Abstract
Sand bedding material is frequently used in dairy operations to reduce the occurrence of mastitis and enhance cow comfort. One objective of this work was to determine if sand-based bedding also supported the microbiologically based suppression of an introduced bacterial pathogen. Bedding samples were collected in summer, fall, and winter from various locations within a dairy operation and tested for their ability to suppress introduced populations of Escherichia coli O157:H7. All sources of bedding displayed a heat-sensitive suppressiveness to the pathogen. Differences in suppressiveness were also noted between different samples at room temperature. At just 1 day postinoculation (dpi), the recycled sand bedding catalyzed up to a 1,000-fold reduction in E. coli counts, typically 10-fold greater than the reduction achieved with other substrates, depending on the sampling date. All bedding substrates were able to reduce E. coli populations by over 10,000-fold within 7 to 15 dpi, regardless of sampling date. Terminal restriction fragment length polymorphism (T-RFLP) analysis was used to identify bacterial populations potentially associated with the noted suppression of E. coli O157:H7 in sand bedding. Eleven terminal restriction fragments (TRFs) were overrepresented in paired comparisons of suppressive and nonsuppressive specimens at multiple sampling points, indicating that they may represent environmentally stable populations of pathogen-suppressing bacteria. Cloning and sequencing of these TRFs indicated that they represent a diverse subset of bacteria, belonging to the Cytophaga-Flexibacter-Bacteroidetes, Gammaproteobacteria, and Firmicutes, only a few of which have previously been identified in livestock manure. Such data indicate that microbial suppression may be harnessed to develop new options for mitigating the risk and dispersal of zoonotic bacterial pathogens on dairy farms.
Escherichia coli O157:H7 is a food-borne pathogen of global public health significance (39), so understanding the factors affecting its survival in the environment is critical to minimize its impact on human health. Cattle manure is a major reservoir of Escherichia coli O157:H7. However, the factors contributing to bovine colonization and shedding of E. coli O157:H7 are poorly understood, and there are still very few tools available to control these zoonotic bacteria in cattle populations (23). Early epidemiological studies in livestock populations showed that individual farms often tended to maintain either a high or a low prevalence of E. coli O157:H7 over time (18, 45). This relative stability of pathogen prevalence on individual farms has been interpreted to be an indication for a role of stable farm management factors in governing the relative abundance of E. coli O157:H7 in cattle populations. In addition, the prevalence of E. coli O157:H7 is seasonally modulated. Most studies indicate a peak in prevalence in cattle that occurs in summer and is coincident with the seasonal peak in human illnesses (3, 5, 11, 12, 17).
Interestingly, there are several similarities in the patterns of E. coli O157:H7 carriage by dairy cattle and cases of mastitis caused by E. coli (and other coliforms) in cows. Stanford et al. (46) demonstrated an association between somatic cell counts, a mastitis indicator, and E. coli O157:H7 prevalence on dairy farms. As with the E. coli O157:H7 prevalence in cattle, the incidence of coliform mastitis also fluctuates in the same seasonal manner (44). The primary source of mastitis-causing coliforms is the bedding material, and the incidence of coliform mastitis is influenced by the type of bedding material used (sand or sawdust) (19, 33, 36, 56). Moreover, LeJeune and Kauffman (22) demonstrated that dairy herds bedded on sawdust showed a significantly greater prevalence of E. coli O157:H7 than herds bedded on sand. Thus, because contaminated livestock bedding can significantly increase E. coli O157:H7 carriage in cattle, successful interventions targeted at this stage of production could reduce prevalence on the farm and have positive downstream impacts on human health.
Pathogen suppression can be mediated by a variety of biotic and abiotic soil factors. In solid-phase media, pathogens are the target of inhibitory soil functions that prevent them from expressing pathological effects to their full potential (3). General suppression of plant pathogens is mediated by the quantity and quality of soil organic matter that supports the resident microbial populations (20, 32). Indeed, judicious applications of soil organic matter can result in the suppression of diverse plant diseases in field and greenhouse systems (43, 47). However, complex interactions in the environment can significantly affect the suppressiveness of such amendments (6, 41). It is supposed that competition for nutrients is the primary driver of general suppression. In contrast, specific suppression is mediated by one or just a few microbial populations that antagonize resident pathogens (49). Production of antibiotics is considered a key feature of microbes associated with specific suppression (37, 38). Studies of suppressive soils have led to the identification of useful biological control strategies for suppressing plant pathogens. For example, culture-dependent (52) and culture-independent (54, 55) methods have been used to identify and isolate multiple microbial antagonists that can effectively suppress soilborne pathogens in field trials (34, 35). As a result, studies of soil microbial community structure are considered useful for developing innovative methods for pathogen and disease suppression (26).
Using concepts garnered from microbially based disease suppression in soils, we hypothesized that suppression of E. coli O157:H7 would be mediated by the predominant microorganisms present in the bedding in and around the cattle. The objectives of the current study were to determine (i) the degree to which biologically based suppression of E. coli O157:H7 occurred in bedding samples taken from different points on the farm and (ii) which bacteria present in such samples might contribute to the noted suppression. We focused on bacteria because previous studies had demonstrated that an overwhelming proportion of the microbial communities of manure were expected to be prokaryotic.
MATERIALS AND METHODS
Livestock bedding sources.
Samples were taken on18 August 2008, 10 November 2008, and 23 March 2009 (subsequently referred to as August, November, and March, respectively) from a dairy operation near Orrville, OH. At this operation, about 2,500 head of cattle were kept for milk production under free-stall housing conditions, where animals had free access to enter and leave resting areas in the barn that were lined at the bottom with a sand mix as bedding material. This dairy used a sand-manure separator (an auger 90 cm in diameter by 9.9 m long; McLanahan Corporation, Hollidaysburg, PA). Three times per week, a mix of fresh and recycled sand (1:1) was supplied as bedding material to the resting boxes with a mechanical distribution wagon. Through the action of the cows entering and exiting the stall, the sand was pushed from the front to the back end of the stall and eventually into the gutter, where it was mechanically collected and transferred to a washing facility for recycling. In this process, the material was moved by a separator auger from a collection pit into a temporary storage pile while it was being washed with water to separate the organic material from the sand. After it was air dried for 2 or more days in an open stockpile, the used sand was mixed with freshly purchased commercial clean sand and distributed in the livestock stalls to start the cycle anew. In this study, samples were collected from four sources within the recycling process of the livestock bedding: (i) fresh, consisting of recently applied bedding sand collected from the front of the resting stall (bedding 1); (ii) in use, consisting of bedding from the rear of the resting stall (bedding 2); (iii) washed, consisting of used livestock bedding collected immediately after separation in the sand-manure separator (bedding 3); and (iv) recycled, consisting of washed bedding material rested for 2 or more days in a pile (bedding 4).
Samples of 1,200 ml were taken and placed in plastic bags for transport to the laboratory; beddings 1 and 2 were collected from eight stall locations (four in the August experiment) from opposite sides of the barns. Samples of bedding 3 were collected as far distant as possible in a circular pattern from around a pile of 1.5 m in height and 2.5 m in diameter 90 cm off the ground. Samples of bedding 4 were collected from a pile 2.0 m in height and 5 m in diameter 90 cm off the ground. Samples were transported to the laboratory in ice coolers.
Chemical analysis of the bedding material.
A portion of each bedding sample was submitted to the Service Testing and Research Laboratory (STAR lab) at the Ohio State Agricultural Research and Development Center (OARDC), Wooster, OH, for analysis of soil chemical parameters. The organic matter content was determined by the loss-on-ignition method (13), the pH was determined by the method by Thomas (48), nitrogen was quantified as described by Mulvaney (30), and carbon was quantified by the ISO 10694:1995(E) method (2).
Heat treatment and inoculation of bedding samples with E. coli O157:H7.
After overnight storage in the laboratory, two 150-g portions of each of the samples were weighed into 250-ml Erlenmeyer flasks (250-ml beakers for the August experiment) and covered with aluminum foil. One set remained nonheated, and a second set was heated for 30 min at 80°C. For the latter, glass beakers with the contained bedding material were submerged in the preheated water of a 20-liter-capacity water bath and kept there for 30 min after a monitoring sample had reached the target temperature (experiments 1 and 2) or were kept in the water bath for 30 min total (experiment 3). This heat treatment was chosen because most bacteria should be severely reduced in number if they were not killed off at this temperature; such heat treatments had been used to disrupt the microbial communities of container substrates and suppressive soils (4, 52).
All inoculations were done with a green fluorescent protein-labeled E. coli O157:H7 strain (American Type Culture Collection strain ATCC 43888). Such transformation was previously demonstrated to be stable for at least 24 days when survival in fruit juices was monitored (16), and the nontoxigenic strain was expected to act similar to the toxigenic equivalent (21). After cooling of the heated bedding samples to room temperature, all samples were inoculated with suspensions of overnight cultures of this strain that had been spun down to remove spent medium, resuspended in 1× phosphate-buffered saline (PBS), and adjusted to deliver concentrations of 107 CFU per gram of bedding material. This was determined spectrophotometrically at 0.16 to 0.35 optical density (OD) units/ml. The containers with the inoculated bedding samples were closed with their corresponding aluminum foil and incubated at room temperature (21°C) in the laboratory in the dark. Samples were taken after vigorous mixing with a spatula for enumeration of the CFU of E. coli O157:H7 and for DNA extraction en route to terminal restriction fragment length polymorphism (T-RFLP) analysis.
Enumeration of CFU of E. coli O157:H7.
In all three experiments, samples were taken 1 day, 1 week, and 2 weeks after inoculation (experiment 1, at 1, 8, and 15 days postinoculation [dpi]; experiment 2, at 1, 7, and 15 dpi; and experiment 3, at 1, 10, and 17 dpi). Depending on the expected numbers of CFU of E. coli O157:H7, samples of 1 to 10 g were taken from the incubated bedding sources and suspended in 20 to 50 ml of 1× PBS (8.00 g NaCl, 0.20 g KCl, 1.44 g Na2HPO4, and 0.24 g KH2PO4 adjusted to pH 7.4) by vortexing 4 times for 15 s each time, processing in a sonicator (Aquasonic ultrasonic cleaner 75HT; VWR, West Chester, PA) for 1 min at 35 kHz, and vortexing at room temperature for 15 s. These suspensions were either plated directly onto the medium plates or entered into a 5-fold dilution series in 1× PBS in microtiter plates and then plated. The medium plates were LB medium (10 g Bacto tryptone, 5 g Bacto yeast extract, 5 g NaCl, and 1 liter double-distilled water adjusted to pH 7; 15 g agar was added and the medium was autoclaved for 20 min) amended, after it was cooled to 50°C, with ampicillin at 96 μg/ml and cycloheximide at 50 μg/ml on 10-cm-diameter agar plates.
After incubation at 37°C for 24 h, colonies were counted over UV light, and the terminal dilution factor and frequency of the green fluorescent colonies were recorded at the proper dilutions. The numbers of CFU per gram bedding material were calculated by a most-probable-number approach using the original terminal dilution factors. The terminal dilution factors for each sample source were converted to log-transformed data [log10(x) CFU per gram] by the proper formulas, considering the different dilution ratios, and the medians were presented in box plots; statistical groupings were derived from the analysis of the terminal dilution factors of the 5-fold dilution series.
DNA isolation.
On the days of dilution plating, 0.25-g subsamples (0.25- to 0.35-g subsamples in experiment 1) of the incubated material were weighed into centrifuge tubes and frozen at −80°C for at least 24 h before DNA was extracted with a PowerSoil DNA extraction kit (MO BIO Laboratories, Carlsbad, CA). DNA extraction was done according to the manufacturer's protocol, with two exceptions: first, centrifugation was at 9,000 × g (10,000 rpm) whenever 10,000 × g was suggested, and second, 450 μl was collected from each of the samples in the extraction tubes at the first collection step where the protocol suggested 400 to 500 μl to improve sample size uniformity. After extraction, DNA was either used immediately (being stored at 4°C between daily operations) or stored at −20°C.
DNA amplification and digestion and fragment analysis.
The 16S rRNA genes in the DNA extracts of the bedding material were amplified with primer 8F (5′-AGAGTTTGATCCTGGCTCAG-3′) labeled with WellRED dye D4 (Sigma, Proligo, St. Louis, MO) and primer 1492R (5′-ACGGCTACCTTGTTACGACTT-3′), on the basis of a previous description (49). A 25-μl reaction mixture with final concentrations of 1.8 mM MgCl2, 0.2 mM deoxynucleoside triphosphate mix, 0.8 pmol μl−1 each of primer 8F-D4 and primer 1492R, 0.04 mg μl−1 of RNase (Novagen, EMD Chemicals, Darmstadt, Germany), 1× PCR buffer of the GoTaq Flexi kit (Promega Co., Madison, WI), and 0.06 U μl−1 GoTaq Flexi (Promega) in sterile water (Sigma, St. Louis, MO), including 2.5 μl of the DNA extract (1:20 dilution of the original DNA extract), was submitted to amplification.
Amplification reactions were conducted in a PTC-200 thermal cycler (Peltier thermal cycler; MJ Research Inc., Waltham, MA) with the following program: after an initial 5 min at 95°C, 28 cycles of 1 min at 94°C, 45 s at 54°C, and 60 s at 70°C and then 8 min at 70°C. The resulting amplification product was cooled to 4°C and digested either immediately or after it was frozen once at −20°C.
For restriction digestion, reaction mixtures of a total of 10 μl containing 0.3 μl of the enzyme MspI (10 U/μl), 0.5 μl of the corresponding buffer B (Promega), 3.5 μl of the PCR product, and 5.7 μl of sterile purified water were incubated for 4 h at 37°C and 25 min at 65°C and were stored initially at 10°C and later at 4°C.
For purification, according to a modified protocol of the Molecular and Cellular Imaging Center (MCIC), 10 μl of the digest was amended with 1 μl of 3 M sodium acetate (pH 5.2) and 27.5 μl of 95% ethanol (cooled at −20°C), mixed, and incubated at −80°C for at least 10 min. After centrifugation at 6,130 × g in a S5700 swinging-bucket rotor (Allegra 25R centrifuge; Beckman Coulter, Fullerton, CA) for 15 min at 4°C, the pellet was rinsed twice with 70% ethanol (chilled at −20°C). After each rinse and careful blotting on paper towels, the plates were spun at 6,130 × g for 2 min at 4°C. After the rinses, the plates were reversed in the centrifuge holders, spun at 180 × g for 1 min to remove excess rinsate, and then dried in a laminar-flow hood. The pellets were resuspended in 15 μl of sterile purified water. A 5-μl mix (1:1) of the purified digest with sterile purified water was submitted for fragment size analysis at the MCIC. There, 0.1 μl of the sample was mixed with 0.5 μl 600-nucleotide (nt) size standard (CEQ DNA size standard kit 600) and 40 μl formamide solution. Samples were analyzed with a CEQ 8800 genetic analysis system (Beckman Coulter).
Fragment length data handling.
Data from the fragment analysis obtained from MCIC were entered into CEQ8000 software and analyzed in the mode for amplified fragment length polymorphism analysis of the D4 signal with a dye mobility calibration and a size standard of 600 nt in a quartic model; the bin size was 1 nt. Fluorescence signals at intensities of ≤300 were excluded from the data to reduce background signals.
After export to a tabular processing program (Excel, Microsoft Office), the data were processed before statistical analysis, as follows. Binning of the terminal restriction fragments (TRFs) was first conducted. For TRFs of ≤300 nt in length, TRF size classes (bins) of ±1 nt were combined, when several of the signals appeared in multiple bins under consideration. For TRFs of >300 nt, such combining was done for ±2 nt. In a second step, bins with <4 signals across the entire data set were eliminated from consideration, as no significant differences would be attributable to such rare events. TRFs with 4 to 8 observations were kept if the majority of the fluorescence was associated with specific treatments and were eliminated if the signals were randomly scattered among treatments.
A tiered method of screening the data was used to identify fragments associated with E. coli O157:H7 suppression. In the first step, data were mined for differences between the fluorescence intensities of a specific TRF in nonheated versus heat-treated bedding samples. In the second step, signal strengths were interval plotted to allow comparison of the fluorescence of nontreated and heated samples within the bedding sources. For these candidate TRFs, signal strengths were compared. In a third step, the numbers of CFU were plotted over the fluorescence in the nonheated bedding sample. Thus, TRFs that fulfilled two parameters were recorded: (i) significantly (P = 0.05) stronger fluorescence in the suppressive than the conducive bedding sample and (ii) a negative regression between the fluorescence of the TRFs and the numbers of CFU of E. coli O157:H7 within a specific nonheated source. If both parameters were fulfilled, the TRF was recorded as being potentially important in suppression of E. coli O157:H7. When the numbers of CFU were below the detection limit, only the first parameter was applied.
Cloning and sequencing of MspI-generated 16S rRNA gene TRFs.
Terminal restriction fragments were isolated as described above and then cloned and sequenced according to the protocol of Benítez and McSpadden Gardener (7). Briefly, samples were selected for identification on the basis of the high-peak intensity of the TRF of interest. One microliter of a double-stranded asymmetrical adapter was ligated to 2 μl MspI-digested fragment using 4.5 U T4 DNA ligase and 1 μl 10× ligase buffer (Promega). The reaction mixture was incubated at 16°C for 12 h. Following incubation, the fragments were gel extracted (UltraClean GelSpin DNA purification kit; MO BIO) after separation by agarose gel electrophoresis. Twenty-four samples were enriched for the 16S rRNA gene by amplification with primer 8F (5′-AGAGTTTGATCCTGGCTCAG-3′) and the MspI adapter primer (5′-GATGAGTCCTGAGTACCG-3′). Amplification was performed in 25-μl reaction volumes containing 1× Mg-free buffer, 1.8 mM MgCl2, 0.2 mM dideoxynucleoside triphosphates, 1 pmol μl−1 of each primer, 0.04 mg ml−1 RNase A (Novagen), 0.06 U μl−1 GoTaq Flexi DNA polymerase (Promega), and 2.5 μl template. Cycling parameters were as follows: 95°C for 5 min; 27 cycles of 94°C for 1 min, 54°C for 45 s, and 70°C for 1 min; and 70°C for 8 min. The TRF-enriched samples were pooled by sample month, ligated into the pGEM-T Easy vector (Promega), and transformed into E. coli JM109 competent cells (Promega). A total of 52 transformants were selected, and plasmids were isolated by a Pure Yield miniprep kit (Promega) and submitted for sequencing (ABI Prism 3100xl genetic analyzer system; MCIC).
Statistics.
Statistical analyses were conducted using Minitab software (version 15.1; Minitab Inc., State College, PA). Median values of the samples were compared by Mood's median test, and pair-wise comparisons were based on nonoverlapping 95% confidence intervals for the medians.
Nucleotide sequence accession numbers.
The sequences of the cloned TRF sequences can be found in GenBank as accession numbers HM146088 through HM146121.
RESULTS
Soil physical and chemical properties.
At initiation of the three laboratory assays, significant variation in the total organic carbon and nitrogen levels of the samples was noted (Table 1). Overall, the range of values varied by no more than a factor of 3 or 4 (i.e., 0.8 to 2.4% C and 0.04 to 0.14% N). In-use samples had higher carbon and nitrogen contents than the washed bedding material, and the other samples had intermediate levels of both C and N. In the three experiments, the pH was alkaline, ranging from pH 8.8 to 10.0. However, no significant differences in average pH were detected among the sources (data not shown).
TABLE 1.
Soil chemical parameters of samples at initiation of laboratory assays for survival of Escherichia coli O157:H7 in livestock bedding in August 2008, November 2008, and March 2009a
| Sourceb | % organic matterc |
% nitrogend |
||||
|---|---|---|---|---|---|---|
| August | November | March | August | November | March | |
| Fresh | 1.54 a | 1.63 b | 1.82 a | 0.06 b | 0.09 b | 0.13 a |
| In use | 1.84 a | 2.15 a | 2.43 a | 0.07 a | 0.11 a | 0.14 a |
| Washed | 1.15 b | 0.86 c | 0.81 c | 0.04 c | 0.04 d | 0.04 c |
| Recycled | 1.53 ab | 0.99 c | 1.21 b | 0.05 b | 0.05 c | 0.05 b |
| P | 0.046 | <0.01 | <0.01 | 0.019 | <0.01 | <0.01 |
Medians are presented; medians within one column followed by the same letter were not significantly different at P equal to 0.05 when they were tested with Mood's median test. n = 4 for August 2008, n = 8 for November 2008, and n = 8 for March 2009.
Livestock bedding samples were collected from (i) front of the cow box (fresh), (ii) back of the cow box (in use), (iii) used material from the pile following sand-manure separation (washed), and (iv) rested (≥2 days) in a pile of washed material (recycled).
Organic matter content determined by loss of weight on ignition (LOI) (13).
Nitrogen was quantified as described by Mulvaney (30).
Detection of microbially based suppression of E. coli O157:H7 in different bedding samples.
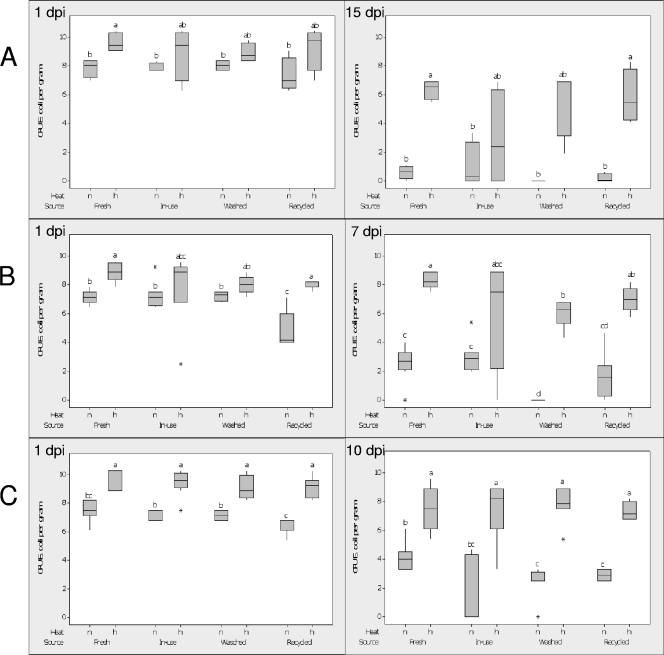
At all sampling times, counts of the inoculated E. coli O157:H7 were numerically larger in all samples that had been heated to 80°C and then cooled to room temperature prior to inoculation than in the nonheated equivalents of samples from the same bedding source. Such differences in population size were significant (P < 0.05) in three of eight contexts in the August experiment (Fig. 1 A), five of eight contexts in the November experiment (Fig. 1B), and eight of eight contexts in the March experiment (Fig. 1C). These data indicate that all samples harbored heat-sensitive factors that contribute to the suppression of E. coli O157:H7 in sand bedding samples.
FIG. 1.
Suppression of E. coli O157:H7 in four livestock bedding sources. Bedding samples were acquired on August 2008 (n = 4) (A), November 2008 (n = 8) (B), and March 2009 (n = 8) (C). Sources were as follows: fresh, front of the cow box; in use, back of the cow box; washed, used material from the pile following use of the sand-manure separator; and recycled, rested (≥2 days) pile of washed bedding material. Heat indicates that fresh livestock bedding samples were left either nonheated (n) or heated (h) at 80°C for 30 min to kill off the majority of endogenous microbes before they were subsequently cooled to room temperature. E. coli O157:H7 was then added at 107 CFU g−1 to all samples and incubated at room temperature. The numbers of CFU of E. coli O157:H7 were measured by dilution plating on LB medium amended with ampicillin (50 μg ml−1) and cycloheximide (96 μg ml−1) at 1 dpi or at 7, 10, and 15 dpi. Within each time point, the values for bars indexed with the same letter were not significantly different at P equal to 0.05 when the terminal dilution factors of a 5-fold dilution series were analyzed with Mood's median test. The log-transformed [log10(x)] numbers of CFU of E. coli O157:H7 are presented.
The median numbers of CFU of E. coli O157:H7 declined in all contexts (nonheated or heated) over time (Fig. 1A to C), suggesting that inhibitory factors in addition to the microbial suppressiveness were not destroyed by the heat treatments. The dynamics of the decay curves varied somewhat from experiment to experiment. However, initial levels of suppressiveness (i.e., decay during the first 24 h) were most dramatic in the recycled bedding samples (sample 4) in all three experiments. Such differences were significant (P < 0.05) in two of the three experiments.
In the August experiment (Fig. 1A) at 1 dpi, the numbers of CFU of E. coli O157:H7 were lower for all nonheated bedding samples than the equivalent heated samples; these differences were statistically significant for the source fresh only. Within the nonheated or heated sample group, the numbers of CFU were similar for the different sources. At 15 dpi, most of the CFU of E. coli O157:H7 had declined; more E. coli O157:H7 cells survived only in samples from the heated fresh and recycled sources than in the nonheated equivalents; within the nonheated or heated samples, the numbers of CFU were similar.
In the November experiment (Fig. 1B) at 1 dpi, the numerical values for the numbers of CFU of E. coli O157:H7 were lower for all nonheated bedding samples than the heated equivalents; these differences were statistically significant for the fresh and recycled bedding. Recycled bedding had the fewest CFU of all nonheated samples, whereas the numbers of CFU were similar for all heated samples. At 7 dpi, the numbers of CFU of the nonheated samples were lower than those of the heated ones, except for the in-use source; CFU were not detected in nonheated samples from the washed source, and this source had the lowest numbers of CFU of all nonheated bedding samples.
In the March experiment (Fig. 1C) at 1 dpi, there were significantly fewer CFU of E. coli O157:H7 in all nonheated bedding samples than in the heated equivalents; the numbers of CFU for all of the latter samples were similar. The recycled bedding had fewer CFU than the nonheated in-use and washed samples. At 10 dpi, the numbers of CFU in all nonheated samples were lower than the numbers in the heated ones; the numbers of CFU for the nonheated washed and recycled samples were lower than the numbers for the fresh material, whereas the number of CFU in the in-use material was intermediate.
T-RFLP analyses and association of heat-sensitive bacterial TRFs with suppressiveness.
In order to determine what components of the bacterial communities present in the bedding samples might contribute to the patterns of observed suppressiveness, we conducted T-RFLP analyses of amplified 16S rRNA gene sequences from all samples. Interval plotting and regression analyses of the relative abundance of different TRFs were conducted to identify candidate TRFs that were more abundant in more suppressive sample pairings in each experiment. For example, in the nonheated versus heated sample pairings of each bedding source, 31 of the 100 total TRFs in the August experiment (see Table S1 in the supplemental material), 67 of the 149 total TRFs in the November experiment (see Table S2 in the supplemental material), and 78 of the 131 total TRFs in the March experiment (see Table S3 in the supplemental material) were tentatively identified to differ in abundance. Because the two mathematical approaches identified different subsets of TRFs of interest, we focused our efforts on identifying those TRFs repeatedly observed to differ using both approaches (boldface values in Tables S1 to S3 in the supplemental material). Thus, while there was a high degree of sample-to-sample variation in the profiles, some TRFs were repeatedly observed to occur in comparisons of more suppressive sample pairings than less suppressive sample pairings in individual experiments.
In the August experiment (see Table S1 in the supplemental material) at dpi 1, a total of 22 TRFs had greater fluorescence in the nonheated than the heated portions of the bedding sources; 27 TRFs had a negative relationship of the numbers of CFU of E. coli O157:H7 over fluorescence, but only 4 TRFs in fresh bedding fulfilled both selection requirements simultaneously. At 15 dpi, there were 25 TRFs with higher fluorescence in nonheated than heated soil, and 51 exhibited a negative relationship with the numbers of CFU of E. coli O157:H7; TRFs M130, M139, and M156 fulfilled both selection criteria and occurred in fresh and in-use bedding materials. Additional TRFs fulfilled both selection criteria but occurred only in one source.
In the November experiment (see Table S2 in the supplemental material) at 1 dpi, the numbers of TRFs of the candidates were higher in the nonheated samples 59 times, and 35 negative regressions with the numbers of CFU of E. coli O157:H7 were detected; but TRFs fulfilled both selection criteria only 12 times when the information for in-use bedding, washed, and recycled bedding is summarized; none were detected in fresh bedding. At 15 dpi, the numbers of TRFs of the candidates were higher in nonheated portions than heated portions 126 times, and a negative regression of the TRF fluorescence with the numbers of CFU of E. coli O157:H7 was detected 84 times. TRFs fulfilled both parameters 65 times. Several TRFs occurred in different sources at the same sampling time; some occurred repeatedly within one sampling time and at both samplings.
In the March experiment (see Table S3 in the supplemental material) at dpi 1, fluorescence was stronger in the nonheated than in the heated equivalents 102 times and there was a negative regression of the numbers of CFU and TRF fluorescence 109 times. TRF fulfilled both requirements a total of 50 times. At dpi 10, the fluorescence was higher in nonheated than in heated equivalents 86 times; there was a negative regression 83 times. Both parameters were fulfilled 21 times.
Identification of bacterial TRFs enriched in unheated bedding samples with the highest initial suppressiveness.
A similar screening approach was used to identify TRFs that were more abundant in the unheated bedding samples which displayed the greatest initial decay of inoculated E. coli O157:H7 populations (Fig. 1A to C). Comparisons of the T-RFLP profiles of the washed and/or recycled bedding to those of the other samples were conducted for the two experiments where significant differences in suppressiveness were noted, i.e., in the November experiment (see Table S4 in the supplemental material) and the March experiment (see Table S5 in the supplemental material).
At dpi 1, fluorescence was stronger in the nonheated recycled bedding than in the nonheated fresh, in-use, or washed material 29 times; there were negative regressions 27 times (see Table S4 in the supplemental material). Four to six TRFs that fulfilled both requirements were detected. At dpi 7, TRFs of nonheated washed samples had higher fluorescence than nonheated fresh, in-use, or recycled material 32 times; no regression analysis could be conducted for the washed samples because no E. coli O157:H7 was detected. TRF M118 occurred in all these comparisons, TRF M299 and TRF M508 occurred at 1 dpi, and TRF M202 occurred only at 7 dpi.
When samples from the recycled source were compared to those from the fresh, in-use, and washed sources, fluorescence was higher in recycled samples 17 times and the regression was negative 10 times. Both parameters were fulfilled 14 times (see Table S5 in the supplemental material). The washed samples had more fluorescent TRFs five times and a negative regression six times, but no TRFs fulfilled both requirements. Similarly, recycled samples had more fluorescence than fresh or in-use samples five times, but none had a negative regression.
Identification of bacterial populations most commonly associated with suppressiveness to E. coli O157:H7.
In the total of 37 comparisons of the August, November, and March experiments outlined above, elevated levels of TRF M299 occurred in 9 comparisons spread over different experiments and sampling times (Table 2). Similarly, TRF M118 was elevated eight times. TRF M142 occurred a total of seven times at various sampling times in all three experiments. TRF M89 and TRF M156 occurred six times: the former in the November and March experiments and the latter in all three experiments at late or early times. TRF M96, TRF M99, TRF M202, TRF M488, TRF M492, and TRF M508 all occurred in five contexts, typically restricted to two of the three experiments.
TABLE 2.
Occurrence of TRFs associated with E. coli O157:H7 suppression across all three experiments (August 2008, November 2008, and March 2009)a
| Parameter | Result for the following TRFs generated by MspI digestion of amplified 16S sequencesb |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| M89 | M96 | M99 | M118 | M142 | M156 | M202 | M299 | M488 | M492 | M508 | |
| Sum parameter A + parameter Bc | 6 | 5 | 5 | 8 | 7 | 6 | 5 | 9 | 5 | 5 | 5 |
| Sample timed | |||||||||||
| Early | 3 | 2, 3 | 2, 3 | 2 | 3 | 3 | 2, 3 | 1, 2, 3 | 3 | 2, 3 | |
| Late | 2 | 2 | 2 | 2 | 1, 2, 3 | 1, 2 | 2, 3 | 2 | 2, 3 | 1, 3 | |
| Abundance in bedding source: | |||||||||||
| 4 > 1, 2, 3e | 3 | 2 | 2 | 2, 3 | |||||||
| 3 > 1, 2, 4f | 2 | 2 | |||||||||
Livestock bedding samples from four distinct sources (fresh, in use, washed, recycled) were split into one nonheated (NH) and one heated (HT) sample, TRFs were recorded for higher fluorescence in nonheated than heat-treated samples (parameter A) and negative regression of the surviving numbers of CFU of E. coli O157:H7 over fluorescence strength or between nonheated samples of different levels of suppressiveness (parameter B).
TRFs of <300 nt were at an accuracy of ±1 nt; TRFs of >300 nt were binned at ±2 nt.
Frequency of occurrence of TRFs with both parameters A and B fulfilled in a total of 37 contexts.
Sampling times: early, 1 dpi; late, 15 dpi (experiment 1), 7 dpi (experiment 2), or 10 dpi (experiment 3). The numbers in the table body indicate the experiment(s) in which the TRFs were detected.
These comparisons among nonheated bedding samples 1 through 4 were conducted only in experiments 2 and 3; in experiment 1, the samples from the recycled source did not have significantly more CFU than those from the fresh source. The numbers in the table indicate the experiment(s) in which the TRFs were detected.
These comparisons among nonheated bedding samples 1 through 4 were conducted only in experiments 2 and 3. In experiment 2, samples from the washed source (bedding 3) did not have significantly fewer CFU of E. coli O157:H7 than those from the recycled bedding source 4. Also, in experiment 3, samples from the washed source (bedding 3) did not have significantly fewer CFU of E. coli O157:H7 than those from the in-use bedding source 2. The numbers in the table indicate the experiment(s) in which the noted TRFs were detected.
In order to identify the bacteria giving rise to these TRFs, we size selected and sequenced clones amplified from the original samples. The sequences were trimmed from the 8F primer to the MspI site and subjected to BLAST analysis (1). Small to medium-sized TRFs were more often successfully cloned than larger TRFs. To assess the novelty of the bacterial species identified, we evaluated the percent identity across the full length of the fragment for both the best overall match and the best published match with a named genus or species designation (Table 3). The smaller TRFs belong to a variety of genera in the diverse Bacteroidetes (previously known as the Cytophaga-Flexibacter-Bacteroidetes group), which have 99 to 100% identity to uncultured bacteria from agricultural environments. The medium-sized TRFs were mainly associated with two bacterial groups, the gammaproteobacteria and the firmicutes. The larger TRFs were associated with the gamma- and betaproteobacteria. No clones corresponding to either TRF M118 or M202 were identified.
TABLE 3.
Sequence-based identifications of bacterial TRFs associated with E. coli O157:H7 suppression across all three experiments
| TRFa | Highest BLAST hitb | Highest published BLAST hit with named genus or genus and species designationb | Bacterial group |
|---|---|---|---|
| M89 | Uncultured bacterium from manure, potato plant root bacterium | Cellulophaga tyrosinoxydans, Cytophaga spp. | Cytophaga-Flexibacter-Bacteroidetes |
| M96 | Uncultured compost bacterium, uncultured bacterium from beetle hindgut, uncultured bacterium from cattle feedlot, uncultured Fluviicola | Proteiniphilum acetatigenes, Fluviicola spp. | Cytophaga-Flexibacter-Bacteroidetes |
| M99 | Uncultured bacterium from cattle feedlot | Bacteroides spp. | Cytophaga-Flexibacter-Bacteroidetes |
| M118c | |||
| M142 | Uncultured bacterium from wetland soil, Cellulosimicrobium cellulans | Marinomonas spp., Cellulosimicrobium cellulans | Gammaproteobacteria, Actinomycetales |
| M156 | Uncultured bacterium from cattle feedlot, uncultured bacterium from hyena feces | Lysobacter spp., Caryophanon spp. | Gammaproteobacteria, Firmicutes |
| M202c | |||
| M299 | Uncultured bacterium from bovine feces, uncultured bacterium from cattle feedlot, Marinobacter spp., Halomonas spp. | Clostridium spp., Faecalibacterium prausnitzii, Marinobacter excellens, Halomonas venusta | Firmicutes, Gammaproteobacteria |
| M488 | Uncultured bacterium from biogas reactor | Oenothera phytoplasma | Mollicutes |
| M492 | Uncultured bacterium from wastewater | Arenimonas donghaensis | Gammaproteobacteria |
| M510 | Uncultured compost bacterium | Bordetella petrii | Betaproteobacteria |
TRF is from the 8F primer site to the first MspI site.
BLAST analysis (1) was used to compare TRF sequences to sequences in GenBank. Putative identifications are highlighted if they possess a ≥99% (boldface) or <89% (underlined) sequence identity across the cloned TRFs.
No clones containing the target fragment length were identified.
DISCUSSION
Here we present the first evidence that the suppression of E. coli O157:H7 in the environment is mediated by heat-sensitive microorganisms (Fig. 1). Other work has established the use of such heat treatments for the determination of biologically based pathogen suppressiveness (42, 51, 52, 53). LeJeune and Kauffman (22) tested the ability of E. coli O157:H7 to survive in sawdust and sand bedding materials collected from dairy operations. The authors showed that E. coli O157:H7 survived at higher densities and for longer periods of time in used sawdust bedding than in used sand bedding. No mechanism for that apparent suppression was elucidated; however, they posited that the differences in survival were influenced by the presence of toxic substances and the lack of water, organic matter, and/or nutrients in sand bedding. While physical and chemical properties of the matrices might affect the survival of the inoculated pathogen, we found no associations between the survival of E. coli O157:H7 and the sample pH or the amount of carbon or nitrogen in this study. Effects of soil moisture differences, though not recorded in detail, were also discounted as a possible source of variable survival because water potential was previously demonstrated to have limited effects on survival of E. coli O157:H7 (40). These findings further expand the collective understanding of pathogen population dynamics on the farm by illustrating that zoonotic bacterial pathogens can be suppressed in situ by other bacterial populations.
It has long been known that diverse oomycete, fungal, and nematode plant pathogens can be suppressed in some agricultural soils (9, 20, 50), and in some instances, agricultural soils can be suppressive to a bacterial plant pathogen (e.g., 31). Such examples of pathogen-suppressive phenomena are known to be mediated by various numbers and types of microorganisms present in agricultural soils. Because of this, we hypothesized that the observed suppression of E coli O157:H7 in sand livestock bedding was mediated by a subset of the microorganisms present in and around the cattle on the farm. The characterization of this hypothesized subset of microorganisms was carried out using the microbial community profiling approach described by Borneman et al. (10). From the results obtained using this approach, we assert that bacteria marked by the 11 TRFs in Table 2 are potentially involved in the E. coli O157:H7 suppressiveness. While this study focused on bacterial populations, it is possible that other microorganisms (e.g., phage, archeae, oomycetes, and fungi) could have also contributed to the noted suppressiveness.
Because these noted TRFs were observed more frequently and/or more abundantly in the suppressive than the nonsuppressive bedding samples, the bacteria giving rise to these TRFs were associated with pathogen suppressiveness. It has previously been shown that there is a monotonic relationship between TRF signal strength and the abundance of the bacteria giving rise to such a signal (24, 27). Thus, the relative abundance of any given TRF will be correlated to the relative abundance of the bacteria giving rise to that signal, and because this numerical relationship is likely weakened by confounding factors, such as the stochastic occurrence of multiple bacteria giving rise to the same-sized TRF signals, any noted correlation between suppression and TRF abundance is likely to be more significant than that calculated using this methodology. The validity of this TRF-based approach was further supported by the initial identification (8) and subsequent recovery (7) of novel bacterial groups belonging to the Genera incertae associated with the suppression of plant pathogens. Some of the TRFs identified here may also come from novel species, on the basis of the low sequence levels of identity of some TRFs to the database sequences (e.g., M488 and M510; Table 3). Similar community profiling approaches have also been used successfully to first identify and subsequently recover microbial antagonists of plant pathogenic nematodes (9, 54, 55). Still, the ultimate proof that the 11 TRFs mark bacteria that actually suppress E. coli O157:H7 in situ will depend on culturing representative bacteria harboring those sequences. Unfortunately, the dairy farm from which the samples were taken in this study is no longer operational (a victim of the recent economic crisis), so sampling for recovery from the same materials is not possible. However, it seems likely that similar, if not clonal, strains exist on other farms, given the cosmopolitan nature of most bacterial species. Subsequent efforts to recover and phenotypically characterize bacteria harboring the identified TRF tags are under way and could lead to the development of an inoculant-based strategy to suppress E. coli O157:H7, in a manner analogous to applied biocontrol in the field of plant pathology.
Previously, Benítez et al. (8) used two separate statistical approaches to identify candidate TRFs associated with (plant) pathogen suppression. Here, we improved on that screening approach by using an additional statistical criterion (regression analysis) to further restrict the number of candidate TRFs considered to be associated with suppression, thereby reducing the potential number of false-positive associations of phenomenological significance. Remarkably, the more restrictive selection criteria applied in this study resulted in an even greater number of candidate TRFs than that previous study examining root disease suppression (i.e., 11 candidates here [Table 2] versus 8 previously [8]). The significance of this is not clear, but it might indicate that bacterium-mediated suppression of strain O157:H7 is less specific than the general suppressiveness to soilborne plant pathogens noted in the earlier study. Such a conclusion is further supported by the observation that many more TRFs were associated with E. coli suppression at different sampling times (Table 2), but only 11 TRFs were observed repeatedly in different samplings (Table 2). Also, the results presented do not preclude other organisms from being involved in suppressiveness.
Because they were found in most all of the samples to various degrees, it is likely that the bacteria giving rise to the 11 noted TRFs were likely present in the feces of the cattle themselves. However, because the greatest suppressiveness was observed in the recycled sand pile (bedding source 4, 1 dpi; Fig. 1), such populations are probably subdominant members of the community until some curing of the bedding takes place, allowing them to become more abundant and/or more active. Interestingly, changes in the relative abundance of specific antagonists, such as phlD-positive Pseudomonas species, have also been associated with the suppression of some plant pathogens (28, 29).
The identified subset of 11 TRFs appears to have been amplified from a variety of bacterial groups (Table 3). These include the diverse Cytophaga-Flexibacter-Bacteroidetes group, the well-known Gammaproteobacteria, and the primarily Gram-positive, spore-forming Firmicutes. Although some 16S rRNA TRF sequences showed greater than 99% identity to sequences in the database, there were some sequences whose best match showed less than 90% identity across the length of the TRF. In these cases of lower sequence identity to database entries, we may have found markers for previously unidentified species of bacteria that are particularly suppressive to E. coli O157:H7 in situ. This hypothesis is supported by previous work where exploitation of similar correlations (8) led to the marker-assisted recovery of two bacterial species that displayed the predicted function of suppressiveness to the plant pathogens present in the tested soils (7).
The ecology of E. coli O157:H7 in the farm environment impacts the prevalence of O157:H7 on the farm and beyond. Franz and van Bruggen (15) recently reviewed the ecology of enteric pathogens and described a number of strategies to reduce the risk of spread of the pathogen on and off farms. Among these were strategies related to altering the manure composition by altering feeding regimes and composting the waste. Indeed, it has long been known that composting can reduce enteric pathogen populations (14, 19, 25). This work adds to previous studies by highlighting how the use of recycled sand bedding might lead to the sustenance of microbially based suppression, an approach that would likely reduce the prevalence of E. coli O157:H7 and possibly other enteric pathogens in and around dairy cattle. By identifying management strategies for dairy farms that select for microbial populations that suppress E. coli O157:H7 in the environment, the contribution of the farm environment to E. coli O157:H7 persistence is effectively reduced and the risk of food-borne contamination is minimized.
Supplementary Material
Acknowledgments
This project was supported by an OARDC Interdisciplinary SEEDS grant to J.T.L. and B.B.M.G.
The first author especially thanks S. Bénitez, C. Cao, and M. Kauffmann for help with conducting this research.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 21 January 2011.
REFERENCES
- 1.Altschul, S. F., et al. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anonymous. 1995. International standard, ISO 10694:1995(E). Soil quality—determination of organic and total carbon after dry combustion (elementary analysis). International Organization for Standardization, Geneva, Switzerland.
- 3.Baker, K. F., and R. J. Cook. 1974. Biological control of plant pathogens. W. H. Freeman & Co., San Francisco, CA.
- 4.Baker, K. F., and C. N. Roistacher. 1957. Heat treatment of soil, p. 123-137. In K. F. Baker (ed.), The UC system for producing healthy container-grown plants. Division of Agricultural Sciences, University of California, Oakland, CA.
- 5.Barkocy-Gallagher, G. A., et al. 2003. Seasonal prevalence of Shiga toxin-producing Escherichia coli, including O157:H7 and non-O157:H7 serotypes, and Salmonella in commercial beef processing plants. J. Food Prot. 66:1978-1986. [DOI] [PubMed] [Google Scholar]
- 6.Baysal, F., M.-S. Benítez, M. D. Kleinhenz, S. A. Miller, and B. Gardener. 2008. Field management effects on damping-off and early season vigor of crops in a transitional organic cropping system. Phytopathology 98:562-570. [DOI] [PubMed] [Google Scholar]
- 7.Benítez, M.-S., and B. McSpadden Gardener. 2009. Linking sequence to function in soil: sequence-directed isolation of novel bacteria contributing to soilborne disease suppression. Appl. Environ. Microbiol. 75:915-924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Benítez, M.-S., et al. 2007. Multiple statistical approaches of community fingerprint data reveal bacterial populations associated with general disease suppression arising from the application of different organic field management strategies. Soil Biol. Biochem. 39:2289-2301. [Google Scholar]
- 9.Borneman, J., and J. O. Becker. 2007. Identifying microorganisms in specific pathogen suppression in soil. Annu. Rev. Phytopathol. 45:153-172. [DOI] [PubMed] [Google Scholar]
- 10.Borneman, J., et al. 2007. Identifying microorganisms involved in specific in situ functions: experimental design considerations for rRNA gene-based population studies and sequence-selective PCR assays, p. 748-757. In Manual of environmental microbiology, 3rd ed. ASM Press, Washington, DC.
- 11.Chapman, P. A., C. A. Siddons, A. T. Gerdan Malo, and M. A. Harkin. 1997. A 1-year study of Escherichia coli O157:H7 in cattle, sheep, pigs and poultry. Epidemiol. Infect. 119:245-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cobbold, R. N., et al. 2007. Rectoanal junction colonization of feedlot cattle by Escherichia coli O157:H7 and its association with supershedders and excretion dynamics. Appl. Environ. Microbiol. 73:1563-1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Combs, S. M., and M. V. Nathan. 1998. Soil organic matter, p. 53-58 In Recommended chemical soil test procedures for the north central region. NCR publication no. 221. Missouri Agricultural Experiment Station, Columbia, MO.
- 14.Forshell, L. P., and I. Ekesbo. 1993. Survival of Salmonellas in composted and not composted solid animal manure. Zentralbl. Veterinarmed. B 40:654-658. [DOI] [PubMed] [Google Scholar]
- 15.Franz, E., and A. H. C. van Bruggen. 2008. Ecology of E. coli O157:H7 and Salmonella enterica in the primary vegetable production chain. Crit. Rev. Microbiol. 34:143-161. [DOI] [PubMed] [Google Scholar]
- 16.Fratamico, P. M., M. Y. Deng, T. P. Strobaugh, and S. A. Palumbo. 1997. Construction and characterization of Escherichia coli O157:H7 strains expressing firefly luciferase and green fluorescent protein and their use in survival studies. J. Food Prot. 60:1167-1175. [DOI] [PubMed] [Google Scholar]
- 17.Freeman, J. T., D. J. Anderson, and D. J. Sexton. 2009. Seasonal peaks in Escherichia coli infections: possible explanations and implications. Clin. Microbiol. Infect. 15:951-953. [DOI] [PubMed] [Google Scholar]
- 18.Hancock, D. D., T. E. Besser, D. H. Rice, D. E. Herriott, and P. I. Tarr. 1997. A longitudinal study of Escherichia coli O157:H7 in fourteen cattle herds. Epidemiol. Infect. 118:193-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hogan, J. S., et al. 1999. Bacterial counts associated with sawdust and recycled manure bedding treated with commercial conditioners. J. Dairy Sci. 82:1690-1695. [DOI] [PubMed] [Google Scholar]
- 20.Hoitink, H., and M. Boehm. 1999. Biocontrol within the context of soil microbial communities: a substrate-dependent phenomenon. Annu. Rev. Phytopathol. 37:427-446. [DOI] [PubMed] [Google Scholar]
- 21.Kudva, I. T., K. Blanch, and C. J. Hovde. 1998. Analysis of Escherichia coli O157:H7 survival in ovine or bovine manure and manure slurry. Appl. Environ. Microbiol. 64:3166-3174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.LeJeune, J. T., and M. Kauffman. 2005. Effect of sand and sawdust bedding materials on the fecal prevalence of Escherichia coli O157:H7 in dairy cows. Appl. Environ. Microbiol. 71:326-330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.LeJeune, J. T., and A. N. Wetzel. 2007. Preharvest control of Escherichia coli O157 in cattle. J. Anim. Sci. 85:E73-E80. [DOI] [PubMed] [Google Scholar]
- 24.Liu, W. T., T. L. Marsh, H. Cheng, and L. J. Forney. 1997. Characterization of microbial diversity by determining restriction fragment length polymorphisms of genes encoding 16S rRNA. Appl. Environ. Microbiol. 63:4516-4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lung, A. J., et al. 2001. Destruction of Escherichia coli O157:H7 and Salmonella enteritidis in cow manure composting. J. Food Prot. 64:1309-1314. [DOI] [PubMed] [Google Scholar]
- 26.Mazzola, M. 2004. Assessment and management of soil microbial community structure for disease suppression. Annu. Rev. Phytopathol. 42:35-59. [DOI] [PubMed] [Google Scholar]
- 27.McSpadden Gardener, B. B. 1998. Examination of soil and rhizosphere bacterial communities using a modified T-RFLP analysis of amplified 16S rDNA. .In Assessing the potential of creating biased rhizospheres based on inositol rhizopines. Ph.D. thesis. Michigan State University, East Lansing, MI.
- 28.McSpadden Gardener, B. B., and D. M. Weller. 2001. Changes in populations of rhizosphere bacteria associated with take-all disease of wheat. Appl. Environ. Microbiol. 67:4414-4425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McSpadden Gardener, B., L. Gutierrez, R. Joshi, R. Edema, and E. Lutton. 2005. Distribution and biocontrol potential of phlD+ pseudomonads in corn and soybean fields. Phytopathology 95:715-724. [DOI] [PubMed] [Google Scholar]
- 30.Mulvaney, R. L. 1996. Nitrogen-inorganic forms, p. 1162-1171. In Methods of soil analysis. Part 3. Chemical methods. Soil Science Society of America, Madison, WI.
- 31.Niushiyama, M., Y. Shiomi, S. Suzuki, and T. Marumoto. 1999. Suppression of growth of Ralstonia solanacearum, tomato bacterial wilt agent, on/in tomato seedlings cultivated in a suppressive soil. Soil Sci. Plant Nutr. 45:79-87. [Google Scholar]
- 32.Noble, R., and E. Coventry. 2005. Suppression of soil-borne plant diseases with composts: a review. Biocontrol Sci. Technol. 15:3-20. [Google Scholar]
- 33.Norring, M., et al. 2008. Effects of sand and straw bedding on the lying behavior, cleanliness, and hoof and hock injuries of dairy cows. J. Dairy Sci. 91:570-576. [DOI] [PubMed] [Google Scholar]
- 34.Olatinwo, R., J. O. Becker, and J. Borneman. 2006. Suppression of Heterodera schachtii populations by Dactylella oviparasitica in four soils. J. Nematol. 38:345-348. [PMC free article] [PubMed] [Google Scholar]
- 35.Olatinwo, R., J. O. Becker, and J. Borneman. 2006. Induction of beet-cyst nematode suppressiveness by the fungi Dactylella oviparasitica and Fusarium oxysporum in field microplots. Phytopathology 96:855-859. [DOI] [PubMed] [Google Scholar]
- 36.Paulin-Curlee, G. G., et al. 2008. Molecular subtyping of mastitis-associated Klebsiella pneumoniae isolates shows high levels of diversity within and between dairy herds. J. Dairy Sci. 91:554-563. [DOI] [PubMed] [Google Scholar]
- 37.Raaijmakers, J. M., and D. M. Weller. 1998. Natural plant protection by 2,4-diacetylphloroglucinol-producing Pseudomonas spp. in take-all decline soils. Mol. Plant Microbe Interact. 11:144-152. [Google Scholar]
- 38.Ramette, A., Y. Moenne-Loccoz, and G. Defago. 2003. Prevalence of fluorescent pseudomonads producing antifungal phloroglucinols and/or hydrogen cyanide in soils naturally suppressive or conducive to tobacco black root rot. FEMS Microbiol. Ecol. 44:35-43. [DOI] [PubMed] [Google Scholar]
- 39.Riley, L. W., et al. 1983. Hemorrhagic colitis associated with a rare Escherichia coli serotype. N. Engl. J. Med. 308:681-685. [DOI] [PubMed] [Google Scholar]
- 40.Ritchie, J. M., et al. 2003. A stable bioluminescent construct of Escherichia coli O157:H7 for hazard assessments of long-term survival in the environment. Appl. Environ. Microbiol. 69:3359-3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rotenberg, D., L. R. Cooperband, and A. Stone. 2005. Dynamic relationships between soil properties and foliar disease as affected by annual additions of organic amendment to a sandy-soil vegetable production system. Soil Biol. Biochem. 37:1343-1357. [Google Scholar]
- 42.Rouxel, F., C. Alabouvette, and J. Louvet. 1977. Recherches sur la résistance des sols aux maladies II—incidence de traitements thermiques sur la résistance microbiologique d'un sol à la Fusariose vasculaire du melon. Ann. Phytopathol. 9:183-192. [Google Scholar]
- 43.Scheuerell, S. J., D. M. Sullivan, and W. F. Mahaffee. 2005. Suppression of seedling damping-off caused by Pythium ultimum, P. irregulare, and Rhizoctonia solani in container media amended with a diverse range of Pacific Northwest compost sources. Phytopathology 95:306-315. [DOI] [PubMed] [Google Scholar]
- 44.Smith, K. L., D. A. Todhunter, and P. S. Schoenberger. 1985. Environmental mastitis: cause, prevalence, prevention. J. Dairy Sci. 68:1531-1553. [DOI] [PubMed] [Google Scholar]
- 45.Smith, D., et al. 2001. Ecological relationships between the prevalence of cattle shedding Escherichia coli O157:H7 and characteristics of the cattle or conditions of the feedlot pen. J. Food Prot. 64:1899-1903. [DOI] [PubMed] [Google Scholar]
- 46.Stanford, K., et al. 2005. Ecology of Escherichia coli O157:H7 in commercial dairies in southern Alberta. J. Dairy Sci. 88:4441-4451. [DOI] [PubMed] [Google Scholar]
- 47.Stone, A. G., et al. 2003. Effect of organic amendments on soilborne and foliar diseases in field-grown snap bean and cucumber. Plant Dis. 87:1037-1042. [DOI] [PubMed] [Google Scholar]
- 48.Thomas, G. W. 1996. Soil pH and soil acidity, p. 475-490. In Methods of soil analysis. Part 3. Chemical methods. Soil Science Society of America, Madison, WI.
- 49.Weisburg, W. G., S. M. Barns, D. A. Pelletier, and D. J. Lane. 1991. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173:697-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Weller, D. M., J. M. Raaijmakers, B. B. Gardener, and L. S. Thomashow. 2002. Microbial populations responsible for specific soil suppressiveness to plant pathogens. Annu. Rev. Phytopathol. 40:309-348. [DOI] [PubMed] [Google Scholar]
- 51.Westphal, A. 2005. Detection and description of soils with specific nematode suppressiveness. J. Nematol. 37:121-130. [PMC free article] [PubMed] [Google Scholar]
- 52.Westphal, A., and J. O. Becker. 2001. Components of soil suppressiveness against Heterodera schachtii. Soil Biol. Biochem. 33:9-16. [Google Scholar]
- 53.Westphal, A., G. T. Browne, and S. Schneider. 2002. Evidence for biological nature of the grape replant problem in California. Plant Soil 242:197-203. [Google Scholar]
- 54.Yin, B., L. Valinsky, X. Gao, J. O. Becker, and J. Borneman. 2003. Bacterial rRNA genes associated with soil suppressiveness against the plant-parasitic nematode Heterodera schachtii. Appl. Environ. Microbiol. 69:1573-1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yin, B., L. Valinsky, X. Gao, J. O. Becker, and J. Borneman. 2003. Identification of fungal rDNA associated with soil suppressivenesss against Heterodera schachtii using oligonucleotide fingerprinting. Phytopathology 93:1006-1013. [DOI] [PubMed] [Google Scholar]
- 56.Zdanowicz, M., J. A. Shelford, C. B. Tucker, D. M. Weary, and M. A. von Keyserlingk. 2004. Bacterial populations on teat ends of dairy cows housed in free stalls and bedded with either sand or sawdust. J. Dairy Sci. 87:1694-1701. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.