Abstract
Cell-to-cell communication, or quorum sensing (QS), enables cell density-dependent regulation of bacterial gene expression which can be exploited for the autonomous-signal-guided expression of recombinant proteins (C. Y. Tsao, S. Hooshangi, H. C. Wu, J. J. Valdes, and W. E. Bentley, Metab. Eng. 12:291-297, 2010). Earlier observations that the metabolic potential of Escherichia coli is conveyed via the QS signaling molecule autoinducer-2 (AI-2) suggested that the capacity for protein synthesis could also be affected by AI-2 signaling (M. P. DeLisa, J. J. Valdes, and W. E. Bentley, J. Bacteriol. 183:2918-2928, 2001). In this work, we found that simply adding conditioned medium containing high levels of AI-2 at the same time as inducing the synthesis of recombinant proteins doubled the yield of active product. We have hypothesized that AI-2 signaling “conditions” cells as a natural consequence of cell-to-cell communication and that this could tweak the signal transduction cascade to alter the protein synthesis landscape. We inserted luxS (AI-2 synthase) into vectors which cosynthesized proteins of interest (organophosphorus hydrolase [OPH], chloramphenicol acetyltransferase [CAT], or UV-variant green fluorescent protein [GFPuv]) and evaluated the protein expression in luxS-deficient hosts. In this way, we altered the level of luxS in the cells in order to “tune” the synthesis of AI-2. We found conditions in which the protein yield was dramatically increased. Further studies demonstrated coincident upregulation of the chaperone GroEL, which may have facilitated higher yields and is shown for the first time to be positively regulated at the posttranscriptional level by AI-2. This report is the first to demonstrate that the protein synthesis capacity of E. coli can be altered by rewiring quorum sensing circuitry.
Quorum sensing (QS) enables population density-based regulation of gene expression, whereby a single cell senses and communicates with a minimal population unit (or quorum) needed for orchestrating population behavior (12, 13, 22, 35). While there is intense interest in understanding the mechanisms of QS signal transduction, there have been few technological or commercial applications that have resulted directly from adapting or rewiring this signaling process. One of the most striking targets is in the field of metabolic engineering, where signaling modules can be constructed to alter phenotype and aid in the synthesis of recombinant gene products (30, 44, 45). For example, Bulter et al. (5) created an artificial genetic switch using acetate for modulating cell-to-cell signaling in Escherichia coli. Neddermann et al. (31) developed a hybrid expression system by incorporating the quorum circuitry of Agrobacterium tumefaciens (e.g., TraR) into a eukaryotic transcriptional controller for HeLa cells. Weber et al. (49) utilized the Streptomyces bacterial QS system for initiating heterologous protein expression in mammalian cell cultures and mice (human primary and mouse embryonic stem cells). Tsao et al. (45) demonstrated autoinduced heterologous protein expression in E. coli by rewiring the native autoinducer-2 (AI-2) signal transduction cascade.
The ability of bacteria, such as E. coli, to produce the AI-2 quorum signal has been attributed to the LuxS protein, a homodimeric zinc metalloenzyme originally identified in Vibrio harveyi (27, 41). AI-2 signal generation results from LuxS-catalyzed cleavage of S-ribosylhomocysteine (SRH), yielding homocysteine and 4,5-dihydroxy-2,3-pentanedione (DPD), which is cyclized into AI-2 (37, 52). The specific genes, proteins, pathways, and functions attributed to AI-2 signaling in E. coli, while described to be widespread (8, 10), are not fully understood and are continually emerging (1, 24, 46). For example, the genes regulated via phosphorylated AI-2 and those regulated by unphosphorylated AI-2 are different (28). Notably, important phenotypes have been attributed to AI-2 signaling (e.g., virulence, biofilm formation, etc.) (11, 17). We have demonstrated that AI-2 also communicates the “metabolic potential” of E. coli cells, particularly when they are expressing recombinant proteins (8, 9). The signal level in the extracellular milieu decreased precipitously upon the overexpression of recombinant proteins, at a rate proportional to their rate of synthesis. This observation was independent of the protein, whether of viral, bacterial, or eukaryotic cell origin (8, 9). We subsequently hypothesized that the protein synthesis landscape (e.g., chaperone, protease, and polymerase activities) could be altered by shifting the window of quorum-dependent gene regulation through the addition of exogenous AI-2 or modulation of AI-2 production via the regulation of luxS.
While metabolic engineering studies often target, via complementation or mutation, the proteins or enzymes directly involved in a particular pathway of interest, such as TraR-mediated expression in eukaryotic hosts (31), an approach described here targets the native signal transduction pathway to alter the global landscape necessary for the desired objective. That is, we describe the intentional manipulation of AI-2 synthase, LuxS, in order to alter QS signaling and improve the synthesis of recombinant proteins. We have confirmed that the approach is general by testing several proteins of interest. Moreover, we attribute this enhancement to increased levels of active GroEL, the chaperone, which, in turn, is shown for the first time to be posttranscriptionally modulated by AI-2. Such QS-mediated posttranscriptional modulation of protein levels in E. coli has never been reported.
MATERIALS AND METHODS
Bacterial strains and plasmid construction.
The strains and plasmids used in this study are listed in Table 1. Chloramphenicol acetyltransferase (CAT) (3) and organophosphorus hydrolase (OPH) (50) were expressed using pTrcHisB (Invitrogen). In luxS coexpression experiments, plasmid pBO was constructed by digestion of the opd gene encoding organophosphorus hydrolase with NcoI and HindIII from pTO (39) and insertion into pBADHisA (Invitrogen). The luxS gene, after amplification by PCR from genomic DNA of strain W3110 using primers LuxSF and LuxSR (Table 2), containing EcoRI restriction sequences, was inserted into pKK223-3 (Amersham Pharmacia), yielding pKKluxS. Plasmid pBOL was constructed by PCR amplification of the tac promoter-luxS fusion from pKKluxS using primers pkk223LuxSF and pkk223LuxSR (Table 2), followed by ligation into NdeI-digested pBO. Plasmid pBOL-LacIq was built by PCR amplification of lacIq encoding and overproducing the Lac repressor from the vector pTrcHisB (Invitrogen), using primers LacIqF and LacIqR (Table 2). The PCR product was blunt cloned into BstZ17I-digested pBOL. Two additional sets of plasmids were derived from pBO, pBOL, and pBOL-LacIq to express two other recombinant proteins, CAT and the UV-variant green fluorescent protein (GFPuv). Plasmids pBC, pBCL, and pBCL-LacIq, carrying the PCR-amplified cat gene from pTrcHisCAT (Invitrogen), used similar methods and primers FCAT and RCAT (Table 2). Likewise, pBG, pBGL, and pBGL-LacIq were constructed to express GFPuv using pTrcHisGFPuv (6) and primers FCAT and RGFPuv (Table 2). All plasmids were transformed into TOP10 competent cells (Invitrogen) for sequencing (DNA sequencing facility, University of Maryland Biotechnology Institute) and later transformed into strain W3110 or MDAI2. The recombinant model proteins were under the control of the arabinose-inducible araBAD promoter, and the luxS gene was controlled by the isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible ptac promoter. In vitro-synthesized AI-2 was made by His6-LuxS and His6-Pfs, which were overproduced by the host, E. coli BL21 (Novagen) cells bearing plasmids pTrcHis-luxS and pTrcHis-pfs individually (17). Vibrio harveyi strains BB170 (luxN::Tn5 sensor 1− sensor 2+) and BB152 (luxL::Tn5 autoinducer-1− autoinducer-2+) (40) were used for AI-2 activity assays (kindly provided by B. Bassler). Transformations, cloning procedures, and DNA isolation were performed using standard protocols (36).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant genotype or description | Source or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| W3110 | Κ-12 strain, wild type, λ− F− IN(rrnD-rrnE)1 rph-1s | CGSCa |
| MDAI2 | W3110 luxS::Tcr W3110-derived luxS mutant strain | 8 |
| BL21 | F−ompT [dcm][lon]hsdS(rB− mB−)gal | Novagen |
| V. harveyi | ||
| BB152 | BB120 luxL::Tn5 (AI-1− AI-2+) Kmr | 40 |
| BB170 | BB120 luxN::Tn5 (sensor 1− sensor 2+) Kmr | 2 |
| Plasmids | ||
| pKK223-3 | Cloning vector, Apr | Pharmacia Biotech |
| pTrcHisA,B,C | Cloning vector, Apr | Invitrogen |
| pBADHisA | Cloning vector, Apr | Invitrogen |
| pTrcHisCAT | pTrcHis derivative, Apr | Invitrogen |
| pKKluxS | pKK223-3 derivative, luxS+ Apr | This study |
| pTO | pTrcHisA derivative, containing opd, Apr | 39 |
| pBO | pBADHisA derivative, containing opd, Apr | This study |
| pBOL | pBO derivative, containing luxS from W3110, Apr | This study |
| pBOL-LacIq | pBO derivative, containing luxS from W3110 and lacIq, Apr | This study |
| pBC | pBO derivative, containing cat, Apr | This study |
| pBCL | pBC derivative, containing luxS from W3110, Apr | This study |
| pBCL-LacIq | pBC derivative, containing luxS from W3110 and lacIq, Apr | This study |
| pTrcHisGFPuv | pTrcHisB derivative, containing GFPuv gene, Apr | 6 |
| pBG | pBO derivative, containing GFPuv gene, Apr | This study |
| pBGL | pBG derivative, containing luxS from W3110, Apr | This study |
| pBGL-LacIq | pBG derivative, containing luxS from W3110 and lacIq, Apr | This study |
| pTrcHis-LuxS | pTrcHisC derivative, containing luxS from W3110, Apr | 17 |
| pTrcHis-Pfs | pTrcHisC derivative, containing pfs from W3110, Apr | 17 |
Coli Genetic Stock Center, Yale University, New Haven, CT.
TABLE 2.
Oligonucleotide primers used in this study
| Name | Sequence | Relevant description |
|---|---|---|
| LuxSF | CCTTGAATTCAGGATGCCGTTGTTAGATAGC | Upstream primer for cloning luxS from W3110 |
| LuxSR | AACTGAATTCCGGCTAGATGTGCAGTT | Downstream primer for cloning luxS from W3110 |
| RTLuxSF | GATGCCGTTGTTAGATAGCTTCAC | Upstream primer for luxS RT-PCR |
| RTLuxSR | CTAGATGTGCAGTTCCTGCAAC | Upstream primer for luxS RT-PCR |
| pkk223LuxSF | ACGCATATGTCCTACTCAGGAGAGCGTTCA | Downstream primer for cloning tac promoter-luxS fusion from pKKluxS |
| pkk223LuxSR | AGCCATATGTCGCTCAAGGCGCACTCCCG | Downstream primer for cloning tac promoter-luxS fusion from pKKluxS |
| LacIqF | GGAGCTGCATGTGTCAGAGGTT | Upstream primer for cloning lacIq from pTrcHisB |
| LacIqR | CAAAAAACATTATCCAGAACGGGAG | Downstream primer for cloning lacIq from pTrcHisB |
| FCAT | TAAAAGACATGTGGGGTTCTCATCATCATC | Upstream primer for cloning cat and the GFPuv gene from pTrcHisCAT and pTrcHisGFPuv, respectively |
| RCAT2 | TTAATGTTTAGCGGCCGCTTAAAAAAATTACGC | Downstream primer for cloning cat from pTrcHisCAT |
| RGFPuv | TTAATGTTTAGCGGCCGCCAGCTTTCATTATTT | Downstream primer for cloning the GFPuv gene from pTrcHisGFPuv |
| RTgroELF | GGCAGCTAAAGACGTAAAATTCGG | Upstream primer for groEL RT-PCR |
| RTgroELR | CATGCATTCGGTGGTGATCATC | Downstream primer for groEL RT-PCR |
| RTdnaKF | GGGTAAAATAATTGGTATCGACCTGGG | Upstream primer for dnaK RT-PCR |
| RTdnaKR | GTCTTTGACTTCTTCAAATTCAGCGTC | Downstream primer for groEL RT-PCR |
| 16S-2F | AGCGCAACCCTTATCCTTTGTTGG | Upstream primer for 16S rRNA RT-PCR internal control |
| 16S-2R | TCGCGAGGTCGCTTCTCTTTGTAT | Downstream primer for 16S rRNA RT-PCR internal control |
Growth media.
Luria-Bertani (LB) medium contained 5 g liter−1 yeast extract (Sigma), 10 g liter−1 Bacto tryptone (Difco), and 10 g liter−1 NaCl. E. coli defined growth medium was prepared according to the protocol of Riesenberg et al. (33) and supplemented with 0.8% glucose (Sigma). Autoinducer bioassay (AB) medium was made according to the protocol of Greenberg et al. (19).
Culture conditions.
Primary E. coli inoculums, consisting of LB medium, glucose (0.8%), ampicillin (100 μg ml−1; Sigma), and E. coli frozen stock, were grown for 4 h at 37°C with 250 rpm shaking, and then 1% (vol/vol) was inoculated into overnight cultures in defined medium (∼16 h at 30°C and 250 rpm) (9). To initiate experimental cell growths, overnight cultures were inoculated into 40 ml of defined medium, and volumes were adjusted to achieve similar initial cell densities (optical density at 600 nm [OD600] = 0.10). For conditioning experiments (Fig. 1), mid-log phase (OD600 ∼ 0.25) cells were spun down gently (2,500 × g for 5 min at 4°C) and resuspended in either fresh defined medium, defined medium plus 10% (vol/vol) conditioned medium (CM), or defined medium plus 50% (vol/vol) CM. For coexpression experiments, arabinose (Sigma) or arabinose and IPTG (Sigma) were added to mid-log phase cultures (OD600 ∼ 0.40).
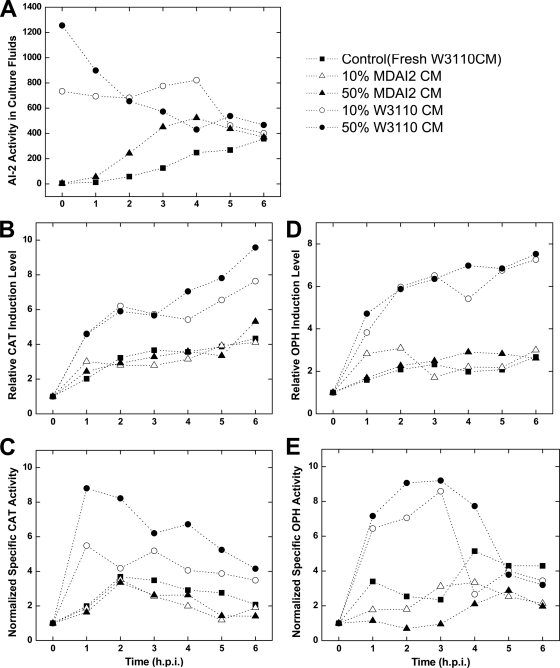
FIG. 1.
Supplementation with AI-2-containing conditioned medium enhances recombinant protein production. The AI-2 level in W3110/pTrcHis-CAT or -OPH cell cultures was modulated by resuspending cells in CM containing AI-2 activity (circles; generated from wild-type strain W3110) or lacking AI-2 activity (triangles; generated from luxS-deficient MDAI2 cells). Recombinant protein expression was induced at t = 0 h (1 mM IPTG). (A) AI-2 activity in W3110/pTrcHis-CAT culture fluids. (B and C) Relative CAT induction level (normalized by total protein concentration) (B) and normalized specific CAT activity (C). These results demonstrate that exogenously added AI-2 enhances CAT production. (D and E) Similar results were found for W3110/pTrcHis-OPH. Induction levels (normalized by total protein concentration) and normalized specific activities are reported as the Western blot band intensity and specific activity, respectively, of each sample relative to the preinduction (t = 0 h) value. The reported blot intensities and activity levels are the average results of duplicate experiments and agree to within 15%.
Preparation of cell-free culture fluids and CM.
Cell-free culture fluids were prepared by centrifugation of 1-ml E. coli whole-broth samples for 10 min (10,000 × g at 4°C). Cleared supernatants were passed through 0.22-μm sterile Millex filters (Millipore) and stored at −20°C. V. harveyi strain BB152 cell-free culture fluids were prepared likewise to obtain positive-control samples as reported previously. CM was prepared by growing E. coli strain W3110 or MDAI2 in LB plus 50 mM glucose or defined medium plus 50 mM glucose to an OD600 of 3.0 (∼6 to 8 h), followed by centrifugation (10 min at 10,000 × g and 4°C) and filtering of cleared supernatants by vacuum-driven filter (Corning). Details of the preparation of cell-free culture fluids for AI-2 activity assays and for conditioning experiments were also described previously (9, 47).
Analytical measurements of AI-2 activity.
The AI-2 activity assay was based on the reports of Surette and Bassler and Surette et al. (40, 41). Luminescence was measured hourly as a function of V. harveyi cell density by quantitating light production with a luminometer (EG&G Berthold). Data, reported as fold activation, were obtained by dividing the light produced by the reporter cells after the addition of E. coli cell-free culture fluids by the light output from the reporter cells when growth medium alone was added.
Western blot and protein activity assays.
Culture volumes equivalent to 2 ml at an OD600 of 1.0 were withdrawn from experiments and centrifuged at 10,000 × g for 10 min. The cell pellets were resuspended and lysed in 300 μl BugBuster protein extraction reagent (Novagen) at room temperature for 30 min and then centrifuged again at 10,000 × g for 10 min to separate soluble and insoluble cell extracts. We found this lysis method to be complete, systematic, and reproducible. The protein concentration of soluble cell extracts was determined by using a protein assay kit (Bio-Rad). Insoluble cell debris was resuspended with 0.1 ml resuspension buffer (0.06 M Tris-HCl [pH 6.8]). The soluble cell extracts or insoluble debris were mixed 1:1 (vol/vol) with sodium dodecyl sulfate (SDS) sample buffer (12.5% 0.5 M Tris-HCl [pH 6.8], 10% glycerol, 2% SDS, 5% β-mercaptoethanol, 0.0025% bromophenol blue), heated at 100°C for 5 min, and centrifuged for 1 min. Samples with identical protein content were loaded onto 12.5% SDS-polyacrylamide gels for electrophoresis and blotted onto nitrocellulose membranes (Bio-Rad) using a Mini Trans-Blot cell (Bio-Rad) and Bjerrum and Schafer-Nielsen transfer buffer (48 mM Tris, 29 mM glycine, 20% methanol) for 30 min at 20 V. Monoclonal antipolyhistidine (Sigma), polyclonal anti-OPH (kindly provided by J. Grimsley), monoclonal anti-GroEL, and monoclonal anti-DnaK (Stressgen) were diluted 1:4,000 in antibody buffer (0.5% Tween 20 [vol/vol], Tris-buffered saline with 1% [wt/vol] nonfat dry milk) to probe recombinant proteins. The membranes were then transferred to a 1:4,000-diluted goat anti-mouse or goat anti-rabbit antibody conjugated with alkaline phosphatase (Sigma). Membranes were developed with 1:50-diluted Nitro Blue tetrazolium-5-bromo-4-chloro-indolyl phosphate (NBT-BCIP) solution (Roche Molecular Chemicals). Lastly, the membranes were scanned and the images were analyzed using ImageJ software (http://rsb.info.nih.gov/ij/). The activities of soluble CAT within crude cell extracts were measured according to the method of Rodriguez and Tait (34), OPH activities were measured according to the method of Wu et al. (50), and the GFP activities of 1-ml whole-cell samples were measured using a Perkin-Elmer LS-3B fluorescence spectrometer at excitation and emission wavelengths of 395 and 509 nm, respectively. Finally, specific CAT and OPH activities were reported as activity divided by total protein concentration (34, 50).
RT-PCR.
To determine the relative transcription levels of the genes of interest (i.e., luxS, groEL, and dnaK), cell pellets were lysed and RNA extracted using an RNAqueous kit (Ambion) according to the manufacturer's instructions. The total RNA concentration was determined by measuring the absorbance of a diluted sample at 260 nm using a UV spectrophotometer (Beckman). To synthesize cDNA, 300 ng total RNA was subjected to reverse transcription (RT) using gene-specific primers. The cDNA was used as the template in PCR with gene-specific primers. The primer sets used for RT-PCR are listed in Table 2. PCR products were run on a 1% agarose gel to compare band intensities using ImageJ software (http://rsb.info.nih.gov/ij/). All data were normalized to the levels of the internal control, endogenous 16S rRNA.
Synthesis and fractionation of in vitro AI-2.
His6-Pfs and His6-LuxS were overexpressed (17, 37) under 1 mM IPTG induction of BL21(pTrcHis-pfs) and BL21(pTrcHis-luxS) as cell densities were grown to OD600 values of ∼0.4 to 0.6 at 37°C. The cells were harvested 4 h postinduction (h.p.i.) by centrifugation at 14,000 × g for 20 min at 4°C. After lysis using BugBuster solution (Novagen) at room temperature for 40 min, the soluble cell extracts were mixed with Co2+ affinity resin (BD Talon; BD Biosciences), and the bound His6-Pfs and His6-LuxS were washed three times using phosphate buffer (pH 7.4) (Sigma) to remove nonspecifically bound proteins. The purified enzymes were eluted (125 mM imidazole in phosphate buffer, pH 7.4) and used to synthesize AI-2 from 1 mM S-adenosylhomocysteine (SAH) in 50 mM Tris-HCl (pH 7.8) at 37°C for 4 h (17). The enzymatic reaction product was extracted twice with chloroform and recovered from the aqueous phase. To remove unreacted SAH substrate and the by-products adenine and homocysteine, the in vitro AI-2 reaction product was fractionated by high-performance liquid chromatography (HPLC) with a preparative silica reverse-phase column (25 by 10 cm), using 90% water-10% acetonitrile eluent at a flow rate of 3 ml/min (Dynamax SD-200 pumps; Varian, Inc., Walnut Creek, CA). Absorbance at 210 nm and 260 nm was recorded using a UV-D II dual-wavelength UV-visible light detector (see Fig. S2 in the supplemental material). After fractionation, acetonitrile was evaporated from each aliquot for 1.5 h by using a CentriVap concentrator (Labconco) and analyzed for AI-2. Fractionated in vitro AI-2 was further confirmed by mass spectrometry using JEOL AccuTOF-CS ESI-TOF mass spectrometers (dual electrospray ionization; mass ranges from 100 to 1,000 m/z were monitored) (see Fig. S3 in the supplemental material) and AI-2 activity bioassay.
RESULTS
“AI-2-conditioned cultures” exhibit increased chloramphenicol acetyltransferase and organophosphorus hydrolase.
In earlier studies, we observed significant drops in AI-2 levels after the induction of recombinant proteins (9). Here, a simple study was performed in which conditioned medium (CM) with or without AI-2 was added to cultures at the same time as the inducer (IPTG). W3110/pTrcHis-CAT and -OPH cells were cultured to mid-log phase and resuspended in various concentrations of conditioned media (10% and 50%) from AI-2-producing (+AI-2) or luxS mutant (−AI-2) cells and then immediately induced with 1 mM IPTG. In CAT-producing cultures, AI-2 was initially highest under the 10% and 50% CM (+AI-2) conditions and progressively dropped to the control levels thereafter (Fig. 1A). Similar results were obtained for E. coli cultures producing OPH (not included here). W3110 produces AI-2 via the normal metabolic pathways, and MDAI2 is an isogenic luxS mutant. Our results suggest behavior due to an imposed large differential in AI-2 activity with, presumably, few other differences in the CM (10).
Remarkably, the expression levels of CAT (25 kDa) and OPH (36 kDa) both increased 2- to 4-fold relative to the levels in control cells identically resuspended in CM from MDAI2 cells (−AI-2) (Fig. 1B and D). In both cases, the specific activities increased concomitantly, with activities in +AI-2 CM cultures reaching 4-fold higher than in controls (Fig. 1C and E). The specific activities reported are the activities of the enzymes normalized by the mass of each protein expressed, obtained via Western blot. The cell growth rates were unaffected by the conditioned media during the times indicated. We also note that the enhancements observed were typically greatest during the periods when the AI-2 levels were most disparate (first 3 h).
Construction of controllable LuxS coexpression system.
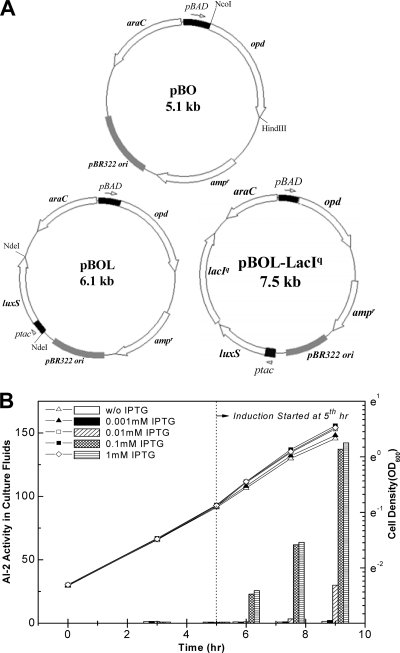
Because CM is poorly defined, we designed a system to link enhanced yield to AI-2. We constructed LuxS coexpression vectors for in vivo generation of AI-2, as well as recombinant proteins, wherein LuxS and the product proteins were independently controlled under different controllable promoters. MDAI2, a luxS null mutant host, was used as the background host to enable a full range of AI-2 “tuning” (from near zero [mutant] to high levels [LuxS overexpression]). In addition, the MDAI2 background eliminates interplay between genomic luxS and genome-synthesized AI-2 and that produced via the plasmids. Organophosphorus hydrolase (OPH) was selected as a model product because its expression in E. coli has proved difficult (50). For coexpression, an IPTG-inducible luxS sequence was inserted into pBO to make pBOL, which produces OPH under the control of the arabinose-inducible araBAD promoter (Fig. 2 A). Further, to minimize background luxS transcription, lacIq was inserted into pBOL, yielding pBOL-LacIq (Fig. 2A). These vectors enable independent control of luxS and opd.
FIG. 2.
LuxS and recombinant protein coexpression vectors. (A) pBO expresses opd under the control of the arabinose-inducible promoter araBAD. An expression cassette of the IPTG-inducible promoter ptac and the luxS gene was inserted into pBO, yielding pBOL. To more effectively regulate luxS expression, LacIq was inserted into pBOL, yielding pBOL-LacIq. (B) Modulation of AI-2 via varied luxS expression was carried out by the addition of different concentrations of IPTG to MDAI2 (pBOL-LacIq) cultures. At different time points during cell growth, aliquots were collected for measurement of cell density (lines) and AI-2 activity (bars). The AI-2 values shown here are representative of three independent experiments. The replicate assays agreed within 15%. w/o, without.
To determine whether luxS expression could modulate the AI-2 levels measured in extracellular medium, MDAI2(pBOL-LacIq) cells were grown to mid-log phase (OD600 ≈ 0.4) in defined minimal medium supplemented with 0.8% glucose which, in turn, ensures high AI-2 activity (40, 47, 51). IPTG was added at various levels (0 to 1 mM) after 5 h of growth; the AI-2 levels in the extracellular medium spanned a 150-fold range after an additional 4 h. The immediate AI-2 activity differences (<1 h) were substantially smaller, but a 25-fold difference was ultimately observed between the 0 and 1 mM IPTG conditions. These results, not surprisingly, confirm that LuxS expression encoded by these luxS coexpression vectors within the luxS mutant can modulate the AI-2 level in the extracellular medium.
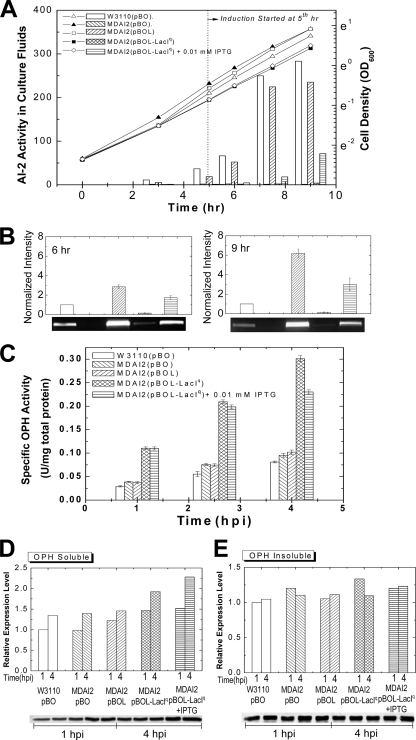
Coexpression of LuxS improves recombinant OPH protein production in strain MDAI2.
Wild-type E. coli W3110 and the luxS isogenic knockout MDAI2 were transformed with plasmids pBO, pBOL, and pBOL-LacIq and grown to mid-log phase (OD600 ≈ 0.4) in defined minimal medium containing 0.8% glucose. Arabinose (0.2%) was added to each culture to induce opd. Additionally, the cultures containing pBOL-LacIq were grown with and without 0.01 mM IPTG, the inducer of luxS. We found that 0.01 mM IPTG was sufficient to generate but not rapidly accumulate AI-2 in the extracellular fluids (Fig. 3 A, similar to Fig. 2). Figure 3B depicts the levels of luxS mRNA, which were highest for the pBOL vector in the luxS mutant, lowest for the pBO vector in the luxS mutant, very low for the uninduced lacIq-repressed vector, and somewhat higher for the same vector minimally induced (0.01 mM IPTG). The results obtained by image analysis suggest a linear correlation between luxS mRNA and AI-2 levels within MDAI2, with the highest level of AI-2 corresponding to the highest level of mRNA (pBOL). In wild-type cells, we found more extracellular AI-2 per luxS mRNA and have no explanation other than perhaps an alternative metabolic effect associated with the luxS mutation (29). The growth rates of MDAI2(pBOL-LacIq) with and without luxS induction were both slightly lower than the growth rates of MDAI2(pBO) and MDAI2(pBOL) (Fig. 3A).
FIG. 3.
OPH accumulation and activity are both enhanced significantly by modulating LuxS expression in a coexpression system. (A) OPH was expressed in E. coli W3110 (wild type) and MDAI2 (luxS deficient) by 0.2% arabinose induction and altered AI-2 signaling. That is, MDAI2(pBOL-LacIq) cultures with and without 0.01 mM IPTG were compared with W3110(pBO), MDAI2(pBO), and MDAI2(pBOL) cultures when identical levels of arabinose (0.2%) were added. Throughout, the cell densities (lines) and AI-2 activities (bars) were observed. (B) Transcriptional analysis of luxS for OPH expression in the coexpression system. The RNA was extracted from 1-h.p.i. and 3-h.p.i. samples, and an agarose gel was run to show luxS mRNA levels from RT-PCR using luxS gene-specific primers. (C) After induction, samples were collected and lysed. The OPH activity in each sample was measured and divided by the total protein concentration to derive the specific OPH activity. The data shown here are representative of two independent experiments. The errors shown are standard errors from triplicate OPH activity and total-protein assays. (D and E) OPH accumulation in the soluble (D) and insoluble (E) fractions of cell extracts was examined 1 h.p.i. and 4 h.p.i. by Western blotting. The results shown here are not pooled but, instead, are representative of triplicate experiments (which agreed to within 20%).
The OPH yield in the MDAI2(pBO) culture was unchanged relative to that of the W3110(pBO) culture. Restoring luxS under ptac promoter control on pBOL resulted in slightly less AI-2 than for W3110 and a relatively unchanged level of OPH. Remarkably, for MDAI2(pBOL-LacIq) under both conditions, a 3- to 4-fold increase in specific OPH activity was observed (Fig. 3C). In the experiment whose results are shown in Fig. 3D, we found an appreciable increase (∼1.5-fold) in OPH in the soluble fraction of cell extracts. The level of OPH found in the insoluble fractions was similar among all cultures (Fig. 3E). The nearly 1.5-fold increase in soluble OPH at 4 h.p.i., however, was insufficient to account for the increased activity per mg protein (4-fold) (Fig. 3C). Thus, the OPH was of higher specific activity (quality) and higher yield (quantity). The results depicted in Fig. 3 demonstrate that within MDAI2 cells, luxS expression led to increased AI-2 accumulation and altered OPH yield and activity. Presumably there was a relationship between luxS expression and the protein synthesis machinery. In order to test whether the enhanced yield was OPH specific, we repeated these experiments with additional recombinant proteins (9).
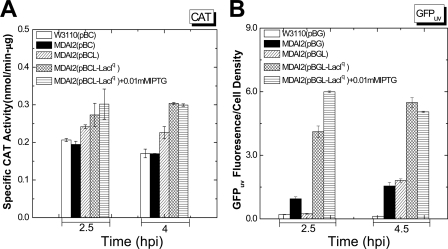
CAT and GFPuv coexpressed with LuxS.
We replaced the opd gene in the plasmids noted above (pBO, pBOL, and pBOL-LacIq) with cat or the GFPuv gene, respectively, for the overexpression of CAT and GFPuv. Again, W3110 and MDAI2 were transformed with the expression plasmids, and LuxS coexpression experiments were executed under the same conditions as described above. In all cases, coexpression of LuxS increased the specific activities of the recombinant model proteins, CAT (∼1.5-fold) and GFPuv (∼4- to 6-fold) (Fig. 4). The protein expression levels were also investigated via Western blot analysis. Both CAT and GFPuv were found to increase in both soluble and insoluble fractions (not shown). These data support the conclusion that enhanced yield via LuxS coexpression is protein independent.
FIG. 4.
Specific activities of CAT (A) and GFPuv (B) are enhanced in the LuxS coexpression system. CAT (A) and GFPuv (B) were expressed in E. coli W3110 and MDAI2 by 0.2% arabinose induction and at different AI-2 levels (by varied IPTG concentrations). CAT activities were divided by the total protein level of each cell extract to generate the specific CAT activities. However, in order to derive specific GFPuv activities, the fluorescence results for GFPuv were divided by the cell density (OD600) directly instead of by the total protein concentration of each sample. Both the CAT and the GFPuv coexpression experiments were duplicated to confirm reproducibility; the data shown here are representative, and the standard errors from triplicate assays are shown.
The chaperone protein GroEL is affected by luxS coexpression.
Protein chaperones, including GroEL, play key roles in the assembly and folding of heterologous proteins expressed in E. coli (18, 42). Coexpression of GroEL is often used to improve folding and enhance yield (16). It is also recognized that the abundance of heat shock proteins (HSPs, including chaperones and proteases) is influenced by heterologous protein overexpression and, in turn, can affect the protein yield (4, 14, 21, 23, 32, 42). We have previously demonstrated that avoiding (32) or intentionally downregulating (39) the heat shock response coincident with protein overexpression, as well as stimulation of HSPs prior to induction (14), can facilitate increased yield and activity of CAT (14) and OPH (39). To ascertain whether luxS coexpression leads to increased yield through the pleiotropic regulation of HSPs, we measured the levels of two important heat shock proteins, GroEL and DnaK, as well as the transcription of these and several other proteins in the presence and absence of varied LuxS expression (48).
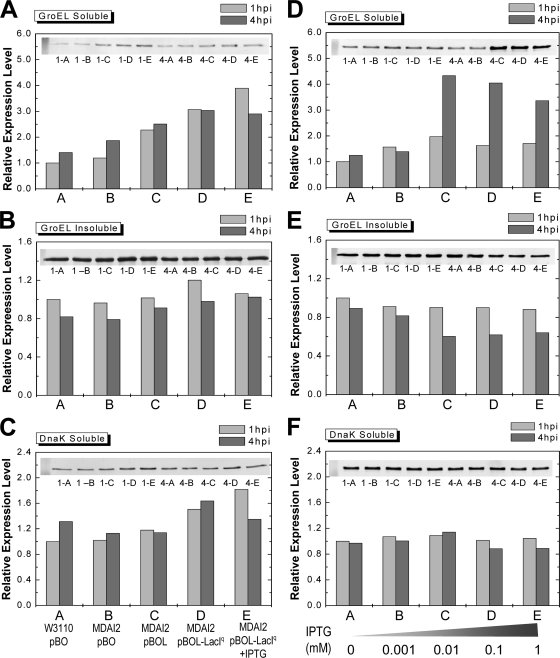
The amounts of GroEL and DnaK in MDAI2 cultures induced with arabinose to synthesize OPH were examined by Western blotting at both 1 and 4 h.p.i. (Fig. 5). In all cases where luxS was introduced in trans, the GroEL levels in the soluble fractions were higher (up to ∼3- to 4-fold) than in controls [W3110(pBO) and MDAI2(pBO)] (Fig. 5A). The GroEL levels in the insoluble fractions of all cultures were similar in all cases (Fig. 5B). The DnaK levels in the soluble fractions were typically unchanged, although there was a 60% increase in the cases where LuxS was regulated by LacIq (Fig. 5C). There was no detectable DnaK in any of the insoluble fractions (not shown). Importantly, in the cases where soluble GroEL increased the most (MDAI2 with lacIq, with or without IPTG), we found the highest and most active levels of OPH (Fig. 3C and D).
FIG. 5.
The expression levels of chaperone protein GroEL in soluble cell extracts were significantly higher than in controls in the luxS-modulated system. (A, B, and C) The amounts of GroEL and DnaK in cultures induced with arabinose to synthesize OPH were examined at both 1 and 4 h.p.i. by Western blotting. MDAI2(pBOL-LacIq) cultures with and without 0.01 mM IPTG were compared with W3110(pBO), MDAI2(pBO), and MDAI2(pBOL) cultures when identical levels of arabinose (0.2%) were added. The lanes are labeled with 1 or 4 for the time (h) postinduction and A, B, C, D, or E for the strains and plasmids, indicated below the panels. (D, E, and F) MDAI2(pBOL-LacIq) cultures were supplemented with different concentrations of IPTG to vary LuxS expression in the absence of recombinant protein synthesis. GroEL and DnaK were examined by Western blotting. In this case, A, B, C, D, and E correspond to different concentrations of IPTG, shown below the panels.
While the overexpression of nonnative proteins has previously been shown to increase the levels of GroEL and DnaK in E. coli (23, 39), we attempted to explore whether LuxS and/or AI-2 had an independent effect on these important chaperones, irrespective of the recombinant product. Hence, MDAI2(pBOL-LacIq) cultures were supplemented with different levels of IPTG to vary LuxS expression, and the two chaperones were examined by Western blotting (Fig. 5D to F). Results for AI-2 in these experiments are also shown in Fig. 2 and show altered levels of luxS induction with no background opd expression (as confirmed by activity measurements; data not shown). Interestingly, GroEL was notably upregulated in the soluble fractions in cultures with IPTG at or above 0.01 mM (Fig. 5D) and was moderately downregulated in the insoluble fractions of the same cultures (Fig. 5E). There were no significant differences in the levels of in DnaK found in the soluble fractions (Fig. 5F), and there was no observable DnaK in the insoluble fractions (not shown). These results demonstrate that LuxS expression in a luxS-deficient host can modulate the levels of GroEL in both the soluble and insoluble fraction and suggest that an appropriate LuxS expression level could be found that is coincident with an appropriate GroEL level that facilitates the folding of target proteins in E. coli.
Does AI-2 communicate with GroEL?
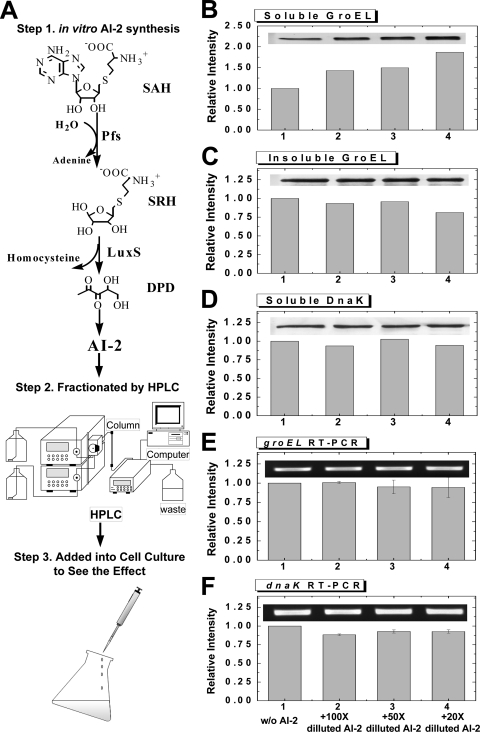
In these experiments, LuxS expression was altered and the yields of several recombinant proteins were increased. Moreover, we observed that the chaperone GroEL was upregulated, both in response to the addition of arabinose and IPTG for the expression of recombinant protein products and LuxS and in response to IPTG for the expression of LuxS alone. To investigate whether the expression of LuxS led to increased GroEL (as a stress response) or whether AI-2 signaling played a role, we added in vitro-synthesized AI-2 (Fig. 6 A) (17, 38) or mock-synthesized-AI-2 synthesis buffer (negative control) to MDAI2 cells. The AI-2 levels in treated MDAI2 cultures decreased steadily, and growth rates were unaffected (data not shown). The two chaperones, GroEL and DnaK, were observed by Western blot analysis (Fig. 6B to D shows the results at 1 h.p.i.). GroEL increased ∼1.5- to 2-fold in the soluble fractions for the first hour when AI-2 was added (20× to 100× dilutions). Moreover, GroEL appeared to increase with AI-2 in a concentration-dependent manner. A slight but statistically insignificant decrease in the level of GroEL in the insoluble fraction was also observed (Fig. 6C). There was no observable trend in soluble DnaK (Fig. 6D), and no insoluble DnaK was detected under any of the conditions (data not shown). Additionally, the results at 2 h.p.i. showed no conclusive changes in GroEL or DnaK levels in response to AI-2. In the experiments whose results are shown in Fig. 6E and F, the corresponding levels of mRNA were measured, and no changes due to the addition of AI-2 were found. We have previously performed genome-spanning DNA microarray analyses on W3110 and MDAI2 (luxS-deficient) cells grown with and without glucose, as well as LsrK mutants exhibiting no phosphorylated AI-2, and found no significant differences in the transcription of GroEL or DnaK. Conversely, Kendall and coworkers (24) found increased transcription of GroEL in a luxS mutant relative to that in its isogenic parent but no further increase in groEL mRNA upon the addition of DPD.
FIG. 6.
In vitro-synthesized AI-2 increases soluble GroEL level. (A) The scheme depicts the synthesis and fractionation of AI-2 and its addition to cell cultures of MDAI2 (no plasmid). First, AI-2 was synthesized in vitro from substrate SAH. Second, any unreacted SAH and by-products homocysteine and adenine were removed by HPLC. After mobile-phase solvent removal via vacuum pump, the fractionated AI-2 was added to MDAI2 cell cultures. (B to D) Samples were taken at 1 h.p.i. Chaperones GroEL (soluble [B] and insoluble [C] fractions) and DnaK (soluble fraction [D]) were analyzed by Western blotting. The results shown here are representative of duplicate experiments and triplicate assays. (E and F) Transcriptional analysis of groEL (E) and dnaK (F) indicating no change due to the addition of AI-2. Error bars show standard error. Western blotting and RT-PCR were performed within the linear ranges of the assays.
In summary, our results shown in Fig. 6 demonstrate an increased level of soluble GroEL in a luxS mutant supplemented with in vitro-synthesized AI-2. Our results shown in Fig. 6E and F, confirming the results of Kendall et al. (24), demonstrate a negligible increase in groEL transcription in luxS mutants supplemented with AI-2. Because the QS signal molecule AI-2 affects the level of GroEL in the soluble fraction of E. coli, we suggest that LuxS expression in luxS mutants can alter the levels of chaperone GroEL in soluble fractions through AI-2-mediated signaling.
DISCUSSION
Studies of AI-2-mediated QS suggest that quorum signaling may communicate the prevailing metabolic condition (8, 9) and that a tweaked signaling process may potentially enable improved recombinant protein production. By the results shown in Fig. 1, we demonstrate for the first time that the addition of exogenous AI-2-containing CM enhances CAT and OPH production both in quantity (protein yield) and quality (protein activities). Recognizing the possibility that many metabolites may have altered concentrations in CM from luxS-deficient versus luxS-positive strains (7, 25), we developed a controlled study to investigate luxS/AI-2 QS during recombinant protein overexpression. Furthermore, because commercial bioprocesses are unlikely to allow the addition of uncharacterized CM to bioreactors, we developed the “tunable” dual-controlled expression vector in which AI-2 synthesis and product synthesis are uncoupled and independently exogenously regulated.
Both the expression level and activity of the recombinant product were increased when luxS-deficient (MDAI2) cells were complemented with luxS under lacIq control. The luxS expression levels in these cultures were in the middle of our tested range [from none in the MDAI2(pBO) cells to maximum levels in the MDAI2(pBOL) cells]. In Fig. S1 in the supplemental material, we demonstrate that increased yield was not due to LacIq; rather, our results suggest that an intermediate level of luxS expression (obtained by luxS expression under LacIq control) was optimal. It is interesting to note that this “optimal” level actually led to intermediate levels of AI-2, as well (compare Fig. 2 to Fig. 3). Hence, the main contributor to the benefits in yield and activity was the manipulation of luxS. We also demonstrated that for all cases of dramatically improved recombinant protein production, the GroEL level was increased in the soluble fractions (Fig. 3 and 5, respectively).
Upregulation of HSPs, including GroEL and DnaK, is commonly observed to accompany recombinant protein overexpression, due to an upregulated heat shock response (4, 8, 21, 23, 32, 42). That is, increases in both groEL and dnaK transcription (8) and GroEL and DnaK protein levels (4, 21, 23, 32) are typically observed. The influence of luxS coexpression in the LacIq experiments (Fig. 5A and C, bars D and E) points to the coordinate change in GroEL and DnaK when there is an abundance of recombinant protein overexpression. This is likely coincident with an upregulated stress response. For this reason, we undertook the systematic study of GroEL and DnaK in the absence of protein overexpression (Fig. 5D to F). In these experiments, there was a decoupling of GroEL from DnaK, suggesting that the differences observed were due to luxS abundance and, perhaps, AI-2 signaling (the luxS-related influence is likely obscured in the overexpression experiments). This conclusion is strengthened by the apparent exchange between soluble and insoluble GroEL in Fig. 5D and E. Indeed, throughout the study, DnaK levels exhibited no systematic trend.
Finally, dnaK and groEL transcription in W3110 and MDAI2 are unaltered by luxS mutation (48) and AI-2 signaling, as demonstrated by our analysis of LsrRK mutations (28) and microarray data for the addition of exogenous AI-2 (24). Their transcription rates are seemingly uncorrelated with QS. Hence, the apparent decoupling of (i) the GroEL level in the soluble fraction from its transcription level and (ii) the levels of GroEL protein and DnaK suggested that the enhanced levels of GroEL were due to other mechanisms than the classic heat shock-like response (15, 18, 23, 38). Since groEL transcription is apparently unaffected by AI-2, we suggest that the apparent linkage between the GroEL level and luxS coexpression is at the posttranscriptional level. We are aware of only one report in which an AI-2-mediated process was found to affect the level of a protein in a manner other than transcriptional regulation. In that report, the AI-2 phosphorelay system of V. harveyi is shown to affect endogenous lux enzyme activity by modulating translation through the recruitment of small RNAs (sRNA) and the RNA chaperone Hfq (26). Interestingly, Guisbert et al. (20) demonstrated Hfq functions in E. coli and the Hfq-mediated decoupling of GroEL and DnaK translation. In the same report, they showed reduced GroEL translation in an Hfq mutant, suggesting that the GroEL-mediated negative-feedback control of σ32 was preserved, as well as long-term adaptation. They also reported that DnaK translation was suppressed by Hfq. Our results, with unchanged DnaK and upregulated GroEL, are seemingly contradictory, assuming that Hfq acts in concert with QS-regulated sRNA in a manner exactly analogous to that in V. harveyi. We have previously shown that AI-2 signaling influences sRNA (28), but there remains no evidence that any QS-regulated sRNA interacts with Hfq. That these components all seem to be functioning in E. coli does suggest that further work is warranted.
While there have been no reports of posttranscriptional regulation in E. coli that are attributed to QS, we found that the level of GroEL in the soluble fraction increased significantly within the first hour in experiments where purified AI-2 was added to cultures of MDAI2. Also, we found in several cases that the increase was accompanied by a decrease in the level in the insoluble fraction, again suggesting that there was no apparent linkage between AI-2 and groEL transcription. That DnaK has no insoluble reservoir in our experiments reinforces the notion of differential AI-2-mediated regulation. Irrespective of the exact cause (via sRNA, Hfq, or other factors), we believe this is the first demonstration that AI-2 alters the level of GroEL in the soluble fraction in E. coli. Moreover, it is well known that GroEL assists in the production of properly folded recombinant proteins (16, 18, 42, 43). The results of this study suggest that altered AI-2 signaling (by luxS coexpression) can be used to improve recombinant protein yield in E. coli. While there are many functions that are altered by luxS coexpression, we observed AI-2-mediated posttranscriptional modulation of GroEL and hypothesize that this was a contributing factor and could have been a very significant factor in the increased yield and activities observed.
Supplementary Material
Acknowledgments
Partial support of this work was provided by the National Science Foundation (grant no. BES-0222687 and BES-0124401) and the U.S. Army.
Footnotes
Published ahead of print on 28 January 2011.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Ahmer, B. M. M. 2004. Cell-to-cell signalling in Escherichia coli and Salmonella enterica. Mol. Microbiol. 52:933-945. [DOI] [PubMed] [Google Scholar]
- 2.Bassler, B. L., M. Wright, R. E. Showalter, and M. R. Silverman. 1993. Intercellular signalling in Vibrio harveyi: sequence and function of genes regulating expression of luminescence. Mol. Microbiol. 9:773-786. [DOI] [PubMed] [Google Scholar]
- 3.Bentley, W. E., R. H. Davis, and D. S. Kompala. 1991. Dynamics of induced CAT expression in E. coli. Biotechnol. Bioeng. 38:749-760. [DOI] [PubMed] [Google Scholar]
- 4.Bentley, W. E., N. Mirjalili, D. C. Andersen, R. H. Davis, and D. S. Kompala. 1990. Plasmid-encoded protein: the principal factor in the metabolic burden associated with recombinant bacteria. Biotechnol. Bioeng. 35:668-681. [DOI] [PubMed] [Google Scholar]
- 5.Bulter, T., et al. 2004. Design of artificial cell-cell communication using gene and metabolic networks. Proc. Natl. Acad. Sci. U. S. A. 101:2299-2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cha, H. J., C. F. Wu, J. J. Valdes, G. Rao, and W. E. Bentley. 2000. Observations of green fluorescent protein as a fusion partner in genetically engineered Escherichia coli: monitoring protein expression and solubility. Biotechnol. Bioeng. 67:565-574. [PubMed] [Google Scholar]
- 7.DeLisa, M. P., and W. E. Bentley. 2002. Bacterial autoinduction: looking outside the cell for new metabolic engineering targets. Microb. Cell Fact. 1:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DeLisa, M. P., J. J. Valdes, and W. E. Bentley. 2001. Mapping stress-induced changes in autoinducer AI-2 production in chemostat-cultivated Escherichia coli K-12. J. Bacteriol. 183:2918-2928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DeLisa, M. P., J. J. Valdes, and W. E. Bentley. 2001. Quorum signaling via AI-2 communicates the “metabolic burden” associated with heterologous protein production in Escherichia coli. Biotechnol. Bioeng. 75:439-450. [DOI] [PubMed] [Google Scholar]
- 10.DeLisa, M. P., C. F. Wu, L. Wang, J. J. Valdes, and W. E. Bentley. 2001. DNA microarray-based identification of genes controlled by autoinducer 2-stimulated quorum sensing in Escherichia coli. J. Bacteriol. 183:5239-5247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Domka, J., J. Lee, and T. K. Wood. 2006. YliH (BssR) and YceP (BssS) regulate Escherichia coli K-12 biofilm formation by influencing cell signaling. Appl. Environ. Microbiol. 72:2449-2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fuqua, C., and E. P. Greenberg. 1998. Cell-to-cell communication in Escherichia coli and Salmonella typhimurium: they may be talking, but who's listening? Proc. Natl. Acad. Sci. U. S. A. 95:6571-6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fuqua, C., and E. P. Greenberg. 1998. Self perception in bacteria: quorum sensing with acylated homoserine lactones. Curr. Opin. Microbiol. 1:183-189. [DOI] [PubMed] [Google Scholar]
- 14.Gill, R. T., M. P. DeLisa, J. J. Valdes, and W. E. Bentley. 2001. Genomic analysis of high-cell-density recombinant Escherichia coli fermentation and “cell conditioning” for improved recombinant protein yield. Biotechnol. Bioeng. 72:85-95. [DOI] [PubMed] [Google Scholar]
- 15.Gill, R. T., J. J. Valdes, W. E. Bentley, and W. E. Bentley. 2000. Analysis of differential stress gene transcription in response to recombinant protein over-expression and high cell density fermentation in Escherichia coli, abstr. BIOT159. In Abstr. 219th ACS National Meeting, San Francisco, CA, 26 to 30 March 2000. American Chemical Society, Washington, DC.
- 16.Goloubinoff, P., A. A. Gatenby, and G. H. Lorimer. 1989. Groe heat-shock proteins promote assembly of foreign prokaryotic ribulose bisphosphate carboxylase oligomers in Escherichia coli. Nature 337:44-47. [DOI] [PubMed] [Google Scholar]
- 17.González Barrios, A. F., et al. 2006. Autoinducer 2 controls biofilm formation in Escherichia coli through a novel motility quorum-sensing regulator (MqsR, B3022). J. Bacteriol. 188:305-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gragerov, A., et al. 1992. Cooperation of GroEL/GroES and DnaK/DnaJ heat-shock proteins in preventing protein misfolding in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 89:10341-10344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Greenberg, E. P., J. W. Hastings, and S. Ulitzur. 1979. Induction of luciferase synthesis in Beneckea-Harveyi by other marine bacteria. Arch. Microbiol. 120:87-91. [Google Scholar]
- 20.Guisbert, E., V. A. Rhodius, N. Ahuja, E. Witkin, and C. A. Gross. 2007. Hfq modulates the σE-mediated envelope stress response and the σ32-mediated cytoplasmic stress response in Escherichia coli. J. Bacteriol. 189:1963-1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harcum, S. W., and W. E. Bentley. 1993. Detection, quantification, and characterization of proteases in recombinant Escherichia coli. Biotechnol. Tech. 7:441-447. [Google Scholar]
- 22.Hastings, J. W., and E. P. Greenberg. 1999. Quorum sensing: the explanation of a curious phenomenon reveals a common characteristic of bacteria. J. Bacteriol. 181:2667-2668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kanemori, M., H. Mori, and T. Yura. 1994. Induction of heat shock proteins by abnormal proteins results from stabilization and not increased synthesis of σ32 in Escherichia coli. J. Bacteriol. 176:5648-5653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kendall, M. M., D. A. Rasko, and V. Sperandio. 2007. Global effects of the cell-to-cell signaling molecules autoinducer-2, autoinducer-3, and epinephrine in a luxS mutant of enterohemorrhagic Escherichia coli. Infect. Immun. 75:4875-4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee, C. W., and M. L. Shuler. 2000. The effect of inoculum density and conditioned medium on the production of ajmalicine and catharanthine from immobilized Catharanthus roseus cells. Biotechnol. Bioeng. 67:61-71. [DOI] [PubMed] [Google Scholar]
- 26.Lenz, D. H., et al. 2004. The small RNA chaperone Hfq and multiple small RNAs control quorum sensing in Vibrio harveyi and Vibrio cholerae. Cell 118:69-82. [DOI] [PubMed] [Google Scholar]
- 27.Lewis, H. A., et al. 2001. A structural genomics approach to the study of quorum sensing: crystal structures of three LuxS orthologs. Structure 9:527-537. [DOI] [PubMed] [Google Scholar]
- 28.Li, J., et al. 2007. Quorum sensing in Escherichia coli is signaled by AI-2/LsrR: effects on small RNA and biofilm architecture. J. Bacteriol. 189:6011-6020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li, J., et al. 2006. A stochastic model of Escherichia coli AI-2 quorum signal circuit reveals alternative synthesis pathways. Mol. Syst. Biol. 2:67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.March, J. C., and W. E. Bentley. 2004. Quorum sensing and bacterial cross-talk in biotechnology. Curr. Opin. Biotechnol. 15:495-502. [DOI] [PubMed] [Google Scholar]
- 31.Neddermann, P., et al. 2003. A novel, inducible, eukaryotic gene expression system based on the quorum-sensing transcription factor TraR. EMBO Rep. 4:159-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ramirez, D. M., and W. E. Bentley. 1995. Fed-batch feeding and induction policies that improve foreign protein synthesis and stability by avoiding stress responses. Biotechnol. Bioeng. 47:596-608. [DOI] [PubMed] [Google Scholar]
- 33.Riesenberg, D., et al. 1991. High cell-density cultivation of Escherichia coli at controlled specific growth rate. J. Biotechnol. 20:17-28. [DOI] [PubMed] [Google Scholar]
- 34.Rodriguez, R. L., and R. C. Tait. 1983. Recombinant DNA techniques: an introduction. Benjamin/Cummings Publishing Company, Inc., Menlo Park, CA.
- 35.Salmond, G. P., B. W. Bycroft, G. S. Stewart, and P. Williams. 1995. The bacterial “enigma”: cracking the code of cell-cell communication. Mol. Microbiol. 16:615-624. [DOI] [PubMed] [Google Scholar]
- 36.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 37.Schauder, S., K. Shokat, M. G. Surette, and B. L. Bassler. 2001. The LuxS family of bacterial autoinducers: biosynthesis of a novel quorum-sensing signal molecule. Mol. Microbiol. 41:463-476. [DOI] [PubMed] [Google Scholar]
- 38.Schweder, T., et al. 2002. Role of the general stress response during strong overexpression of a heterologous gene in Escherichia coli. Appl. Microbiol. Biotechnol. 58:330-337. [DOI] [PubMed] [Google Scholar]
- 39.Srivastava, R., H. J. Cha, M. S. Peterson, and W. E. Bentley. 2000. Antisense downregulation of σ32 as a transient metabolic controller in Escherichia coli: effects on yield of active organophosphorus hydrolase. Appl. Environ. Microbiol. 66:4366-4371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Surette, M. G., and B. L. Bassler. 1998. Quorum sensing in Escherichia coli and Salmonella typhimurium. Proc. Natl. Acad. Sci. U. S. A. 95:7046-7050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Surette, M. G., M. B. Miller, and B. L. Bassler. 1999. Quorum sensing in Escherichia coli, Salmonella typhimurium, and Vibrio harveyi: a new family of genes responsible for autoinducer production. Proc. Natl. Acad. Sci. U. S. A. 96:1639-1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thomas, J. G., and F. Baneyx. 1996. Protein folding in the cytoplasm of Escherichia coli: requirements for the DnaK-DnaJ-GrpE and GroEL-GroES molecular chaperone machines. Mol. Microbiol. 21:1185-1196. [DOI] [PubMed] [Google Scholar]
- 43.Thomas, J. G., and F. Baneyx. 1996. Protein misfolding and inclusion body formation in recombinant Escherichia coli cells overexpressing heat-shock proteins. J. Biol. Chem. 271:11141-11147. [DOI] [PubMed] [Google Scholar]
- 44.Tjalsma, H., et al. 2004. Engineering of quorum-sensing systems for improved production of alkaline protease by Bacillus subtilis. J. Appl. Microbiol. 96:569-578. [DOI] [PubMed] [Google Scholar]
- 45.Tsao, C. Y., S. Hooshangi, H. C. Wu, J. J. Valdes, and W. E. Bentley. 2010. Autonomous induction of recombinant proteins by minimally rewiring native quorum sensing regulon of E. coli. Metab. Eng. 12:291-297. [DOI] [PubMed] [Google Scholar]
- 46.Vendeville, A., K. Winzer, K. Heurlier, C. M. Tang, and K. R. Hardie. 2005. Making “sense” of metabolism: autoinducer-2, LuxS and pathogenic bacteria. Nat. Rev. Microbiol. 3:383-396. [DOI] [PubMed] [Google Scholar]
- 47.Wang, L., Y. Hashimoto, C. Y. Tsao, J. J. Valdes, and W. E. Bentley. 2005. Cyclic AMP (cAMP) and cAMP receptor protein influence both synthesis and uptake of extracellular autoinducer 2 in Escherichia coli. J. Bacteriol. 187:2066-2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang, L., J. Li, J. C. March, J. J. Valdes, and W. E. Bentley. 2005. luxS-dependent gene regulation in Escherichia coli K-12 revealed by genomic expression profiling. J. Bacteriol. 187:8350-8360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Weber, W., et al. 2003. Streptomyces-derived quorum-sensing systems engineered for adjustable transgene expression in mammalian cells and mice. Nucleic Acids Res. 31:e71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wu, C. F., H. J. Cha, G. Rao, J. J. Valdes, and W. E. Bentley. 2000. A green fluorescent protein fusion strategy for monitoring the expression, cellular location, and separation of biologically active organophosphorus hydrolase. Appl. Microbiol. Biotechnol. 54:78-83. [DOI] [PubMed] [Google Scholar]
- 51.Xavier, K. B., and B. L. Bassler. 2005. Regulation of uptake and processing of the quorum-sensing autoinducer AI-2 in Escherichia coli. J. Bacteriol. 187:238-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhu, J., R. Patel, and D. Pei. 2004. Catalytic mechanism of S-ribosylhomocysteinase (LuxS): stereochemical course and kinetic isotope effect of proton transfer reactions. Biochemistry 43:10166-10172. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.