Abstract
Listeria monocytogenes strains (n = 117) were screened for the presence of stress survival islet 1 (SSI-1). SSI-1+ strains (32.5%) belonged mainly to serotypes 1/2c, 3b, and 3c. All sequence type 121 (ST-121) strains included (n = 7) possessed homologues to Listeria innocua genes lin0464 and lin0465 instead of SSI-1.
The genus Listeria comprises eight species: L. monocytogenes, L. innocua, L. ivanovii, L. welshimeri, L. seeligeri, L. grayi, and the newly described L. rocourtiae and L. marthii (7, 11, 18). From a human perspective, the food-borne pathogen L. monocytogenes is the most important species of this genus. Recently, a stress survival islet (stress survival islet 1 [SSI-1]) has been identified in L. monocytogenes (29). Deletion mutants showed impaired growth at low pH and high salt concentrations. The islet was present in about 50% of tested L. monocytogenes strains, mainly those not belonging to serogroup 4. According to the sequence of L. monocytogenes EGD-e (serotype 1/2a), the islet contains five genes, lmo0444, lmo0445, lmo0446 (pva), lmo0447 (gadD1), and lmo0448 (gadT1), positioned between two genes highly conserved across different Listeria spp. An open reading frame (ORF) transcribed in the opposite direction is present at this location in L. monocytogenes strains without the islet (e.g., strain F2365, serotype 4b), and in L. innocua CLIP 11262, two genes transcribed in opposite directions (lin0464 and lin0465) are present at the same location (29). L. welshimeri SLCC 5334 does not contain any genes at this position. In L. seeligeri SLCC 3954, this region was similar to that in L. welshimeri SLCC 5334, with no open reading frame and with small differences in the lengths of the intergenic region, i.e., 281 bp (L. welshimeri FN557490) and 166 bp (L. seeligeri AM263198). There was 63% identity between a 46-bp stretch at the 5′ end in L. welshimeri and that in L. seeligeri, both of which were followed by an L. welshimeri-specific portion of 112 bp and a 123-bp stretch at the 3′ end with 73.2% identity.
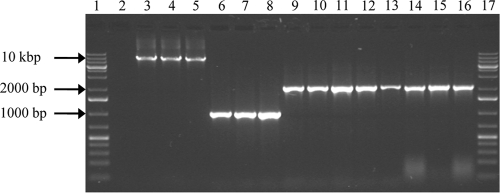
In the present study, a collection of 117 L. monocytogenes strains, including isolates from cheese dairies, meat and meat products, fish, and veterinary and human infections, was screened by PCR for the presence of SSI-1 (Table 1; also see Table S1 in the supplemental material) (29). Serotypes were determined by classical serotyping and by a multiplex PCR method for grouping strains into four major serotype-related groups (9, 31). The predicted 9.7-kbp (SSI-1+) and 1.1-kbp (SSI-1−) fragments were observed in 32.5% and 61.5% of strains, respectively (Fig. 1; Table 2). SSI-1 was present in the majority of 1/2c, 3b, and 3c strains tested but in only one strain each of serotypes 4a and 4b. A single serotype 4c strain was included in the survey, and it contained SSI-1. The majority of serotype 4b, 1/2a, and 4a strains, as well as all strains with ambiguous serotype 4d or 4e, lacked SSI-1. Earlier findings suggest that SSI-1 is more prevalent in non-serogroup 4 strains, which was confirmed by our results: 50% of non-serogroup 4 strains but only 7.5% of serogroup 4 strains contained SSI-1 (χ2 = 20.33, P < 0.0005) (29). However, seven strains yielded PCR fragments of about 2.2 kbp (Table 2), all of them serotype 1/2a and representing 25% of tested strains belonging to this serotype (Fig. 1). The sequence of this 2.2-kbp fragment was determined for one strain (CDL67) by primer walking (Macrogen Inc., Seoul, South Korea) using primers lmo0443 Fwd and lmo0449 Rev (Table 1). A BLAST search revealed 95% identity with L. innocua CLIP 11262 (GenBank accession no. AL596165). The sequence contained homologues to lin0464 (98% identity, 15 substitutions, five of them nonsynonymous leading to five amino acid substitutions) and lin0465 (94% identity, 35 substitutions, 16 of them nonsynonymous leading to 14 amino acid substitutions). Based on sequence information of L. innocua CLIP 11262, primers lmo0443 Fwd and lmo0449 Rev were expected to amplify a 2,166-bp fragment. Indeed, a survey of nine L. innocua strains from culture collections (DSM 20649, NCTC 10528 and 12210, and CIP 78.44, 79.45, 80.11, 80.12, 106065, and 107775) and 27 L. innocua strains obtained from dairy plants yielded a 2.2-kbp fragment in all cases (Fig. 1). Thus, the seven L. monocytogenes strains yielding PCR fragments of about 2.2 kbp contained a region common in L. innocua at that genomic site. PCR analysis targeting the 16S rRNA gene, hly, iap, prfA, inlA, and inlB yielded results typical for L. monocytogenes (1, 2, 14, 28, 32), whereas PCR results for the L. innocua-specific targets lin0198 and the noncoding intergenic region between lin0454 and lin0455 were negative (15). Pulsed-field gel electrophoresis (PFGE) profiles (ApaI and AscI) of these L. monocytogenes strains were similar to each other (Fig. 2) (13). Multilocus sequence typing (MLST) was performed as outlined on the Institute Pasteur Listeria monocytogenes MLST Database website (www.pasteur.fr/mlst) and revealed a profile consistent with sequence type 121 (ST-121): abcZ-7, bglA-6, cat-8, dapE-8, dat-6, ldh-37, and lhkA-1 (26).
TABLE 1.
Oligonucleotides used in this study
| Primer or probe | Sequence (5′-3′) | Target(s) | Reference |
|---|---|---|---|
| lmo0443 Fwd | GGC ACA ATG AGC GAA TTG | SSI-1 | 29 |
| lmo0449 Rev | GTC CTT CTG GAA CAT TGC | SSI-1 | 29 |
| gadD1 F | GGT ATT GTG GGT ATT CTG G | gadD1 | 29 |
| gadD1 R | CTG ACC GAT AAT CTG ACT C | gadD1 | 29 |
| LI1 | CTC CAT AAA GGT GAC CCT | 16S rRNA gene | 1 |
| U1 | CAG CAG CCG CGG TAA TAC | 16S rRNA gene | 1 |
| LM1 | CCT AAG ACG CCA ATC GAA | hly | 1 |
| LM2 | AAG CAC TTG CAA CTG CTC | hly | 1 |
| Siwi2 | TAA CTG AGG TAG CGA GCG AA | iap | 2 |
| Ino2 | ACT AGC ACT CCA GTT GTT AAA C | iap | 2 |
| MonoA | CAA ACT GCT AAC ACA GCT ACT | iap | 2 |
| Mugra | CCA GCA GTT TCT AAA CCT GCT | iap | 2 |
| Lis1B | TTA TAC GCG ACC GAA GCC AAC | iap | 2 |
| inlAF1 | TTA CAT CAG TCC CCT AGC AGG T | inlA | 32 |
| inlAR1 | TCC AAT AGT GAC AGG TTG GCT A | inlA | 32 |
| inlB-forward | CTC GCA CCG CTG TAA AGC T | inlB | 14 |
| inlB-reverse | TTA TTT CTG TGC CCT TAA ATT A | inlB | 14 |
| LIP1 | GAT ACA GAA ACA TCG GTT GGC | prfA | 28 |
| LIP2 | GTG TAA TCT TGA TGC CAT CAG G | prfA | 28 |
| LIP probe 2 | FAM-CAG GAT TAA AAG TTG ACC GCA-MGBa | prfA | 28 |
| Lin0198F | ATG AAC AAA TTA GTT AGT CAA AGT AAT G | lin0198 | 15 |
| Lin0198R | TAT CGA TGT CTT GAG GTC ACA CAA AGT TC | lin0198 | 15 |
| LinNCR1F | GGA TTT GGT AAA TTA TAC AAA GGT TTT AAG | lin0454 and lin0455 | 15 |
| LinNCR1R | TGC TTC TTG GCA TTT TAG TAA TCT TTC | Intergenic region | 15 |
| inlAseqF1 | CAC CAT TGG AAA AGG AAC GA | inlA | This study |
| seq02 | TGT GAC CTT CTT TTA CGG GC | inlA | 28a |
FAM, 6-carboxyfluorescein; MGB, minor groove binder.
FIG. 1.
PCR analysis of the lmo0443 and lmo0449 intergenic region of L. monocytogenes. Lanes 1 and 17, 1-kb Plus DNA ladder (Fermentas International Inc., Burlington, Ontario, Canada); lane 2, no-template control; lanes 3 to 13, L. monocytogenes strains (lane 3, CDL73; lane 4, CDL74; lane 5, CDL75; lane 6, CDL201; lane 7, CDL211; lane 8, CDL218; lane 9, CDL64; lane 10, CDL67; lane 11, L56/65; lane 12, L58/55; lane 13, L37/35); lanes 14 to 16, L. innocua strains (lane 14, CDL25; lane 15, CDL26; lane 16, CDL192).
TABLE 2.
Amplicons observed when screening L. monocytogenes strains of different serotypes for the presence of SSI-1
| Serotype | No. (%) of isolates with: |
||
|---|---|---|---|
| 9.7-kbp fragment | 2.2-kbp fragment | 1.1-kbp fragment | |
| 1/2a | 6 (21.4) | 7 (25.0) | 18 (64.3) |
| 1/2b | 8 (50.0) | 0 (0.0) | 8 (50.0) |
| 1/2c | 11 (78.6) | 0 (0.0) | 3 (21.4) |
| 3a | 2 (50.0) | 0 (0.0) | 2 (50.0) |
| 3b | 3 (60.0) | 0 (0.0) | 2 (40.0) |
| 3c | 4 (80.0) | 0 (0.0) | 1 (20.0) |
| 4a | 1 (20.0) | 0 (0.0) | 4 (80.0) |
| 4b | 1 (3.6) | 0 (0.0) | 27 (96.4) |
| 4c | 1 (100.0) | 0 (0.0) | 0 (0.0) |
| 4d/e | 0 (0.0) | 0 (0.0) | 5 (100.0) |
| 4e | 0 (0.0) | 0 (0.0) | 1 (100.0) |
| 7 | 1 (50.0) | 0 (0.0) | 1 (50.0) |
| All | 38 (32.5) | 7 (6.0) | 72 (61.5) |
FIG. 2.
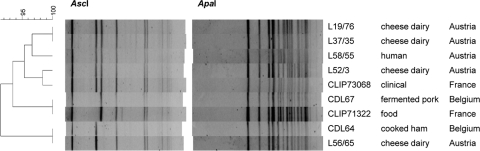
PFGE analysis of L. monocytogenes strains harboring lin0464 and lin0465 homologues. Cluster analysis was performed with Fingerprinting II software (Bio-Rad, Hercules, CA) using the following settings: 0.2% optimization, 0.5% position tolerance, and the Dice similarity coefficient. For dendrogram construction, the UPGMA (unweighted-pair group method using average linkages) algorithm was used. Strain designations, sources, and countries of origin are given on the right, and percentages of similarity are indicated on the left.
The ST-121 strains analyzed in this study were isolated in Austria and Belgium from different ecological niches, including food and human cases, over several years (Table 3). In two instances, two strains each represent clones which were consistently isolated from the same dairy plant over a course of at least 3 years. Isolation of L. monocytogenes ST-121 strains has also been reported in France, Italy, and Spain (25, 26, 30) (Table 3). Two French strains, CLIP 73068 and CLIP 71322, were available for testing and yielded the 2.2-kbp amplicon with primers lmo0443 Fwd and lmo0449 Rev. Both strains had PFGE profiles similar to those of the other ST-121 strains, with those of CLIP 71322 being identical to those of CDL67 (Fig. 2).
TABLE 3.
L. monocytogenes ST-121 strains with lin0464 and lin0465 homologues identified in this study and ST-121 strains reported by others
| Strain | Country of origin | Yr of isolation | Source | Reference |
|---|---|---|---|---|
| CDL64 | Belgium | 2001-2006 | Cooked ham | This study |
| CDL67 | Belgium | 2001-2006 | Salami | This study |
| L52/3 | Austria | 1998 | Cheese dairy | This study |
| L56/65 | Austria | 2000 | Cheese dairy | This study |
| L58/55 | Austria | 2001 | Clinical isolate | This study |
| L19/76 | Austria | 1997 | Cheese dairy | This study |
| L37/35 | Austria | 1999 | Cheese dairy | This study |
| CLIP 71322 | France | 1996 | Food | 26 |
| CLIP 73068 | France | 1996 | Clinical isolate | 26 |
| 829 | Spain | 2000 | Pork | 30 |
| 21-P | Italy | 1993-2004 | Meat product | 25 |
| 174 | Italy | 1993-2004 | Ready-to-eat product | 25 |
| 350 | Italy | 1993-2004 | Meat product | 25 |
| 170 | Italy | 1993-2004 | Meat product | 25 |
| 204 | Italy | 1993-2004 | Meat product | 25 |
| 90-P | Italy | 1993-2004 | Cured fish | 25 |
| 87 | Italy | 1993-2004 | Soil | 25 |
| 206 | Italy | 1993-2004 | Meat product | 25 |
| 159 | Italy | 1993-2004 | Meat product | 25 |
| 115 | Italy | 1993-2004 | Mammalian stool | 25 |
A transition from C to T at nucleotide position 1474 of the inlA gene has been reported in two French ST-121 strains. This transition resulted in a premature stop codon (PMSC) and a truncated form of InlA, which is secreted rather than anchored to the cell wall (22, 26). Sequencing of the respective region in our strains yielded an identical transition (Table 1). Several mutations associated with PMSCs have been described for this gene (26). Although inlA PMSCs yield virulence-attenuated strains, such strains have been isolated from human cases. Immunosuppression has been suggested to be a risk factor present in these cases (21, 22). One of our ST-121 strains is also a human isolate. ST-121 represents an L. monocytogenes clone which is spatially, temporally, and ecologically widespread.
A BLAST search of ST-121 homologues to lin0464 and lin0465 revealed homologues in other Firmicutes as well (Fig. 3). The DNA-based identity ranged from 65% to 79%. Protein-based identities were similar to the DNA-based data (data not shown). In Acetivibrio cellulolyticus, Anaerostipes caccae, Eubacterium limosum, Clostridium asparagiforme, and an Enterococcus faecalis plasmid, homologues to both genes were present. The arrangement of these genes was identical to that of lin0464 and lin0465: two successive ORFs with opposing directions of transcription. lin0464 belongs to the GntR family of transcriptional regulators, with a winged helix-turn-helix DNA-binding domain. lin0465 is characterized by a type 1 glutamine amidotransferase-like domain found in a subgroup of proteins similar to the ATP-independent intracellular protease PfpI from Pyrococcus furiosus (12). The conservation of the arrangement of both genes in different species suggests that they are a functional unit and that lin0464 or the homologues thereof might act as transcriptional regulators for the intracellular protease encoded by lin0465 or the lin0465 homologue. The substrate of the Pyrococcus furiosus protease has not been identified so far. In Pseudomonas aeruginosa, a similar protease provides general stress protection (27).
FIG. 3.
Maximum-parsimony tree of the BLAST hits against the lin0464 homologue (top) and the lin0465 homologue (bottom) of L. monocytogenes ST-12. The tree was generated by using MEGA (v. 4.0) software (16). The scale bar represents a distance of 5%. Accession numbers are from GenBank. Bootstrap values of 500 trees are indicated as percent confidence values for the branching. GenBank accession numbers, query coverages (C), and identities (I) are given in parentheses.
The presence of homologues to lin0464 and lin0465 in the conjugative E. faecalis plasmid pBEE99 hints at the mobility of these genes. In this plasmid, these genes are flanked upstream by a transcriptional regulator also belonging to the GntR family and a transposase and downstream by a site-specific recombinase and a transposase. These genes are located in a 5,000-bp region containing genes with 54 to 60% identity to genes found in species other than E. faecalis, whereas the rest of the plasmid contains almost exclusively genes associated with E. faecalis (6). The sequenced ST-121 strains possess a short inverted repeat 66 bp upstream of the lin0464 homologue (5′-AAGATTTT-3′) and 73 bp downstream of the lin0465 homologue (5′-AAAATCTT-3′), which might indicate mobility of that genomic region. However, this inverted repeat is not present in the L. innocua CLIP 11262 genome.
In Clostridium hathewayi and a Clostridiales and an Erysipelotrichaceae bacterium, a lin0464 homologue was present but was followed by an isochorismatase protein instead of a homologue to lin0465. On the other hand, L. seeligeri SLCC 3954 contains only a lin0465 homologue. The 5′ end of the homologue region is part of lse_0574, and the rest is part of lse_0575. The homologue to lmo_0443 in L. seeligeri is lse_0377. Thus, the lin0465 homologue region is integrated in this L. seeligeri strain in a genomic region different from that in L. innocua or L. monocytogenes ST-121 strains.
Cross-reactivity of a PCR system targeting lin0464, which was initially thought to be specific for L. innocua, has been reported for four L. monocytogenes serotype 4c strains belonging to phylogenetic lineage IIIA (19, 20). Thus, it seems that the presence of a region commonly found in L. innocua at that genomic site in L. monocytogenes is not restricted to ST-121 strains.
With the exception of L. marthii, as the closest relative to L. monocytogenes, L. monocytogenes and L. innocua are more closely related to each other than to the other Listeria species and coexist in various ecological niches (4, 11). Comparative genomics and DNA array data suggest that L. innocua evolved by successive gene loss from an ancestor of L. monocytogenes serogroup 4 strains (10). Recently, L. monocytogenes 4a strains that harbor L. innocua-specific lin0372 and lin1073 but not lin0464 and share some gene deletions with L. innocua have been identified (5). On the other hand, L. innocua strains containing L. monocytogenes-specific genes have been identified as well. These genes include Listeria pathogenicity island 1 (LIPI-1) genes, inlA, partial inlB, and gtcA, which is involved in teichoic acid glycosylation and characteristic of L. monocytogenes serogroup 4 strains (17, 32). In addition, a 10-gene MLST scheme provided evidence for interspecific recombination between these two species (7). Serotype 4c belongs to lineage IIIA, whereas serotype 1/2a belongs to lineage II. Recombination rates are higher in lineage II strains, and a large number of imports from lineage IIIA to lineage II was observed (8, 23, 24). In addition, SSI-1 is located within a 616-kbp region in the first third of the genome, which may be a hot spot for horizontal gene transfer (3). Taking into account the high degree of identity to lin0464 and lin0465, the homologous genes in L. monocytogenes ST-121 strains might have been acquired rather recently from L. innocua. It is tempting to speculate that this might have happened not directly but by recombination with lin0464-positive L. monocytogenes serotype 4c strains belonging to lineage IIIA. An alternative scenario might be independent acquisition from an external source or a common ancestor harboring the insert now common in L. innocua, albeit these do not seem to be very likely. Comparison of the lin0464 and lin0465 homologues and the flanking regions of lin0464-positive L. monocytogenes 4c strains to the respective sequences in L. innocua and L. monocytogenes ST-121 strains might aid in reconstructing the evolutionary events leading to the present situation. The lower degree of identity, the different position in the genome, and the lack of a lin0464 homologue suggest that the lin0465 homologue in L. seeligeri SLCC 3954 might have been acquired in the more distant past or from a different source than the lin0465 homologue in L. innocua and L. monocytogenes ST-121 strains.
In conclusion, Listeria strains harboring lin0464 and lin0465 or their homologues, e.g., L. innocua, L. monocytogenes serovar 4c, and L. monocytogenes serovar 1/2a ST-121 strains, rarely cause diseases and are associated with the environment. It is likely that lin0464 and lin0465 and their homologues contribute to the fitness of these bacteria in the environment. These genes may also have contributed to the broad spatiotemporal distribution of the ST-121 clone. Thus, further investigations should focus on the function of lin0464 and lin0465 in L. innocua and their homologues in L. monocytogenes.
Nucleotide sequence accession number.
The sequence of the 2.2-kbp fragment determined for L. monocytogenes CDL67 was deposited in GenBank under accession number HQ179545.
Supplementary Material
Acknowledgments
This work was supported by the Christian Doppler Laboratory for Molecular Food Analysis, Vienna, Austria.
We thank H. Hof, K. Houf, T. Kostic, J. McLauchlin, M. Schmid, J. Schrenzel, and M. Lecuit for kindly providing and collecting L. monocytogenes strains, and we thank B. Auer for serotyping.
Footnotes
Published ahead of print on 14 January 2011.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Border, P. M., J. J. Howard, G. S. Plastow, and K. W. Siggens. 1990. Detection of Listeria species and Listeria monocytogenes using polymerase chain reaction. Lett. Appl. Microbiol. 11:158-162. [DOI] [PubMed] [Google Scholar]
- 2.Bubert, A., et al. 1999. Detection and differentiation of Listeria spp. by a single reaction based on multiplex PCR. Appl. Environ. Microbiol. 65:4688-4692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buchrieser, C., et al. 2003. Comparison of the genome sequences of Listeria monocytogenes and Listeria innocua: clues for evolution and pathogenicity. FEMS Immunol. Med. Microbiol. 35:207-213. [DOI] [PubMed] [Google Scholar]
- 4.Chen, J., et al. 2010. Internalin profiling and multilocus sequence typing suggests four Listeria innocua subgroups with different evolutionary distances from Listeria monocytogenes. BMC Microbiol. 10:97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen, J., et al. 2009. Listeria monocytogenes serovar 4a is a possible evolutionary intermediate between L. monocytogenes serovars 1/2a and 4b and L. innocua. J. Microbiol. Biotechnol. 19:238-249. [PubMed] [Google Scholar]
- 6.Coburn, P. S., et al. 2010. A novel conjugative plasmid from Enterococcus faecalis E99 enhances resistance to ultraviolet radiation. Plasmid 64:18-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.den Bakker, H. C., B. N. Bundrant, E. D. Fortes, R. H. Orsi, and M. Wiedmann. 2010. A population genetics-based and phylogenetic approach to understanding the evolution of virulence in the genus Listeria. Appl. Environ. Microbiol. 76:6085-6100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.den Bakker, H. C., X. Didelot, E. D. Fortes, K. K. Nightingale, and M. Wiedmann. 2008. Lineage specific recombination rates and microevolution in Listeria monocytogenes. BMC Evol. Biol. 8:277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Doumith, M., C. Buchrieser, P. Glaser, C. Jacquet, and P. Martin. 2004. Differentiation of the major Listeria monocytogenes serovars by multiplex PCR. J. Clin. Microbiol. 42:3819-3822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Doumith, M., et al. 2004. New aspects regarding evolution and virulence of Listeria monocytogenes revealed by comparative genomics and DNA arrays. Infect. Immun. 72:1072-1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Graves, L. M., et al. 2010. Listeria marthii sp. nov., isolated from the natural environment, Finger Lakes National Forest. Int. J. Syst. Evol. Microbiol. 60:1280-1288. [DOI] [PubMed] [Google Scholar]
- 12.Halio, S. B., I. I. Blumentals, S. A. Short, B. M. Merrill, and R. M. Kelly. 1996. Sequence, expression in Escherichia coli, and analysis of the gene encoding a novel intracellular protease (PfpI) from the hyperthermophilic archaeon Pyrococcus furiosus. J. Bacteriol. 178:2605-2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Halpin, J. L., N. M. Garrett, E. M. Ribot, L. M. Graves, and K. L. Cooper. 2010. Re-evaluation, optimization, and multilaboratory validation of the PulseNet-standardized pulsed-field gel electrophoresis protocol for Listeria monocytogenes. Foodborne Pathog. Dis. 7:293-298. [DOI] [PubMed] [Google Scholar]
- 14.Jiang, L., et al. 2008. Virulence characterization and genotypic analyses of Listeria monocytogenes isolates from food and processing environments in eastern China. Int. J. Food Microbiol. 121:53-59. [DOI] [PubMed] [Google Scholar]
- 15.Johnson, J., et al. 2004. Natural atypical Listeria innocua strains with Listeria monocytogenes pathogenicity island 1 genes. Appl. Environ. Microbiol. 70:4256-4266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kumar, S., N. Masatoshi, J. Dudley, and K. Tamura. 2008. MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief. Bioinform. 9:299-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lan, Z., F. Fiedler, and S. Kathariou. 2000. A sheep in wolf's clothing: Listeria innocua strains with teichoic acid-associated surface antigens and genes characteristic of Listeria monocytogenes sergroup 4. J. Bacteriol. 182:6161-6168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leclercq, A., et al. 2010. Listeria rocourtiae sp. nov. Int. J. Syst. Evol. Microbiol. 60:2210-2214. [DOI] [PubMed] [Google Scholar]
- 19.Liu, D., A. J. Ainsworth, F. W. Austin, and M. L. Lawrence. 2003. Identification of Listeria innocua by PCR targeting a putative transcriptional regulator gene. FEMS Microbiol. Lett. 223:205-210. [DOI] [PubMed] [Google Scholar]
- 20.Liu, D., M. L. Lawrence, and A. D. Hitchins. 2008. Molecular characterization of Listeria monocytogenes strains harbouring Listeria innocua putative transcriptional regulator gene lin0464. J. Rapid Methods Automat. Microbiol. 16:412-427. [Google Scholar]
- 21.Nightingale, K. K., et al. 2008. inlA premature stop codons are common among Listeria monocytogenes isolates from foods and yield virulence-attenuated strains that confer protection against fully virulent strains. Appl. Environ. Microbiol. 74:6570-6583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nightingale, K. K., K. Windham, K. E. Martin, M. Yeung, and M. Wiedmann. 2005. Select Listeria monocytogenes subtypes commonly found in foods carry distinct nonsense mutations in inlA, leading to expression of truncated and secreted internalin A, and are associated with a reduced invasion phenotype for human intestinal epithelial cells. Appl. Environ. Microbiol. 71:8764-8772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Orsi, R. H., et al. 2008. Lineage specific recombination and positive selection in coding and intragenic regions contributed to evolution of the main Listeria monocytogenes virulence gene cluster. Infect. Genet. Evol. 8:566-576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Orsi, R. H., Q. Sun, and M. Wiedmann. 2008. Genome-wide analyses reveal lineage specific contributions of positive selection and recombination to the evolution of Listeria monocytogenes. BMC Evol. Biol. 8:233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parisi, A., et al. 2010. Amplified fragment-length polymorphism and multi-locus sequence typing for high-resolution genotyping of Listeria monocytogenes from foods and the environment. Food Microbiol. 27:101-108. [DOI] [PubMed] [Google Scholar]
- 26.Ragon, M., et al. 2008. A new perspective on Listeria monocytogenes evolution. PLoS Pathog. 4:e1000146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rodríguez-Rojas, A., and J. Blàzquez. 2009. The Pseudomonas aeruginosa pfpI gene plays an antimutator role and provides general stress protection. J. Bacteriol. 191:844-850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rossmanith, P., M. Krassnig, M. Wagner, and I. Hein. 2006. Detection of Listeria monocytogenes in food using a combined enrichment/real-time PCR method targeting the prfA gene. Res. Microbiol. 157:763-771. [DOI] [PubMed] [Google Scholar]
- 28a.Rousseaux, S., M. Olier, J. P. Lemaître, P. Piveteau, and J. Guzzo. 2004. Use of PCR-restriction fragment length polymorphism of inlA for rapid screening of Listeria monocytogenes strains deficient in the ability to invade Caco-2 cells. Appl. Environ. Microbiol. 70:2180-2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ryan, S., M. Begley, C. Hill, and C. G. M. Gahan. 2010. A five-gene stress survival islet (SSI-1) that contributes to the growth of Listeria monocytogenes in suboptimal conditions. J. Appl. Microbiol. 109:984-995. [DOI] [PubMed] [Google Scholar]
- 30.Salcedo, C., L. Arreaza, B. Alcalá, L. de la Fuente, and J. A. Vázquez. 2003. Development of a multilocus sequence typing method for analysis of Listeria monocytogenes clones. J. Clin. Microbiol. 41:757-762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Seeliger, H. P. R., and K. Höhne. 1979. Serotyping of Listeria monocytogenes and related species, p. 31-49. In T. Bergan and J. R. Norris (ed.), Methods in microbiology, vol. 13. Academic Press, London, United Kingdom. [Google Scholar]
- 32.Volokhov, D. V., et al. 2007. The presence of the internalin gene in natural atypically hemolytic Listeria innocua strains suggests descent from L. monocytogenes. Appl. Environ. Microbiol. 73:1928-1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.