-field gel electrophoresis patterns of 29 E. coli strains isolated from raw-milk cheese (C), humans (H), cattle feces (F), and raw-milk (M) samples. The dendrogram was constructed using the Dice coefficient with a 2% band position tolerance and the unweighted pair group method using arithmetic averages. The names of strains isolated in this study are indicated in boldface. −, negative; +, positive.
DISCUSSION
The prevalence of the stx-positive cheeses analyzed here was similar to those reported previously (2, 22) but higher than those found in other studies (5.7% to 13%) (42, 50, 53). However, direct comparison of the results is difficult, as the last studies were based on the use of distinct stx detection methods (i.e., conventional PCR) and included analysis of hard cheeses. In contrast, samples selected for this study were uncooked and soft cheeses, all made from raw milk and thus associated with higher risk of STEC infection than other types of cheeses derived from manufacturing processes more effective in eliminating STEC. Only 15 STEC strains could be isolated by colony hybridization, confirming the difficulties frequently encountered in STEC isolation from food (2, 3, 22, 50). Several hypotheses have been proposed to explain this outcome, including the presence of high levels of competing microflora or of natural inhibitors within dairy products that interfere with STEC isolation (22, 50). STEC might also be present in a stressed or injured state that prevents its isolation.
When the presence of the five main pathogenic STEC serotypes was investigated in cheeses based on the simultaneous detection of stx, eae, and at least one of the five STEC O group markers, the prevalence of samples with a presumptive positive result was 15.3%. However, this prevalence was lowered to 6.5% when the associations between the eae variants and the STEC serotypes were taken into account, highlighting the interest of the use of combinations of related STEC genetic alleles or variants for food screening. The most prevalent combination was stx-wzxO26-eae-β1 (4.8%), and the presumptive presence of STEC O26:H11 in stx-wzxO26-eae-β1-positive cheeses could be confirmed by IMS for 7 out of 19 samples. In contrast, no STEC O103:H2, O145:H28, or O157:H7 isolates could be recovered by IMS from the 9 samples that tested positive by PCR, again illustrating the problems associated with STEC isolation. Due to their high prevalence in cheeses, the five STEC fliC alleles were not selective enough to be used as reliable markers for food screening. The prevalence of the ihp1O145 gene was also high, suggesting that, although proposed for specific detection of STEC O145 (38), this marker could be associated with other E. coli serogroups commonly present in raw-milk cheeses. Indeed, ihp1O145 was detected in non-O145 E. coli (i.e., O133 and O137) (6), as was also shown here for STEC O166:[H28] and E. coli O133:H29 isolates. Other real-time PCR assays, such as that targeting the O145-specific O antigen gene cluster (23), might therefore be used as alternative tests in screening for STEC O145 in foods.
E. coli O26:H11 has been frequently detected in calves and cattle (26), and contamination of raw milk and raw-milk cheeses with STEC O26:H11 has also been described (1, 18). The STEC O26:H11 strains isolated here tested positive for stx1 only, except for one strain that also carried stx2. They contained genomic islands OI-71 and OI-122 and showed high genetic diversity by PFGE, as reported previously (17, 30, 52). According to their virulence genetic profiles, these strains should be considered pathogenic for humans. However, their ability to cause severe disease and their potential to cause outbreaks could be questioned. The last outbreak of STEC O26:H11 infection in France occurred in 2005 and was caused by stx2-positive strains (18). The virulence of stx2-positive O26:H11 might differ from that of stx1-positive O26:H11, which seems to be associated with milder clinical presentation even in an outbreak situation (19). Furthermore, the stx2 variant is known to be the most clinically important stx subtype (24), and HUS-associated STEC O26:H11 harboring stx2 (alone or in combination with stx1) has been more frequently isolated in central Europe since 1996 than strains containing stx1 only (52). The fact that stx-negative E. coli O26:[H11] strains were isolated from stx-positive samples also raises some questions about the diagnostic result. As loss of the bacteriophage-associated stx gene was shown to occur frequently both in vitro and in vivo (9), it is tempting to speculate that these strains are derivatives of STEC that have lost their stx genes during the enrichment or isolation procedure. Alternatively, the stx gene detected in cheese samples might have been carried by other bacterial strains. Although colony hybridization could have been useful to test this hypothesis, it was not performed on these samples, as they showed high stx CT values indicative of low STEC concentrations. When isolated from human patients, such stx-negative E. coli O26:H11 strains are referred to as atypical EPEC, as they lack the EAF plasmid (10). Interestingly, these strains were found to produce α-hemolysin and did not cluster with any human strains in the PFGE analysis, as was also reported by others (31, 33). Although α-hemolysin-producing E. coli O26:H11 strains have been isolated from children with diarrhea in the past (8), their virulence potential remains to be clearly defined.
Among the non-O26 STEC strains isolated here, none carried the eae gene, which is a virulence trait of typical EHEC strains. Most of them belonged to serotypes previously found in cheese and cattle, such as O2:H27, O8:H19, and O175:H16 (5, 44, 49, 50). One STEC strain belonged to O91:H10, a serotype of atypical (eae-negative) EHEC strains known to be associated with human disease (12). Finally, one E. coli isolate belonged to O103:H25, a serotype that was responsible for an outbreak in Norway linked to the consumption of contaminated fermented sausages (48). This isolate was eae-γ2/θ positive, as reported for another O103:H25 strain (16), and harbored OI-122. However, as reported for several human O103:H25 isolates (48), the strain was stx negative.
In conclusion, the molecular approach based on real-time PCR assays targeting specific EHEC combinations of genetic markers (such as stx, O groups, and eae variants) represents an interesting and valuable strategy for rapid screening of the most pathogenic STEC strains within food products. In the particular case of STEC O157:H7, this approach could be further evaluated by comparison with standardized or validated methods available for this serotype. In terms of risk to human health, the significance of the prevalence found for typical EHEC combinations of markers in raw-milk cheeses (i.e., 6.5%) deserves further investigation. In particular, the pathogenic potential of cheese STEC isolates belonging to EHEC serotypes, such as O26:H11 and O91:H10, needs to be further examined. The hypothetical loss of stx genes during STEC isolation from foods should also be investigated, as loss of stx could result in foodstuffs being falsely considered uncontaminated by pathogenic STEC and therefore safe.
Acknowledgments
This work was supported by funds from the Ministère de l'Alimentation, de l'Agriculture et de la Pêche and the Association de Coordination Technique pour l'Industrie Agro-Alimentaire (UMT-TERESA), as well as by a Transversalité AFSSA-INRA grant. J.M. is the recipient of a doctoral fellowship (CIFRE no. 241/2007) cofinanced by ACTILAIT and the Association Nationale de la Recherche Technique (ANRT). This study was also supported and coordinated by the National Interprofessional Center for the Dairy Economy (CNIEL), Paris, France.
We are grateful to E. Loukiadis and D. Thévenot (French National Reference Laboratory for E. coli, VetAgro Sup, Marcy l'Etoile, France) for providing 11 O26:H11 strains from cattle and food origins and for helpful discussion. We thank J. Taché (ANSES) for quality assurance assistance.
Footnotes
Published ahead of print on 14 January 2011.
REFERENCES
- 1.Allerberger, F., et al. 2003. Hemolytic-uremic syndrome associated with enterohemorrhagic Escherichia coli O26:H− infection and consumption of unpasteurized cow's milk. Int. J. Infect. Dis. 7:42-45. [DOI] [PubMed] [Google Scholar]
- 2.Auvray, F., C. Lecureuil, F. Dilasser, J. Tache, and S. Derzelle. 2009. Development of a real-time PCR assay with an internal amplification control for the screening of Shiga toxin-producing Escherichia coli in foods. Lett. Appl. Microbiol. 48:554-559. [DOI] [PubMed] [Google Scholar]
- 3.Auvray, F., et al. 2007. Detection, isolation and characterization of Shiga toxin-producing Escherichia coli in retail-minced beef using PCR-based techniques, immunoassays and colony hybridization. Lett. Appl. Microbiol. 45:646-651. [DOI] [PubMed] [Google Scholar]
- 4.Baylis, C. 2009. Raw milk and raw milk cheeses as vehicles for infection by verocytotoxin-producing Escherichia coli. Int. J. Dairy Technol. 62:293-307. [Google Scholar]
- 5.Beutin, L., D. Geier, H. Steinruck, S. Zimmermann, and F. Scheutz. 1993. Prevalence and some properties of verotoxin (Shiga-like toxin)-producing Escherichia coli in seven different species of healthy domestic animals. J. Clin. Microbiol. 31:2483-2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beutin, L., S. Jahn, and P. Fach. 2009. Evaluation of the ‘GeneDisc’ real-time PCR system for detection of enterohaemorrhagic Escherichia coli (EHEC) O26, O103, O111, O145 and O157 strains according to their virulence markers and their O- and H-antigen-associated genes. J. Appl. Microbiol. 106:1122-1132. [DOI] [PubMed] [Google Scholar]
- 7.Beutin, L., G. Krause, S. Zimmermann, S. Kaulfuss, and K. Gleier. 2004. Characterization of Shiga toxin-producing Escherichia coli strains isolated from human patients in Germany over a 3-year period. J. Clin. Microbiol. 42:1099-1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beutin, L., M. Montenegro, S. Zimmermann, and R. Stephan. 1986. Characterization of hemolytic strains of Escherichia coli belonging to classical enteropathogenic O-serogroups. Zentralbl. Bakteriol. Mikrobiol. Hyg. A 261:266-279. [DOI] [PubMed] [Google Scholar]
- 9.Bielaszewska, M., et al. 2007. Shiga toxin gene loss and transfer in vitro and in vivo during enterohemorrhagic Escherichia coli O26 infection in humans. Appl. Environ. Microbiol. 73:3144-3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bielaszewska, M., A. K. Sonntag, M. A. Schmidt, and H. Karch. 2007. Presence of virulence and fitness gene modules of enterohemorrhagic Escherichia coli in atypical enteropathogenic Escherichia coli O26. Microbes Infect. 9:891-897. [DOI] [PubMed] [Google Scholar]
- 11.Blanco, M., et al. 2004. Serotypes, virulence genes, and intimin types of Shiga toxin (verotoxin)-producing Escherichia coli isolates from cattle in Spain and identification of a new intimin variant gene (eae-xi). J. Clin. Microbiol. 42:645-651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bonnet, R., et al. 1998. Non-O157:H7 Stx2-producing Escherichia coli strains associated with sporadic cases of hemolytic-uremic syndrome in adults. J. Clin. Microbiol. 36:1777-1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brooks, J. T., et al. 2005. Non-O157 Shiga toxin-producing Escherichia coli infections in the United States, 1983-2002. J. Infect. Dis. 192:1422-1429. [DOI] [PubMed] [Google Scholar]
- 14.Brunder, W., H. Schmidt, M. Frosch, and H. Karch. 1999. The large plasmids of Shiga-toxin-producing Escherichia coli (STEC) are highly variable genetic elements. Microbiology 145:1005-1014. [DOI] [PubMed] [Google Scholar]
- 15.Brunder, W., H. Schmidt, and H. Karch. 1996. KatP, a novel catalase-peroxidase encoded by the large plasmid of enterohaemorrhagic Escherichia coli O157:H7. Microbiology 142:3305-3315. [DOI] [PubMed] [Google Scholar]
- 16.Cookson, A. L., J. Bennett, F. Thomson-Carter, and G. T. Attwood. 2007. Intimin subtyping of Escherichia coli: concomitant carriage of multiple intimin subtypes from forage-fed cattle and sheep. FEMS Microbiol. Lett. 272:163-171. [DOI] [PubMed] [Google Scholar]
- 17.Coombes, B. K., et al. 2008. Molecular analysis as an aid to assess the public health risk of non-O157 Shiga toxin-producing Escherichia coli strains. Appl. Environ. Microbiol. 74:2153-2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Espie, E., et al. 2006. Shiga-toxin producing Escherichia coli O26 infection and unpasteurised cows cheese, France, 2005, poster. In J. Sofronidis (ed.), Progr. Abstr. 6th Int. Symp. Shiga Toxin (Verocytoxin)-Producing Escherichia coli Infect., Melbourne, Australia, 2006. Cambridge Publishing, West Leederville, W.A., Australia. http://www.invs.sante.fr/surveillance/shu/poster_melbourne_o26.pdf.
- 19.Ethelberg, S., et al. 2009. Outbreak of non-O157 Shiga toxin-producing Escherichia coli infection from consumption of beef sausage. Clin. Infect. Dis. 48:e78-e81. [DOI] [PubMed] [Google Scholar]
- 20.European Food Safety Authority. 2007. Scientific opinion of the panel on biological hazards on a request from EFSA on monitoring of verotoxigenic Escherichia coli (VTEC) and identification of human pathogenic types. EFSA J. 579:1-61. [Google Scholar]
- 21.European Food Safety Authority. 2009. Technical specifications for the monitoring and reporting of verotoxigenic Escherichia coli (VTEC) on animals and food (VTEC surveys on animals and food). EFSA J. 7:1366. [Google Scholar]
- 22.Fach, P., S. Perelle, F. Dilasser, and J. Grout. 2001. Comparison between a PCR-ELISA test and the Vero cell assay for detecting Shiga toxin-producing Escherichia coli in dairy products and characterization of virulence traits of the isolated strains. J. Appl. Microbiol. 90:809-818. [DOI] [PubMed] [Google Scholar]
- 23.Fratamico, P. M., C. DebRoy, T. Miyamoto, and Y. Liu. 2009. PCR detection of enterohemorrhagic Escherichia coli O145 in food by targeting genes in the E. coli O145 O-antigen gene cluster and the Shiga toxin 1 and Shiga toxin 2 genes. Foodborne Pathog. Dis. 6:605-611. [DOI] [PubMed] [Google Scholar]
- 24.Friedrich, A. W., et al. 2002. Escherichia coli harboring Shiga toxin 2 gene variants: frequency and association with clinical symptoms. J. Infect. Dis. 185:74-84. [DOI] [PubMed] [Google Scholar]
- 25.Ito, K., et al. 2007. Intimin types determined by heteroduplex mobility assay of intimin gene (eae)-positive Escherichia coli strains. J. Clin. Microbiol. 45:1038-1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jenkins, C., J. Evans, H. Chart, G. A. Willshaw, and G. Frankel. 2008. Escherichia coli serogroup O26: a new look at an old adversary. J. Appl. Microbiol. 104:14-25. [DOI] [PubMed] [Google Scholar]
- 27.Jenkins, C., et al. 2003. Distribution of the saa gene in strains of Shiga toxin-producing Escherichia coli of human and bovine origins. J. Clin. Microbiol. 41:1775-1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Karch, H., P. I. Tarr, and M. Bielaszewska. 2005. Enterohaemorrhagic Escherichia coli in human medicine. Int. J. Med. Microbiol. 295:405-418. [DOI] [PubMed] [Google Scholar]
- 29.Karmali, M. A., V. Gannon, and J. M. Sargeant. 2010. Verocytotoxin-producing Escherichia coli (VTEC). Vet. Microbiol. 140:360-370. [DOI] [PubMed] [Google Scholar]
- 30.Karmali, M. A., et al. 2003. Association of genomic O island 122 of Escherichia coli EDL 933 with verocytotoxin-producing Escherichia coli seropathotypes that are linked to epidemic and/or serious disease. J. Clin. Microbiol. 41:4930-4940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leomil, L., A. F. Pestana de Castro, G. Krause, H. Schmidt, and L. Beutin. 2005. Characterization of two major groups of diarrheagenic Escherichia coli O26 strains which are globally spread in human patients and domestic animals of different species. FEMS Microbiol. Lett. 249:335-342. [DOI] [PubMed] [Google Scholar]
- 32.Madic, J., et al. 2010. Simplex and multiplex real-time PCR assays for the detection of flagellar (H-antigen) fliC alleles and intimin (eae) variants associated with enterohaemorrhagic Escherichia coli (EHEC) serotypes O26:H11, O103:H2, O111:H8, O145:H28 and O157:H7. J. Appl. Microbiol. 109:1696-1705. [DOI] [PubMed] [Google Scholar]
- 33.Miko, A., B. A. Lindstedt, L. T. Brandal, I. Lobersli, and L. Beutin. 2010. Evaluation of multiple-locus variable number of tandem-repeats analysis (MLVA) as a method for identification of clonal groups among enteropathogenic, enterohaemorrhagic and avirulent Escherichia coli O26 strains. FEMS Microbiol. Lett. 303:137-146. [DOI] [PubMed] [Google Scholar]
- 34.Nataro, J. P., and J. B. Kaper. 1998. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 11:142-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nielsen, E. M., and M. T. Andersen. 2003. Detection and characterization of verocytotoxin-producing Escherichia coli by automated 5′ nuclease PCR assay. J. Clin. Microbiol. 41:2884-2893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ørskov, F., and I. Ørskov. 1984. Serotyping of Escherichia coli, p. 43-112. In T. Bergan (ed.), Methods in microbiology, vol. 14. Academic Press, London, United Kingdom. [Google Scholar]
- 37.Oswald, E., et al. 2000. Typing of intimin genes in human and animal enterohemorrhagic and enteropathogenic Escherichia coli: characterization of a new intimin variant. Infect. Immun. 68:64-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Perelle, S., F. Dilasser, J. Grout, and P. Fach. 2004. Detection by 5′-nuclease PCR of Shiga-toxin producing Escherichia coli O26, O55, O91, O103, O111, O113, O145 and O157:H7, associated with the world's most frequent clinical cases. Mol. Cell Probes 18:185-192. [DOI] [PubMed] [Google Scholar]
- 39.Perelle, S., F. Dilasser, J. Grout, and P. Fach. 2005. Detection of Escherichia coli serogroup O103 by real-time polymerase chain reaction. J. Appl. Microbiol. 98:1162-1168. [DOI] [PubMed] [Google Scholar]
- 40.Perelle, S., F. Dilasser, J. Grout, and P. Fach. 2007. Screening food raw materials for the presence of the world's most frequent clinical cases of Shiga toxin-encoding Escherichia coli O26, O103, O111, O145 and O157. Int. J. Food Microbiol. 113:284-288. [DOI] [PubMed] [Google Scholar]
- 41.Possé, B., L. De Zutter, M. Heyndrickx, and L. Herman. 2008. Novel differential and confirmation plating media for Shiga toxin-producing Escherichia coli serotypes O26, O103, O111, O145 and sorbitol-positive and -negative O157. FEMS Microbiol. Lett. 282:124-131. [DOI] [PubMed] [Google Scholar]
- 42.Pradel, N., et al. 2000. Prevalence and characterization of Shiga toxin-producing Escherichia coli isolated from cattle, food, and children during a one-year prospective study in France. J. Clin. Microbiol. 38:1023-1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ribot, E. M., et al. 2006. Standardization of pulsed-field gel electrophoresis protocols for the subtyping of Escherichia coli O157:H7, Salmonella, and Shigella for PulseNet. Foodborne Pathog. Dis. 3:59-67. [DOI] [PubMed] [Google Scholar]
- 44.Sandhu, K. S., et al. 1996. Prevalence of the eaeA gene in verotoxigenic Escherichia coli strains from dairy cattle in Southwest Ontario. Epidemiol. Infect. 116:1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Scheutz, F. 1997. Vero cytotoxin producing Escherichia coli (VTEC) isolated from Danish patients. Ph.D. thesis. Statens Serum Institut, Copenhagen, Denmark.
- 46.Scheutz, F., T. Cheasty, D. Woodward, and H. R. Smith. 2004. Designation of O174 and O175 to temporary O groups OX3 and OX7, and six new E. coli O groups that include Verocytotoxin-producing E. coli (VTEC): O176, O177, O178, O179, O180 and O181. APMIS 112:569-584. [DOI] [PubMed] [Google Scholar]
- 47.Scheutz, F., and N. A. Strockbine. 2005. Escherichia, p. 607-624. In G. M. Garrity, D. J. Brenner, N. R. Krieg, and J. T. Staley (ed.), Bergey's manual of systematic bacteriology. Springer, New York, NY.
- 48.Sekse, C., et al. 2009. An outbreak of Escherichia coli O103:H25—bacteriological investigations and genotyping of isolates from food. Int. J. Food Microbiol. 133:259-264. [DOI] [PubMed] [Google Scholar]
- 49.Stephan, R., et al. 2008. Prevalence and characteristics of Shiga toxin-producing Escherichia coli in Swiss raw milk cheeses collected at producer level. J. Dairy Sci. 91:2561-2565. [DOI] [PubMed] [Google Scholar]
- 50.Vernozy-Rozand, C., M. P. Montet, M. Berardin, C. Bavai, and L. Beutin. 2005. Isolation and characterization of Shiga toxin-producing Escherichia coli strains from raw milk cheeses in France. Lett. Appl. Microbiol. 41:235-241. [DOI] [PubMed] [Google Scholar]
- 51.Zhang, W., et al. 2007. Structural and functional differences between disease-associated genes of enterohaemorrhagic Escherichia coli O111. Int. J. Med. Microbiol. 297:17-26. [DOI] [PubMed] [Google Scholar]
- 52.Zhang, W. L., et al. 2000. Molecular characteristics and epidemiological significance of Shiga toxin-producing Escherichia coli O26 strains. J. Clin. Microbiol. 38:2134-2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zweifel, C., et al. 2010. Characteristics of Shiga toxin-producing Escherichia coli isolated from Swiss raw milk cheese within a 3-year monitoring program. J. Food Prot. 73:88-91. [DOI] [PubMed] [Google Scholar]