Abstract
A 15-mer fragment that is derived from the helical region in the C-terminal half of pediocin PA-1 inhibited the activity of pediocin PA-1. Of 13 other pediocin-like (hybrid) bacteriocins, only the hybrid bacteriocin Sak/Ped was markedly inhibited by the 15-mer fragment. Sak/Ped was the only one of these bacteriocins that had a sequence (in the C-terminal helix-containing half) identical to that of the 15-mer fragment, indicating that the fragment inhibits pediocin-like bacteriocins in a sequence-dependent manner. By replacing (one at a time) all 15 residues in the fragment with Ala or Leu, five residues (K1, A2, T4, N8, and A15) were identified as being especially important for the inhibitory action of the fragment. The results suggest that the corresponding residues (K20, A21, T23, N27, and A34, respectively) in pediocin PA-1 might be involved in interactions between pediocin PA-1 and its receptor. To characterize the environment surrounding these five residues when pediocin PA-1 interacts with target cells, these residues were replaced (one at a time) with a hydrophobic large (Leu) residue, a hydrophilic charged (Asp or Arg) residue, and a small (Ala or Gly) residue. The results revealed that residues A21 and A34 are in a spatially constrained environment, since the replacement with a small (Gly) residue was the only substitution that did not markedly reduce the bacteriocin activity. The positive charge in K20 and the polar amide group in N27 appeared to interact with electronegative groups, since the replacement of these two residues with a positive (Arg) residue was well tolerated, while replacement with a negative (Asp) residue was detrimental to the bacteriocin activity. K20 was in a less constrained environment than N27, since the replacement of K20 with a large hydrophobic (Leu) residue was tolerated fairly well and to a greater extent than N27. T23 seemed to be in an environment that was not restricted with respect to size, polarity, and charge, since replacements with large (Leu) and small (Ala) hydrophobic residues and a hydrophilic negative (Asp) residue were tolerated fairly well (2- to 6-fold reduction in activity). Moreover, the replacement of T23 with a large positive (Arg) residue resulted in wild-type or better-than-wild-type activity.
Membrane-permeabilizing, cationic, ribosomally synthesized antimicrobial peptides (AMPs) are produced by a wide variety of organisms, from bacteria to humans. When produced by bacteria, such peptides are generally termed bacteriocins, and they are synthesized together with a cognate immunity protein that renders the bacteriocin-producing bacteria immune to their own bacteriocins (6, 8, 13, 26-28). Bacteriocins produced by “food-grade” lactic acid bacteria (LAB) have especially been the focus of extensive studies because of their potential application as nontoxic food preservatives and therapeutic agents for gastrointestinal infections. The LAB bacteriocins nisin and pediocin PA-1 are in fact used as biopreservatives (4, 6), and the potential of LAB bacteriocins in medical applications is exemplified by previously reported results showing that the oral intake of bacteriocin-producing LAB protects mice from lethal doses of Listeria monocytogenes (5).
There are two main classes of LAB AMPs: the lanthionine-containing (class I) bacteriocins, and the non-lanthionine-containing (class II) bacteriocins (6, 8, 13, 26-28). Class II bacteriocins are heterogeneous and may be divided further into four groups: (i) the pediocin-like (class IIa) bacteriocins, (ii) the two-peptide (class IIb) bacteriocins, (iii) the cyclic (class IIc) bacteriocins, and (iv) the nonpediocin one-peptide linear (class IId) bacteriocins (6). The pediocin-like (class IIa) bacteriocins, of which more than 20 have been identified, are perhaps the class II bacteriocins that are best characterized (8, 13, 27). The interest in these bacteriocins is due to their antilisterial activity combined with the fact that they are produced by food-grade bacteria.
All pediocin-like bacteriocins have similar sequences, especially in their N-terminal half, which contains a disulfide bridge and a common YGNGV/L “pediocin box” motif (8, 13, 27). Their well-conserved cationic and rather hydrophilic N-terminal half (about 18 residues) forms an S-shaped β-sheet-like structure, which is followed by a hinge and a somewhat more hydrophobic and diverse helix-containing C-terminal half (14, 16, 33, 35). The hinge enables the N-terminal β-sheet-like region and the C-terminal helix-containing region to move relative to each other and may thus allow the more hydrophobic C-terminal half to extend into the hydrophobic part of target cell membranes, with the cationic and rather hydrophilic N-terminal half remaining in the hydrophilic exterior (8, 13, 27).
The membrane-embedded part (the C and/or D subunit) of the mannose phosphotransferase permease has been shown to act as a receptor or target molecule for the pediocin-like bacteriocins (7). Furthermore, the immunity proteins that protect cells from being killed by pediocin-like bacteriocins interact indirectly with their cognate bacteriocins through the same mannose permease (7). The helix-containing C-terminal half, being the membrane-penetrating part of pediocin-like bacteriocins (10, 27), is thought to interact with the membrane-embedded part of the permease. Consistent with the notion that the C-terminal half is involved in receptor recognition are results showing that the C-terminal half is involved in determining the target cell specificity of pediocin-like bacteriocins and is the half that is recognized by the cognate immunity protein (9, 20). The immunity proteins apparently recognize the C-terminal half of their cognate bacteriocins indirectly, by interacting with the receptor-bacteriocin complex (7).
A 15-mer fragment whose sequence is identical to a sequence in the C-terminal helical region of pediocin PA-1 inhibits the activity of pediocin PA-1 (11). It has been speculated that the fragment binds to the receptor and thus functions as a competitive inhibitor of pediocin-like bacteriocins that have sequence similarity to pediocin PA-1 in the helical region. In this study we have determined the ability of the 15-mer fragment and mutated variants of the fragment to inhibit wild-type, hybrid, and mutated pediocin-like bacteriocins. Residues that seem to be particularly important for the inhibitory action of the fragment, and, thus, possibly for the interaction between pediocin PA-1 and its receptor, have thereby been identified, and the effect on the bacteriocin activity of altering these residues in pediocin PA-1 has been determined.
MATERIALS AND METHODS
Bacterial strains, media, plasmids, and production of peptides.
Escherichia coli DH5α was used for the production and isolation of the plasmids before transferring plasmids to Lactobacillus sakei Lb790 cultures. The E. coli strain was grown at 37°C in LB medium, either with vigorous agitation in liquid medium or on agar plates solidified by the addition of 2% (wt/vol) agar. All lactic acid bacteria were grown without agitation in liquid medium or on agar plates solidified by the addition of 1.5% (wt/vol) agar. Carnobacterium piscicola UI 49 was grown at 30°C in M17 medium (Oxoid) supplemented with Tween 80 (Sigma-Aldrich) and glucose (Sigma-Aldrich) to final concentrations of 0.1% and 0.4%, respectively. Lactic acid bacteria producing wild-type and hybrid bacteriocins were grown at 30°C in MRS medium (Oxoid), while lactic acid bacteria producing pediocin PA-1 mutants were grown at 20°C in either MRS medium or (for the A21L, A21G, T23A, N27L, N27R, A34L, and A34R mutations) in MRS medium supplemented with 4 g/liter yeast extract (Merck).
Three wild-type producer strains were used in this study: Lactobacillus curvatus LTH1174 producing curvacin A (31), Leuconostoc mesenteroides 6 producing leucocin A (34), and Pediococcus acidilactici LMG 2351 producing pediocin PA-1 (24). Wild-type sakacin P, enterocin A, and mutant pediocin PA-1 variants were produced by the two-plasmid heterologous expression systems L. sakei Lb790(pSAK20/pSSP2), L. sakei Lb790(pSAK20/pEI), and L. sakei Lb790(pSAK20/pPED2), respectively, as described previously (2). Wild-type strains were not used for the production of sakacin P and enterocin A because purification was problematic due to protease degradation (in the case of sakacin P) and the production of more than one bacteriocin by the wild-type strain (in the case of enterocin A). Selective antibiotic concentrations of 2 μg/ml erythromycin and 5 μg/ml chloramphenicol were used for these two-plasmid systems. As described previously (20), all hybrid bacteriocins except Ped[1-21]/Sak[21-43] were produced by L. sakei Lb790 transformed with a plasmid containing a gene encoding the hybrid bacteriocin and genes coding for proteins needed for the secretion of and immunity to the hybrid bacteriocin. A sakacin P mutant with an inserted C-terminal disulfide bridge (Sak P Mut) was produced by L. sakei Lb790 transformed with a plasmid containing the mutant bacteriocin gene as described previously (33). A selective antibiotic concentration of 10 μg/ml erythromycin was used when producing the hybrid bacteriocins, whereas concentrations of 2 μg/ml erythromycin and 5 μg/ml chloramphenicol were used when producing the mutant bacteriocins. These systems permitted the routine production of correctly processed and secreted wild-type, hybrid, and mutant bacteriocins, which could be purified by using well-established methods for the purification of these peptides from LAB (see below).
The hybrid bacteriocin Ped[1-21]/Sak[21-43] and the 15-mer fragments derived from the wild-type bacteriocin sequences were synthesized by standard methods for solid-phase multiple-peptide synthesis with a 9-fluorenylmethoxy carbonyl (F-moc) strategy as described previously (11). Pediocin PA-1 15-mer fragments containing point mutations were synthesized by GenScript using their FlexPeptide technology.
Plasmid isolation, preparation of competent cells, and cell transformation.
Plasmids were isolated from E. coli DH5α and LAB using the Nucleo Spin plasmid kit (Macherey-Nagel). To ensure the lysis of LAB, lysozyme was added to resuspension buffer A1, included in the Nucleo Spin plasmid kit, to a final concentration of 5 mg/ml. For the preparation of competent E. coli DH5α cells, the cells were cultured in LB medium to an optical density at 600 nm (OD600) of about 0.3. The culture was cooled on ice for 10 min, and the cells were then washed with ice-cold 0.1 M CaCl2 and suspended in ice-cold 0.1 M CaCl2 containing 15% (vol/vol) glycerol. The cells were ready for transformation after incubation on ice for 45 min and transformed by use of heat shock (42°C for 90 s). L. sakei Lb790/pSAK20 cells were made competent as described previously (1). Briefly, the cells were cultured to an OD600 of between 0.5 and 0.6 in MRS broth containing 10 μg/ml chloramphenicol and 2% (wt/vol) glycine and thereafter washed with 1 mM MgCl2 and then with 30% (wt/vol) polyethylene glycol 1500 (molecular weight range, 1,300 to 1,600). The cells were then transformed by electroporation using a Gene Pulser and a Pulse Controller unit (Bio-Rad Laboratories) as described previously (1). L. sakei Lb790 cells were made competent and transformed by using the same method, but no selective antibiotics were used for initial growth.
Purification of wild-type, hybrid, and mutant bacteriocins.
Wild-type, hybrid, and mutant bacteriocins were purified to homogeneity from 500- or 1,000-ml cultures by applying the bacteria culture directly onto a cation exchanger followed by reverse-phase chromatography, as described previously (32). Briefly, the cells in 500-ml cultures grown overnight were applied onto a 5- to 6-ml SP Sepharose Fast Flow (GE Healthcare) cation-exchange column that had been equilibrated with 50 ml 20 mM sodium phosphate buffer (pH 6). The column was then washed with 100 ml of sodium phosphate buffer, and the peptides were subsequently eluted with 40 ml of sodium phosphate buffer supplemented with NaCl to a final concentration of 1 M. Trifluoroacetic acid (TFA) and 2-propanol were added to the eluent to final concentrations of 0.1% and 5% (vol/vol), respectively, and the eluent was then applied onto a reverse-phase column (Resource RPC; GE Healthcare). The peptides were eluted from the reverse-phase column (equilibrated with distilled water [dH2O] containing 0.1% [vol/vol] TFA) with a linear 2-propanol (containing 0.1% [vol/vol] TFA) gradient. To confirm that the recombinant lactobacilli had correctly produced and processed the bacteriocins, molecular masses of the isolated peptides were determined by mass spectrometry using an Ultraflex matrix-assisted laser desorption ionization-tandem time of flight (MALDI-TOF/TOF) (Bruker Daltonics, Bremen, Germany) instrument, in the positive reflectron mode, with α-cyano-4-hydroxy-cinnamic acid as a matrix. In addition, the pediocin PA-1 mutants were checked for correct disulfide bridge formation by using a bacteriocin assay (see below for a general description of the assay) with and without 2 mM 1,4-dithiothreitol (DTT) added to the indicator strain, as described previously (12). Bacteriocins with correct disulfide bridges had high levels of antimicrobial activity and showed a marked decrease in activity upon treatment with DTT. Bacteriocins with incorrect disulfide bridges had low levels of antimicrobial activity and showed a marked increase in activity upon the same DTT treatment.
The purity of bacteriocins was verified to be greater than 80% by analytical reverse-phase chromatography using a μRPC SC 2.1/10 C2/C18 column (GE Healthcare) on the Smart chromatography system (GE Healthcare), except for the pediocin PA-1 K20L and K20A mutants, which were judged to have a purity of 50% or more. The concentration of purified bacteriocins was determined by measuring the UV absorption at 280 nm, which was converted to a protein concentration by using molecular extinction coefficients, calculated from the contributions of individual amino acid residues.
Bacteriocin assay.
Bacteriocin activity was measured by using a microtiter plate assay system essentially as described previously (25). Each well of a microtiter plate contained 200 μl of culture medium with bacteriocin fractions at 2-fold dilutions and the indicator strain at an OD600 of about 0.01. For inhibition assays with mixtures containing different peptide fragments, the fragments were added together with the indicator strain to a final concentration of 10 μM. The following strains were used as indicator strains: C. piscicola UI 49, Lactobacillus coryniformis subsp. torquens NCDO 2740, Enterococcus faecalis NCDO 581, and L. sakei NCDO 2714. The microtiter plate cultures were incubated overnight (10 to 15 h) at 30°C for all strains except the C. piscicola strain, which was incubated for approximately 8 h, after which the growth of the indicator strains was measured spectrophotometrically at 600 nm with a microtiter plate reader. The MIC was defined as the concentration of bacteriocin that inhibited the growth of the indicator strain by 50%. The MIC values presented are the results of at least 3 independent measurements.
RESULTS
The Ped-15-mer fragment inhibits bacteriocins in which the helix-containing C-terminal half stems from pediocin PA-1.
Consistent with previous results (11), the Ped-15-mer fragment, whose sequence is identical to a sequence in the helical region in the C-terminal half of pediocin PA-1 (Fig. 1 A), inhibited the bacteriocin activity of pediocin PA-1 (Table 1). Although the fragment also inhibited other tested pediocin-like bacteriocins, especially enterocin A and curvacin A, the inhibition was clearly less efficient (Table 1). To determine whether the inhibition depended on the sequence in the helix-containing C-terminal half of these bacteriocins, we tested how well the fragment inhibited hybrid bacteriocins containing N- and C-terminal domains from different pediocin-like bacteriocins. Eight different hybrid bacteriocins (Sak[1-19]/Ped[16-44], Ped[1-21]/Sak[21-43], Cur[1-18]/Sak[18-43], Sak[1-19]/Cur[20-41], Ent[1-23]/Sak[14-43], Sak[1-18]/Ent[19-47], Leu[1-16]/Sak[16-43], and Sak[1-19]/Leu[18-37]) derived from five different wild-type bacteriocins (pediocin PA-1, sakacin P, enterocin A, curvacin A, and leucocin A) were tested (20). The Sak[1-19]/Ped[16-44] hybrid consists of the N-terminal half of sakacin P (residues 1 to 19) and the C-terminal half of pediocin PA-1 (residues 16 to 44). Residues 16 to 19 in sakacin P and pediocin PA-1 are identical (Fig. 1B). The terminology for the other hybrid peptides is analogous (Fig. 1B). Interestingly, only the hybrid bacteriocin (Sak[1-19]/Ped[16-44]) in which the helix-containing C-terminal half stemmed from pediocin PA-1 was consistently inhibited by the Ped-15-mer fragment with the same efficiency or greater efficiency than that of pediocin PA-1 (Table 1).
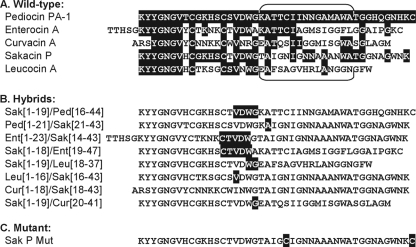
FIG. 1.
(A) Amino acid sequences of the wild-type pediocin-like bacteriocins used in this study. Regions where the sequences are identical to the sequence in pediocin PA-1 are in white with a black background. The brackets above and below the sequences indicate the regions from which the 15-mer fragments were derived. The Ped-15-mer, Sak-15-mer, and Leu-15-mer fragments span residues 20 to 34 of pediocin PA-1, sakacin P, and leucocin A, respectively, whereas the Ent-15-mer fragment spans residues 25 to 34 of enterocin A and the Cur-15-mer fragment spans residues 21 to 35 of curvacin A. (B) Amino acid sequences of the hybrid bacteriocins used in this study. The Sak[1-19]/Ped[16-44] hybrid consists of the N-terminal half of sakacin P (residues 1 to 19) and the C-terminal half of pediocin PA-1 (residues 16 to 44). Residues 16 to 19 of sakacin P and pediocin PA-1 are identical and are in white with a black background. The terminology and use of white with a black background are analogous for the other hybrid bacteriocins. Note that there is a conservative mutation, V17I, in the Cur[1-18]/Sak[18-43] hybrid. (C) Amino acid sequences of the sakacin P variant. This variant has a disulfide bridge in its C-terminal half, since two cysteine residues have been introduced into sakacin P, at positions 24 and 44, both indicated in white with a black background. (The sequences are based on data from references 18, 23, and 24 for pediocin PA-1; reference 3 for enterocin A; references 19 and 31 for curvacin A; reference 30 for sakacin P; and reference 15 for leucocin A.)
TABLE 1.
Inhibition of bactericidal activities of pediocin-like bacteriocins and hybrid variants of these bacteriocins due to the presence of the Ped-15-mer fragment
| Bacteriocin variantb | Mean fold inhibition ± SDa |
|||
|---|---|---|---|---|
| E. faecalis | L. coryniformis | L. sakei | C. piscicola | |
| Pediocin PA-1 | 16 ± 8 | 25 ± 10 | 11 ± 5 | 45 ± 15 |
| Sakacin P | 1.5 ± 0.5 | 2 ± 1 | 1.5 ± 0.5 | 1.0 ± 0.5 |
| Curvacin A | 3 ± 1 | 5 ± 3 | 3 ± 1 | 9 ± 5 |
| Enterocin A | ND | 6 ± 2 | 3 ± 1 | 2 ± 1 |
| Leucocin A | 2 ± 1 | 3 ± 1 | 1.5 ± 0.5 | 2 ± 1 |
| Sakacin P mutantc | 2 ± 1 | 2 ± 1 | 2 ± 1 | 2 ± 1 |
| Sak/Ped | 20 ± 6 | 40 ± 15 | 8 ± 2 | 70 ± 35 |
| Ped/Sak | ND | ND | ND | 1.0 ± 0.5 |
| Cur/Sak | 4 ± 2 | ND | 3 ± 1 | 5 ± 1 |
| Sak/Cur | 2 ± 1 | ND | 1.5 ± 0.5 | ND |
| Ent/Sak | 2 ± 1 | 2 ± 1 | 2 ± 1 | 1.0 ± 0.5 |
| Sak/Ent | 7 ± 3 | 7 ± 5 | 10 ± 7 | 1.0 ± 0.5 |
| Leu/Sak | 1.5 ± 0.5 | ND | 2 ± 1 | 2 ± 1 |
| Sak/Leu | 2 ± 1 | 3 ± 1 | 5 ± 3 | 3 ± 2 |
Fold inhibition is the MIC of the bacteriocin (or hybrid bacteriocin) measured in the presence of a 10 μM concentration of the Ped-15-mer fragment divided by the MIC of the bacteriocin (or hybrid bacteriocin) measured in the absence of the fragment. The inhibition values presented are the results of at least 3 independent measurements. The MICs of the wild-type bacteriocins in the absence of fragment were all between 0.07 and 3 nM (depending on the indicator cell), and those of the hybrid bacteriocins were all between 0.02 and 14 nM. The indicator strains were E. faecalis NCDO 581, L. coryniformis subsp. torquens NCDO 2740, L. sakei NCDO 2714, and C. piscicola UI 49. ND indicates not determined, as reliable values for fold inhibition were not obtained since the MICs obtained in the absence of the fragment were too high (i.e., the potency was too low).
See Fig. 1 for sequences of the hybrid bacteriocins. The terminology for the hybrid bacteriocins in Table 1 is the same as that in Fig. 1, except that the numbering indicating the positions where the N- and C-terminal regions were combined has not been included.
The sakacin P mutant has an inserted C-terminal disulfide bridge (see Fig. 1C for a more detailed specification of this mutant bacteriocin).
The Ent-15-mer, Cur-15-mer, Sak-15-mer, and Leu-15-mer fragments (see Fig. 1A for a more detailed specification of these fragments) were also synthesized and tested for their abilities to inhibit bacteriocin activity. These five fragments correspond to the Ped-15-mer fragment but were derived from enterocin A, curvacin A, sakacin P, and leucocin A, respectively (Fig. 1A). Somewhat surprisingly, none of these fragments inhibited any of the tested bacteriocins effectively, with the highest inhibition being about a 2-fold inhibition of enterocin A by the Ent-15-mer fragment (results not shown).
Residues in the Ped-15-mer fragment which are important for its inhibitory activity.
The Ped-15-mer fragment was mutated and then assayed for its ability to inhibit pediocin PA-1 in order to identify residues that are especially important for the fragment's inhibitory activity. All 15 residues in the Ped-15-mer fragment were (one at a time) replaced with an alanine or leucine residue. The results revealed that there were five residues in the Ped-15-mer fragment where replacements with an alanine or a leucine residue markedly reduced the inhibitory activity of the fragment (Table 2). These residues were K1, A2, T4, N8, and A15, which correspond to K20, A21, T23, N27, and A34 in pediocin PA-1, respectively.
TABLE 2.
Inhibition of pediocin PA-1 by Ped-15-mer fragments in which residues have been replaced with either an alanine or a leucine residue
| Replaced residuea | Mean fold inhibition ± SDb |
|
|---|---|---|
| Alanine replacement | Leucine replacement | |
| K1 | 4 ± 1 | 3 ± 1 |
| A2 | WT | 2 ± 1 |
| T3 | 70 ± 30 | 80 ± 30 |
| T4 | ≥100 | 2 ± 1 |
| C5 | ≥100 | ≥100 |
| I6 | 55 ± 25 | ≥100 |
| I7 | 70 ± 20 | ≥100 |
| N8 | 13 ± 4 | 7 ± 3 |
| N9 | 60 ± 30 | 20 ± 10 |
| G10 | 80 ± 30 | 40 ± 20 |
| A11 | WT | 60 ± 20 |
| M12 | 40 ± 10 | 65 ± 15 |
| A13 | WT | 70 ± 30 |
| W14 | 90 ± 30 | 80 ± 30 |
| A15 | WT | 14 ± 6 |
The residues in the Ped-15-mer fragment which were replaced with alanine and leucine residues are designated by their one-letter abbreviations and a number indicating their position in the fragment, starting from the N terminus. The first residue (K1) in the fragment corresponds to residue 20 (K20) in pediocin PA-1, and so on, with the last residue (A15) in the fragment corresponding to residue 34 (A34) in pediocin PA-1.
Fold inhibition is the MIC of pediocin PA-1 measured in the presence of a 10 μM concentration of the mutant Ped-15-mer fragment divided by the MIC of pediocin PA-1 measured in the absence of the fragment. The inhibition values presented are the results of at least 3 independent measurements using C. piscicola UI 49 as the indicator strain. The MIC of pediocin PA-1 in the absence of the fragment was 0.3 ± 0.1 nM. WT indicates the wild-type Ped-15-mer fragment (i.e., alanine replaced with alanine), and it resulted in a 60- ± 20-fold inhibition.
Effect of altering K20, A21, T23, N27, and A34 in pediocin PA-1 on bacteriocin activity.
The results obtained with the Ped-15-mer fragment and variants of this fragment (Tables 1 and 2) suggest that the Ped-15-mer fragment acts as a competitive inhibitor by binding to the bacteriocin receptor. The results suggest, moreover, that the K20, A21, T23, N27, and/or A34 residue in pediocin PA-1 may be involved in interactions between pediocin PA-1 and its receptor. It was consequently of interest to characterize the environment surrounding each of these five residues by determining the effect that replacements of these residues had on the bacteriocin activity. All five residues were individually replaced with a hydrophobic large residue (leucine) and a hydrophilic negatively (aspartate) or positively (arginine) charged residue (Table 3). In addition, K20, T23, and N27 were replaced with a hydrophobic small residue (alanine), whereas A21 and A34 were replaced with a glycine residue. A total of 19 different pediocin PA-1 variants were thus produced and purified and were analyzed by mass spectrometry to confirm that the correct residue replacements had been made. The activities of all purified mutants were then determined by using four different indicator strains, and the activity relative to that of wild-type pediocin PA-1 was calculated (Table 3). Four different strains were used in order to detect general trends in mutational effects, as such effects may in some cases be indicator strain dependent (12, 17, 21). With just a few exceptions, the effect of the various residue replacements on bacteriocin activity did not depend on the indicator strain (Table 3). Exceptions were the A21L, N27L, and A34R mutations. The A21L and N27L mutations were somewhat less detrimental when assayed using the L. coryniformis and L. sakei strains than when assayed using the E. faecalis and C. piscicola strains (Table 3). The A34R mutation was less detrimental when assayed against the L. sakei strain than when assayed against the three other strains (Table 3).
TABLE 3.
Relative MICs of the pediocin PA-1 variants
| Position | Indicator strain | Mean corresponding relative MIC ± SD for mutation ofa: |
||||
|---|---|---|---|---|---|---|
| Leu | Asp | Arg | Ala | Gly | ||
| K20 | E. faecalis | 4.2 ± 0.9 | 33 ± 10 | 0.8 ± 0.5 | 1.6 ± 0.7 | ND |
| L. coryniformis | 5.7 ± 1.6 | 24 ± 6 | 0.7 ± 0.3 | 2.0 ± 0.4 | ||
| L. sakei | 3.7 ± 0.9 | 21 ± 2 | 0.9 ± 0.4 | 1.1 ± 0.4 | ||
| C. piscicola | 2.3 ± 0.6 | 17 ± 10 | 0.8 ± 0.4 | 1.1 ± 0.5 | ||
| A21 | E. faecalis | 21 ± 8 | ≥500 | ≥150 | WT | 5.1 ± 1.2 |
| L. coryniformis | 3.5 ± 1.1 | ≥250 | ≥120 | 2.6 ± 1.0 | ||
| L. sakei | 3.5 ± 0.2 | ≥500 | ≥100 | 6.5 ± 0.4 | ||
| C. piscicola | 17 ± 3 | ≥150 | 80 ± 15 | 3.3 ± 1.4 | ||
| T23 | E. faecalis | 5.9 ± 1.3 | 2.1 ± 0.9 | 1.0 ± 0.6 | 1.9 ± 0.7 | ND |
| L. coryniformis | 3.4 ± 0.7 | 4.5 ± 2.6 | 0.5 ± 0.4 | 1.8 ± 0.9 | ||
| L. sakei | 2.7 ± 0.1 | 4.5 ± 0.4 | 1.8 ± 0.7 | 3.7 ± 1.5 | ||
| C. piscicola | 4.2 ± 1.0 | 2.5 ± 0.6 | 0.8 ± 0.6 | 1.8 ± 1.1 | ||
| N27 | E. faecalis | 15 ± 6 | 20 ± 8 | 1.7 ± 0.6 | 1.4 ± 0.6 | ND |
| L. coryniformis | 5.8 ± 1.4 | 25 ± 11 | 0.6 ± 0.2 | 0.8 ± 0.4 | ||
| L. sakei | 5.4 ± 1.6 | 12 ± 1 | 2.9 ± 0.8 | 2.2 ± 0.7 | ||
| C. piscicola | 15 ± 2 | 12 ± 7 | 3.0 ± 1.7 | 1.5 ± 1.2 | ||
| A34 | E. faecalis | ND | 30 ± 8 | ≥130 | WT | 1.5 ± 0.6 |
| L. coryniformis | 10 ± 2 | ≥1,000 | 1.0 ± 0.3 | |||
| L. sakei | 26 ± 8 | 30 ± 15 | 1.5 ± 0.7 | |||
| C. piscicola | 15 ± 2 | ≥100 | 1.3 ± 0.3 | |||
The relative MICs are the MICs of the mutant peptides divided by the MICs of wild-type pediocin PA-1. Each relative MIC is the result of at least 3 independent measurements. The MICs of wild-type pediocin PA-1 were between 0.1 and 0.5 nM (depending on the indicator cell). The L. coryniformis and L. sakei strains were 2 to 5 times more sensitive to pediocin PA-1 than the E. faecalis and C. piscicola strains. ND indicates not determined. Replacement with a Gly residue was done only when replacement with an Ala residue was not relevant due to the presence of an Ala residue in pediocin PA-1. The A34L mutation was not analyzed because we were unable to produce this variant, despite several attempts. WT indicates wild-type bacteriocin, and the relative MIC is thus 1.
The replacement of the positively charged (Lys) residue in position 20 with another positively charged (Arg) residue resulted in an activity as good as or better than that of wild-type pediocin PA-1 (Table 3). In contrast, the introduction of a negatively charged (Asp) residue was detrimental, causing a 20- to 30-fold reduction in activity (Table 3). It thus seems that K20 interacts with an electronegative group. The environment surrounding K20 seems not to be highly hydrophilic or sterically restricted, since the replacement of K20 with a large (Leu) or small (Ala) hydrophobic residue was surprisingly well tolerated, causing only a 2- to 4-fold reduction in activity (Table 3). The neighboring residue, A21, seemed to be in a much more restricted environment, as replacement with a hydrophilic positively (Arg) or negatively (Asp) charged residue was highly detrimental (≥100-fold reduction in activity) (Table 3). Moreover, replacement with a large hydrophobic (Leu) residue reduced the activity 3- to 20-fold (depending on the target strain), and replacement with the small Gly residue reduced it 3- to 7-fold (Table 3). T23 seemed in turn to be in a less restrictive environment, since the replacement of this hydrophilic residue with a large (Leu) or small (Ala) hydrophobic residue or a negatively charged residue (Asp) reduced the activity only 2- to 6-fold. Moreover, replacement with a large positively charged residue (Arg) resulted in an activity as good as or better than that of wild-type pediocin PA-1 (Table 3).
The hydrophilic amide group in Asn at position 27 seemed to be involved in an interaction with an electronegative group, since replacement with a large positively charged residue (Arg) did not markedly reduce the activity (0.5- to 3-fold reduction) (Table 3), whereas replacement with a negatively charged Asp residue was detrimental (10- to 30-fold reduction in activity) (Table 3). Replacement with a small hydrophobic residue (Ala) was well tolerated (about the same effect as that of the replacement with an Arg residue), while replacement with a large hydrophobic residue (Leu) was more detrimental (5- to 20-fold reduction in activity) (Table 3). The Ala residue at position 34 seemed to be in a relatively hydrophobic environment, as replacement with a hydrophilic positively (Arg) or negatively (Asp) charged residue was highly detrimental, resulting in a 10-fold to more than a 100-fold reduction in activity (Table 3). Replacement with the small Gly residue did not significantly reduce the activity (Table 3).
DISCUSSION
The Ped-15-mer fragment, whose sequence is identical to a sequence in the helix in the C-terminal half of pediocin PA-1, clearly inhibited pediocin PA-1. The fragment also inhibited the Sak/Ped hybrid, whose N-terminal half is from sakacin P and whose C-terminal helix-containing half is from pediocin PA-1. The 12 other tested hybrids and wild-type bacteriocins, none of which had a C-terminal half identical to that of pediocin PA-1, were not inhibited to the same extent. A previously reported study showed that, of all possible 15-mer fragments that can be derived from pediocin PA-1, only the Ped-15-mer fragment and the 4 adjacent fragments (i.e., fragments covering residues 18 to 32, 19 to 33, 21 to 35, and 22 to 36) inhibit pediocin PA-1 (11). The four adjacent fragments, however, inhibit pediocin PA-1 less efficiently than did the Ped-15-mer fragment (11). Also, the pediocin-like bacteriocins enterocin CRL35 and leucocin A were shown to be inhibited by fragments that are derived from the (putative) helical region in these bacteriocins (29, 36). In contrast, fragments derived from the nonhelical N-terminal part of enterocin CRL35 and the pediocin-like bacteriocin carnobacteriocin B2 did not inhibit these bacteriocins (29, 36). Taken together, the results indicate that peptide fragments that inhibit pediocin-like bacteriocins do so in a specific and sequence-dependent manner in the sense that their sequence must be derived from a sequence in the helical region of the C-terminal half of these bacteriocins.
The helix-containing C-terminal half is an important target cell specificity determinant in these bacteriocins, since hybrid bacteriocins constructed by joining the N- and C-terminal halves from different pediocin-like bacteriocins have target cell specificities similar to those of the bacteriocin from which the C-terminal half is derived (9, 20). The membrane-penetrating C-terminal half is thus thought to interact with the membrane-embedded part (the C and/or D subunit) of the mannose phosphotransferase permease that acts as the receptor for the pediocin-like bacteriocins (7). The specific and sequence-dependent manner by which the Ped-15-mer fragment inhibits bacteriocin activity suggests that the fragment acts as a competitive inhibitor by binding to the bacteriocin receptor and that this binding may involve the five residues (K1, A2, T4, N8, and A15) identified as being the most essential for the fragment's inhibitory activity. The corresponding residues in pediocin PA-1 (K20, A21, T23, N27, and A34, respectively) are thus expected to be involved in the binding of pediocin PA-1 to its receptor. Interestingly, four of these residues (K20, T23, N27, and A34) are positioned on the same side of the membrane-penetrating α-helix and might thus create a receptor-interacting stretch along one side of the helix.
Four of the five residues (K20, A21, N27, and A34) seemed to be in a restricted environment, as expected if they are involved in specific interactions with a receptor. A21 and A34 especially seemed to be in a highly restricted environment due to spatial constraints and hydrophobic surroundings, as the replacement with a small neutral (Gly) residue was the only mutation that was fairly well tolerated. Both the positive charge in K20 and the polar amide group in N27 appeared to interact with electronegative groups, since the replacement of these two residues with a positive Arg residue was well tolerated, while replacement with a negative Asp residue was detrimental. For the amide group in N27, it thus seems that it is the —NH2 with a positive dipole rather than the electronegative =O that is involved in the electrostatic interaction. K20 seemed to be in a somewhat less restricted environment than N27, since K20 tolerated replacement with a large hydrophobic (Leu) residue fairly well and to a greater extent than did N27. In contrast to K20, A21, N27, and A34, the Thr residue at position 23 seemed to be in a remarkably unrestricted environment, with the reduction in activity being only about 2- to 6-fold irrespective of whether T23 was replaced by a large (Leu) or a small (Ala) hydrophobic residue or a hydrophilic negatively charged residue (Asp). Moreover, wild-type or better-than-wild-type activity was obtained upon replacement with a large positively charged residue (Arg). Consequently, T23 does not seem to be involved in receptor interactions that are of great importance for bacteriocin activity. The fact that the introduction of an Arg residue at either position 20, 23, or 27 resulted in a somewhat better-than-wild-type activity against some of the indicator strains suggests that even more potent bacteriocin variants might be constructed by introducing Arg residues at all 3 positions.
The sensitivities of different indicator strains to pediocin PA-1 vary considerably, presumably in part due to the sequence diversity in the mannose phosphotransferase permease. The extent to which a residue in pediocin PA-1 is involved in the binding of the bacteriocin to the permease might consequently be indicator strain dependent. The effect of mutations involving A21, N27, and A34 indeed seemed to show some indicator strain dependency. The A21L and N27L mutations were somewhat less detrimental when assayed against the L. coryniformis and L. sakei strains than when assayed against the E. faecalis and C. piscicola strains, whereas the A34R mutation was less detrimental when assayed against the L. sakei strain than when assayed against the three other strains.
It should be noted that residues not identified as being essential for the fragment's inhibitory activity might also interact with the receptor, since some of the residue replacements were conservative and may consequently not have a marked detrimental effect on the fragment's inhibitory effect on pediocin PA-1. This might, for instance, be the case for replacements involving I6. The conservative I6L replacement increased the fragment's inhibitory effect on pediocin PA-1, whereas the less conservative I6A replacement caused a slight reduction in the fragment's inhibitory effect (Table 2). Also, the N9L replacement reduced the fragment's inhibitory effect (Table 2). These two residues (i.e., I6 and N9) correspond to residues I25 and N28 in pediocin PA-1, respectively, which are on the same side of the membrane-penetrating helix as residue A21, which was clearly essential for the fragment's inhibitory activity (Table 2). These three residues could, in a manner similar to that of K20, T23, N27, and A34, create a receptor-interacting stretch along one side of the helix.
Somewhat surprisingly, the Ent-15-mer, Cur-15-mer, Sak-15-mer, and Leu-15-mer fragments derived from enterocin A, curvacin A. sakacin P, and leucocin A, respectively (Fig. 1A), did not inhibit any of the bacteriocins efficiently. These fragments may perhaps not associate with and/or penetrate into the target cell membrane as efficiently as the Ped-15-mer fragment and may therefore be unable to efficiently interfere with bacteriocin-receptor interactions. The Ped-15-mer fragment is cationic (net charge of 1) and contains a tryptophan residue. The fragment might thus associate more readily with membranes than the anionic Cur-15-mer fragment (net charge of −1), the neutral Sak-15-mer fragment (no net charge), and the Ent-15-mer and Leu-15-mer fragments, which both lack a membrane-interacting tryptophan residue. An alternative explanation for their inefficient inhibition of bacteriocin activity is that these fragments might not cover the relevant helical region to a sufficient extent. The structure of curvacin A indeed reveals that its 13-mer C-terminal helix stretches from Gly29 to the C terminus (16), and thus, only the last 7 residues in the Cur-15-mer fragment span this helical region. A screening of all possible 15-mer fragments that can be derived from enterocin A, curvacin A, sakacin P, and leucocin A may be required in order to identify 15-mer fragments that efficiently inhibit these bacteriocins.
Interactions between the helix-containing C-terminal half of pediocin-like bacteriocins and the membrane-embedded subunits of the mannose permease do not exclude additional bacteriocin-permease interactions. The fact that the fragments alone are not toxic suggests that additional interactions may indeed be required to obtain a bactericidal effect. More importantly, a recently reported study revealed that residues in an extracellular loop in the C subunit of the permease along with residues in a flanking transmembrane segment are required for target cell sensitivity to all of the tested pediocin-like bacteriocins (22). However, residues in other unidentified parts of the C and/or D subunit seemed to also be needed to obtain maximal sensitivity to some of the tested bacteriocins, such as sakacin P and enterocin P, but perhaps not pediocin PA-1 (22). The extracellular loop of the C subunit might be an interaction site for the more hydrophilic and relatively conserved β-sheet-like N-terminal part of these bacteriocins, whereas transmembrane segments of the permease might be interaction sites for the bacteriocin's more diverse and hydrophobic helix-containing C-terminal half that is especially important for the relative potencies that different pediocin-like bacteriocins have toward different sensitive target cells.
Acknowledgments
We thank Line Johnsen for constructing the expression system for the production of the hybrid bacteriocins.
Footnotes
Published ahead of print on 21 January 2011.
REFERENCES
- 1.Aukrust, T. W., M. B. Brurberg, and I. F. Nes. 1995. Transformation of Lactobacillus by electroporation. Methods Mol. Biol. 47:201-208. [DOI] [PubMed] [Google Scholar]
- 2.Axelsson, L., T. Katla, M. Bjornslett, V. G. Eijsink, and A. Holck. 1998. A system for heterologous expression of bacteriocins in Lactobacillus sake. FEMS Microbiol. Lett. 168:137-143. [DOI] [PubMed] [Google Scholar]
- 3.Aymerich, T., et al. 1996. Biochemical and genetic characterization of enterocin A from Enterococcus faecium, a new antilisterial bacteriocin in the pediocin family of bacteriocins. Appl. Environ. Microbiol. 62:1676-1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cleveland, J., T. J. Montville, I. F. Nes, and M. L. Chikindas. 2001. Bacteriocins: safe, natural antimicrobials for food preservation. Int. J. Food Microbiol. 71:1-20. [DOI] [PubMed] [Google Scholar]
- 5.Corr, S. C., et al. 2007. Bacteriocin production as a mechanism for the antiinfective activity of Lactobacillus salivarius UCC118. Proc. Natl. Acad. Sci. U. S. A. 104:7617-7621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cotter, P. D., C. Hill, and R. P. Ross. 2005. Bacteriocins: developing innate immunity for food. Nat. Rev. Microbiol. 3:777-788. [DOI] [PubMed] [Google Scholar]
- 7.Diep, D. B., M. Skaugen, Z. Salehian, H. Holo, and I. F. Nes. 2007. Common mechanisms of target cell recognition and immunity for class II bacteriocins. Proc. Natl. Acad. Sci. U. S. A. 104:2384-2389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Drider, D., G. Fimland, Y. Héchard, L. M. McMullen, and H. Prévost. 2006. The continuing story of class IIa bacteriocins. Microbiol. Mol. Biol. Rev. 70:564-582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fimland, G., et al. 1996. New biologically active hybrid bacteriocins constructed by combining regions from various pediocin-like bacteriocins: the C-terminal region is important for determining specificity. Appl. Environ. Microbiol. 62:3313-3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fimland, G., V. Eijsink, and J. Nissen-Meyer. 2002. Mutational analysis of the role of tryptophan residues in an antimicrobial peptide. Biochemistry 41:9508-9515. [DOI] [PubMed] [Google Scholar]
- 11.Fimland, G., R. Jack, G. Jung, I. F. Nes, and J. Nissen-Meyer. 1998. The bactericidal activity of pediocin PA-1 is specifically inhibited by a 15-mer fragment that spans the bacteriocin from the center toward the C terminus. Appl. Environ. Microbiol. 64:5057-5060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fimland, G., et al. 2000. A C-terminal disulfide bridge in pediocin-like bacteriocins renders bacteriocin activity less temperature dependent and is a major determinant of the antimicrobial spectrum. J. Bacteriol. 182:2643-2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fimland, G., L. Johnsen, B. Dalhus, and J. Nissen-Meyer. 2005. Pediocin-like antimicrobial peptides (class IIa bacteriocins) and their immunity proteins: biosynthesis, structure, and mode of action. J. Pept. Sci. 11:688-696. [DOI] [PubMed] [Google Scholar]
- 14.Gallagher, N. L. F., et al. 1997. Three-dimensional structure of leucocin A in trifluoroethanol and dodecylphosphocholine micelles: spatial location of residues critical for biological activity in type IIa bacteriocins from lactic acid bacteria. Biochemistry 36:15062-15072. [DOI] [PubMed] [Google Scholar]
- 15.Hastings, J. W., et al. 1991. Characterization of leucocin A-UAL 187 and cloning of the bacteriocin gene from Leuconostoc gelidum. J. Bacteriol. 173:7491-7500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haugen, H. S., G. Fimland, J. Nissen-Meyer, and P. E. Kristiansen. 2005. Three-dimensional structure in lipid micelles of the pediocin-like antimicrobial peptide curvacin A. Biochemistry 44:16149-16157. [DOI] [PubMed] [Google Scholar]
- 17.Haugen, H. S., P. E. Kristiansen, G. Fimland, and J. Nissen-Meyer. 2008. Mutational analysis of the class IIa bacteriocin curvacin A and its orientation in target cell membranes. Appl. Environ. Microbiol. 74:6766-6773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Henderson, J. T., A. L. Chopko, and P. D. van Wassenaar. 1992. Purification and primary structure of pediocin PA-1 produced by Pediococcus acidilactici PAC-1.0. Arch. Biochem. Biophys. 295:5-12. [DOI] [PubMed] [Google Scholar]
- 19.Holck, A., L. Axelsson, S. E. Birkeland, T. Aukrust, and H. Blom. 1992. Purification and amino acid sequence of sakacin A, a bacteriocin from Lactobacillus sake Lb706. J. Gen. Microbiol. 138:2715-2720. [DOI] [PubMed] [Google Scholar]
- 20.Johnsen, L., G. Fimland, and J. Nissen-Meyer. 2005. The C-terminal domain of pediocin-like antimicrobial peptides (class IIa bacteriocins) is involved in specific recognition of the C-terminal part of cognate immunity proteins and in determining the antimicrobial spectrum. J. Biol. Chem. 280:9243-9250. [DOI] [PubMed] [Google Scholar]
- 21.Kazazic, M., J. Nissen-Meyer, and G. Fimland. 2002. Mutational analysis of the role of charged residues in target-cell binding, potency and specificity of the pediocin-like bacteriocin sakacin P. Microbiology 148:2019-2027. [DOI] [PubMed] [Google Scholar]
- 22.Kjos, M., Z. Salehian, I. F. Nes, and D. B. Diep. 2010. An extracellular loop of the mannose phosphotransferase system component IIC is responsible for specific targeting by class IIa bacteriocins. J. Bacteriol. 192:5906-5913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marugg, J. D., et al. 1992. Cloning, expression, and nucleotide sequence of genes involved in production of pediocin PA-1, a bacteriocin from Pediococcus acidilactici PAC1.0. Appl. Environ. Microbiol. 58:2360-2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nieto Lozano, J. C., J. Nissen-Meyer, K. Sletten, C. Pelaz, and I. F. Nes. 1992. Purification and amino acid sequence of a bacteriocin produced by Pediococcus acidilactici. J. Gen. Microbiol. 138:1985-1990. [DOI] [PubMed] [Google Scholar]
- 25.Nissen-Meyer, J., H. Holo, L. S. Håvarstein, K. Sletten, and I. F. Nes. 1992. A novel lactococcal bacteriocin whose activity depends on the complementary action of two peptides. J. Bacteriol. 174:5686-5692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nissen-Meyer, J., C. Oppegård, P. Rogne, H. S. Haugen, and P. E. Kristiansen. 2010. Structure and mode-of-action of the two-peptide (class-IIb) bacteriocins. Probiotics Antimicrob. Proteins 2:52-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nissen-Meyer, J., P. Rogne, C. Oppegård, H. S. Haugen, and P. E. Kristiansen. 2009. Structure-function relationships of the non-lanthionine-containing peptide (class II) bacteriocins produced by Gram-positive bacteria. Curr. Pharm. Biotechnol. 10:19-37. [DOI] [PubMed] [Google Scholar]
- 28.Oppegård, C., et al. 2007. The two-peptide class II bacteriocins: structure, production, and mode of action. J. Mol. Microbiol. Biotechnol. 13:210-219. [DOI] [PubMed] [Google Scholar]
- 29.Saavedra, L., C. Minahk, A. P. D. Holgado, and F. Sesma. 2004. Enhancement of the enterocin CRL35 activity by a synthetic peptide derived from the NH2-terminal sequence. Antimicrob. Agents Chemother. 48:2778-2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tichaczek, P. S., R. F. Vogel, and W. P. Hammes. 1994. Cloning and sequencing of sakP encoding sakacin P, the bacteriocin produced by Lactobacillus sake LTH673. Microbiology 140:361-367. [DOI] [PubMed] [Google Scholar]
- 31.Tichaczek, P. S., R. F. Vogel, and W. P. Hammes. 1993. Cloning and sequencing of curA encoding curvacin A, the bacteriocin produced by Lactobacillus curvatus LTH1174. Arch. Microbiol. 160:279-283. [DOI] [PubMed] [Google Scholar]
- 32.Uteng, M., H. H. Hauge, I. Brondz, J. Nissen-Meyer, and G. Fimland. 2002. Rapid two-step procedure for large-scale purification of pediocin-like bacteriocins and other cationic antimicrobial peptides from complex culture medium. Appl. Environ. Microbiol. 68:952-956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Uteng, M., et al. 2003. Three-dimensional structure in lipid micelles of the pediocin-like antimicrobial peptide sakacin P and a sakacin P variant that is structurally stabilized by an inserted C-terminal disulfide bridge. Biochemistry 42:11417-11426. [DOI] [PubMed] [Google Scholar]
- 34.Vaughan, A., V. G. Eijsink, T. F. O'Sullivan, K. O'Hanlon, and D. van Sinderen. 2001. An analysis of bacteriocins produced by lactic acid bacteria isolated from malted barley. J. Appl. Microbiol. 91:131-138. [DOI] [PubMed] [Google Scholar]
- 35.Wang, Y., et al. 1999. Solution structure of carnobacteriocin B2 and implications for structure-activity relationships among type IIa bacteriocins from lactic acid bacteria. Biochemistry 38:15438-15447. [DOI] [PubMed] [Google Scholar]
- 36.Yan, L. Z., A. C. Gibbs, M. E. Stiles, D. S. Wishart, and J. C. Vederas. 2000. Analogues of bacteriocins: antimicrobial specificity and interactions of leucocin A with its enantiomer, carnobacteriocin B2, and truncated derivatives. J. Med. Chem. 43:4579-4581. [DOI] [PubMed] [Google Scholar]