Abstract
Coxiella burnetii is the etiological agent of Q fever. Currently, the Netherlands is facing the largest Q fever epidemic ever, with almost 4,000 notified human cases. Although the presence of a hypervirulent strain is hypothesized, epidemiological evidence, such as the animal reservoir(s) and genotype of the C. burnetii strain(s) involved, is still lacking. We developed a single-nucleotide-polymorphism (SNP) genotyping assay directly applicable to clinical samples. Ten discriminatory SNPs were carefully selected and detected by real-time PCR. SNP genotyping appeared to be highly suitable for discrimination of C. burnetii strains and easy to perform with clinical samples. With this new method, we show that the Dutch outbreak is caused by at least 5 different C. burnetii genotypes. SNP typing of 14 human samples from the outbreak revealed the presence of 3 dissimilar genotypes. Two genotypes were also present in livestock at 9 farms in the outbreak area. SNP analyses of bulk milk from 5 other farms, commercial cow milk, and cow colostrum revealed 2 additional genotypes that were not detected in humans. SNP genotyping data from clinical samples clearly demonstrate that at least 5 different C. burnetii genotypes are involved in the Dutch outbreak.
Coxiella burnetii is a Gram-negative, obligate intracellular bacterium and the causative agent of Q fever, a widespread zoonotic infectious disease (2, 11). Human infections are often associated with affected livestock, especially goats and sheep (20). Its primary route of infection is through inhalation, with less than 10 organisms being sufficient for human infection (2). Because of its high infectivity and extreme resistance to heat, UV radiation, disinfectants, and desiccation, C. burnetii is listed as a category B biowarfare agent (15).
Sixty percent of patients exposed to C. burnetii seroconvert asymptomatically, whereas 40% develop a disease known as acute Q fever, which in general presents as a mild flu-like disease. Some patients develop severe pneumonia and may need hospitalization. The inoculum size has been associated with the incubation time. Primary C. burnetii infection may progress to chronic Q fever, presenting as endocarditis or vascular disease in 78% and 9% of chronic Q fever patients, respectively (19).
Currently, there is a Q fever epidemic of unprecedented magnitude ongoing in the southern part of the Netherlands, with almost 4,000 human cases being diagnosed since 2007. Fifty-nine percent of notified human Q fever cases in 2009 lived within a 3-mile radius of an infected dairy goat or dairy sheep farm, while only 12% (roughly 1 million people) of the Dutch population resided within these zones (26). The most affected province is Noord-Brabant, located in the south of the Netherlands. Noord-Brabant covers 1,900 square miles and has 2.4 million inhabitants and about 6.4 million livestock (80,000 sheep, 135,000 goats, 660,000 cows, and 5.5 million pigs) (3). Q fever was diagnosed at 74 dairy goat farms and 2 dairy sheep farms, out of the total of 360 dairy goat farms and 40 dairy sheep farms with more than 50 animals in the Netherlands (26).
To date, several techniques for genotyping of C. burnetii have been described: pulsed-field gel electrophoresis, plasmid nested PCR, PCR-restriction fragment length polymorphism, infrequent restriction site PCR (IRS-PCR), multispacer sequence typing, multiple-locus variable-number tandem-repeat analysis (MLVA), and, recently, matrix-assisted laser desorption ionization-time-of-flight mass spectrometry (1, 5, 8, 9, 16, 22, 23, 29). Most of these techniques are limited in their sensitivity and rely on enrichment of the bacterium by culture before it can be genotyped. C. burnetii culture is mostly performed by the use of tissue culture under biosafety level 3 conditions (18), for which the necessities are often not available in peripheral hospital laboratories. In addition, culturing of C. burnetii is bound by strict government regulations due to its infectious potential and status as a category B biowarfare agent (15). A drawback of IRS-PCR and other gel-based systems is that these methods are not well-suited for interlaboratory standardization and database building (1). The most sensitive technique is probably MLVA typing, based on amplification and detection—often with a capillary sequencer—of short DNA repeats on different loci (1, 22). These repeats are often hypervariable and can therefore be very informative with regard to strain differentiation. However, the value of small repeat differences is often not clear (1, 17, 25).
In 2009, a small-scale genotyping study of 3 MLVA loci including 5 patients, 3 sheep, and 3 lambs suggested a link between one human Q fever case and a sheep farm. Most of the genotypes in this study differed only in a single repeat. It was therefore suggested that the C. burnetii types might represent microvariants of a hypervirulent strain that has been introduced in the Dutch animal population (4, 12). However, thorough genotyping data to provide epidemiological insight into the Dutch Q fever outbreak are still missing.
A new approach to overcome the problems of currently used typing techniques is single-nucleotide-polymorphism (SNP)-based typing. SNP methods are rapid, sensitive, easy to perform, and unambiguous in result interpretation (6, 7, 10, 14). We assumed that SNP genotyping might be well-suited for genotyping of C. burnetii in an outbreak setting. Therefore, we set out to develop an SNP genotyping assay for C. burnetii directly applicable to human and animal samples.
MATERIALS AND METHODS
SNP selection and real-time PCR design.
Sequences of approximately 100,000 bp of the complete genomes of C. burnetii strains RSA493 (Nine Mile strain, GenBank accession no. NC_002971), RSA331 (Henzerling strain, GenBank accession no. NC_010117), CbuG_Q212 (GenBank accession no. NC_011527), Cbuk_Q154 (GenBank accession no. NC_011528), and 5J108 111 Dugway (GenBank accession no. NC_009727) were aligned using Geneouis software (version 4.8.5; Biomatters Ltd., Auckland, New Zealand). The resulting consensus sequence was screened for strain-specific SNPs. The Basic Local Alignment Search Tool (BLAST) from the National Center for Biotechnology Information was used to confirm whether these SNPs could be of discriminatory value and to determine the SNP locus.
Using the custom TaqMan assay design tool (Applied Biosystems, Foster City, CA), we designed real-time PCRs for amplification and detection of the SNPs (Table 1).
TABLE 1.
Oligonucleotide sequences for real-time SNP amplification
| SNP | Sequencea |
|||
|---|---|---|---|---|
| Forward primer | Reverse primer | Probe 1 (5′-VIC/3′-NFQ-MGB) | Probe 2 (5′-FAM/3′-NFQ-MGB) | |
| 769 | GCA-ACA-CAA-TGC-CAT-GAA-CGA-ATT | AAT-TGA-ATA-TCG-TCA-ATT-AAC-AAC-GCA-TTT | AGA-GCG-GTA-AAA-GC | AAG-AGC-GAT-AAA-AGC |
| 2287 | GCG-TTT-ACT-CGA-GGA-GGA-TGA-AG | GAA-ACA-CCG-ATG-TGA-TTA-TTC-CCA-ATT-C | CTG-GCC-GCT-ATC-C | CTG-GCC-ACT-ATC-C |
| 4439 | TCT-GCC-GCC-GAA-GTT-ATT-ATG-AC | CCG-CCG-GAA-ACT-TTA-TAA-GCA-TTT-T | CTT-CAC-GCA-GGC-GGT | CTT-CAC-GCC-GGC-GGT |
| 4557 | ATC-AGA-AGA-ACT-CTA-TAT-GAC-TAT-TCG-C | GAC-GCC-CAT-TTT-GTA-GGT-TTG-A | CAA-AAC-GGT-AAC-ATC-TAT-AC | AAA-CGG-TAA-CGT-CTA-TAC |
| 4844 | CGG-TAT-AAA-AGC-TTT-CGT-TGA-ACA-TTT | CTT-TTC-AGT-GGA-AAA-ATG-GAA-GAC-GT | AAA-CCC-CCA-TTC-ATT-CA | AAC-CCC-CAT-CCA-TTC-A |
| 5423 | TTC-CTG-GTG-GAA-GGC-GAT-TC | GGC-GAT-CAC-GGG-CTT-GT | TAG-CCG-AAC-CAC-CAG-CC | CCG-AAC-CGC-CAG-CC |
| 6025 | TTG-CAG-ATT-TGA-GTG-AGG-AGG-AAA-A | GTT-TTG-GGT-TCG-TTT-AAC-TGT-TCC-T | CAA-CCG-CTG-TCT-CCA-AG | ACC-GCT-GTG-TCC-AAG |
| 7087 | CTT-CCC-GCG-CCT-CGA-T | GCG-CTT-GAA-CGT-CTT-GTT-GTT | CAC-CGT-TTT-ACC-CTG-CAC-AA | CCG-TTT-TAC-CCC-GCA-CAA |
| 7726 | CTT-GTG-TGG-CGT-TAG-CGA-AT | ACG-TTT-CTT-GGC-GTG-TTA-AAA-GC | AAA-ATG-CGC-CGA-TCA-T | AAT-GCG-CGG-ATC-AT |
| 7974 | CAT-AAT-TCA-TCA-AGG-CAC-CAA-TGG-T | TGA-AAA-GAA-AGC-GGT-TGC-ATT-CG | CGG-CAT-CAC-AAT-TTA | CGG-CAT-CAC-GAT-TTA |
The underlined nucleotides represent the location of the SNP.
C. burnetii genotyping panel. (i) Reference strains.
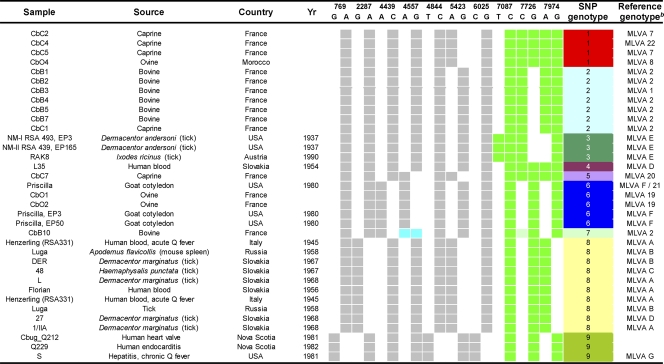
A reference panel consisted of DNA from 28 C. burnetii strains previously typed by MLVA (1, 22) and DNA from commercially obtained strains CbuG_Q212, Priscilla, Henzerling, Luga, and Q229 (Université de la Méditerranée, Marseille, France). The strains were isolated from humans, ticks, and ruminants (ovine, caprine, and bovine strains) in the years from 1937 to 2000 (Table 2). As presented in Table 2, several strains were included multiple times. The two Nine Mile strains (strains RSA493 and RSA439, phases I and II, respectively) represent strains from two different passages (22), the three Priscilla strains are from two different passages (22) and one was commercially obtained, the two Henzerling strains represent one reference strain (22) and one strain obtained commercially, and the two Luga strains have been described previously (1, 22).
TABLE 2.
Panel of reference strains, SNP-based genotyping results, SNP genotypes, and reference MLVA genotypesa
Gray and white squares indicate the presence and absence of the allele for the SNPs located within the single-copy genes, respectively (Fig. 1A). Green and light green squares indicate the presence of the allele for the SNPs located within the multicopy IS1111 element, with dark green squares yielding a different curve than light green squares. White squares indicate the absence of the allele (Fig. 1B). A, adenine; G, guanine; C, cytosine; T, thymine. The blue squares indicated for SNP 4557 strain CbB10 represent the presence of both alleles. Matching colors in the SNP genotype column represent identical SNP profiles.
(ii) Dutch outbreak-related samples.
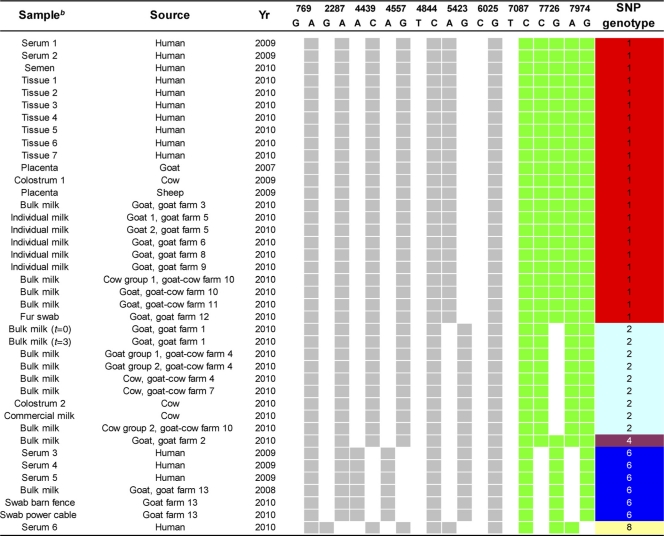
A total of 40 clinical/outbreak-related samples were included: 14 of human origin and 26 of animal origin (Table 3). Six serum samples were obtained from patients (5 males and 1 female) in the early phase of acute Q fever. After 12 months of follow-up, two of these five patients had developed chronic Q fever (defined as high IgG antibody titers against C. burnetii phase I antigen with a positive IS1111 PCR result with a serum or tissue sample), while two other patients successfully cleared their infections. One patient still awaits the 12-month serological follow-up. No clinical data from one patient were available. One semen sample was obtained from a chronic Q fever patient with an aortic prosthesis who suffered from epididymitis. Seven tissue samples were derived from patients (5 males and 2 females) diagnosed with chronic Q fever. Tissue samples 1, 2, 4, 5, and 7 were from an aortic aneurysm. Tissue sample 3 was from a prosthesis of the aorta ascendens. Tissue sample 6 was derived from an iliac artery aneurysm. The mean age of the 14 patients was 59 ± 21 years. Thirteen of the 14 human clinical materials were derived from patients living in Noord-Brabant, the Q fever epidemic center, and 1 (serum sample 6) was from a Dutch outbreak area close to the Belgian-Dutch border. All human samples were obtained for diagnosis-related purposes. The study was carried out with the approval of the Board of Directors and the Scientific Advisory Board of the Jeroen Bosch Hospital.
TABLE 3.
Panel of Dutch Q fever outbreak samples, SNP-based genotyping results, and SNP genotypesa
Gray and white squares indicate the presence and absence of the allele for the SNPs located within the single copy genes, respectively (Fig. 1A). Dark green and white squares indicate the presence and absence of the allele for the SNPs located within the multicopy IS1111 element, respectively (Fig. 1B). A, adenine; G, guanine; C, cytosine; T, thymine. Matching colors in the SNP genotype column represent identical SNP profiles.
bt, time (days).
The veterinary samples included one goat placenta sample, two cow colostrum samples, one commercial cow milk sample, one sheep placenta sample, and samples from 13 dairy farms (9 goat farms and 4 goat/cow farms). From the dairy farms, 9 bulk goat milk and 4 bulk cow milk samples from tanks and 5 individual goat milk samples were obtained. In addition, 2 swabs (from a barn fence and from power cables) from goat farm 13 and a hand swab obtained after a goat from farm 12 was petted (fur swab) were included. Farms 1, 4, and 9 were located outside the Q fever epidemic center.
DNA extraction. (i) Human serum.
DNA was extracted from 500 μl of serum using a NucliSens EasyMAG extraction system (bioMérieux, Boxtel, Netherlands). Five hundred microliters of serum was added to 2 ml NucliSens lysis buffer and incubated for 10 min at room temperature. Magnetic silica particles were diluted 1:1 (vol/vol) with ultrapure water. One hundred microliters of diluted magnetic silica particles was added to each lysate. The samples were further processed according to the manufacturer's instructions and eluted in 60 μl of elution buffer. TE buffer (1 mmol/liter EDTA, 10 mmol/liter Tris-HCl buffer, pH 8.0) was added to a final volume of 180 μl.
(ii) Human tissue and veterinary samples.
To efficiently extract DNA, proteinase K digestion was performed prior to DNA extraction. Two hundred fifty microliters of liquid sample or approximately 50 to 100 mm3 of tissue was digested with 25 μl of proteinase K (20 mg/ml; Roche Diagnostics GmbH, Mannheim, Germany) and 225 μl digestion solution (produced by adding 1 ml of 1 M Tris-HCl buffer, pH 8.0, and 2.5 ml 10% SDS to 44 ml of ultrapure water). Digestion was performed in a thermoshaker at 55°C and 1,400 rpm overnight. DNA was extracted as described above.
SNP real-time PCR.
SNP amplification assays were used according to the manufacturer's instructions. The 25-μl PCR mixture contained 20 mmol/liter Tris-HCl, pH 8.4, 50 mmol/liter KCl, 5 mmol/liter MgCl2 (prepared from 10× PCR buffer [Invitrogen BV, Breda, Netherlands] and 50 mmol/liter MgCl2 solution [Applied Biosystems]), 0.75 U of Ex Taq HS polymerase (TaKaRa Bio, Otsu, Japan), 0.01% bovine serum albumin (molecular biology grade; Westburg BV Benelux, Leusden, Netherlands), 200 μmol/liter of each deoxynucleoside triphosphate (Invitrogen BV), 0.5 μl of carboxy-X-rhodamine reference dye (Invitrogen BV), 1.25 μl of an SNP primer-probe pool (Applied Biosystems) containing two primers and two MGB TaqMan probes (5′ VIC for allele 1, 5′ 6-carboxyfluorescein [FAM] for allele 2, and a 3′ nonfluorescent quencher [NFQ]), and 11.25 μl of eluted target DNA. An ABI Prism 7500 Fast sequence detection system (Applied Biosystems) was used for amplification and detection (10 min at 95°C, 45 cycles of 3 s at 95°C and 30 s at 60°C, and an infinite hold step at 25°C in Fast 7500 mode). Data analysis was performed using SDS software (version 1.3.1). Linear amplification plots were analyzed with a baseline from cycle 6 to cycle 15 for both reporters.
RESULTS
SNP selection and real-time PCR design.
SNPs were selected on the basis of both the consensus sequence (data not shown) generated from 100,000 bp of the 5 known whole-genome sequences of C. burnetii (RSA493, RSA331, CbuG_Q212, Cbuk_Q154, and 5J108 111 Dugway) and an in silico investigation of their discriminatory power using BLAST. A total of 10 specific real-time PCR SNP genotyping assays were designed: 7 based on different single-copy genes and 3 based on the multicopy insertion sequence IS1111 (21). The SNPs and the oligonucleotides designed for the real-time amplification and detection are presented in Table 1. In silico, no additional SNPs were present in the target sequences.
C. burnetii genotyping panel. (i) Reference strains.
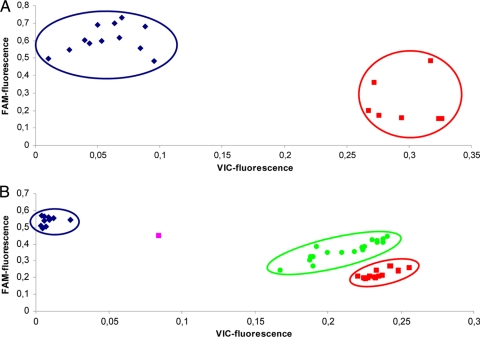
SNPs 769, 2287, 4439, 4557, 4844, 5423, and 6025 (positions indicated in reference sequence RSA493, GenBank accession no. AE016828.2) are located within single-copy genes, and the results are therefore binary in nature. Allelic discrimination of SNP 2287, a representative of the 7 SNPs located within the single-copy genes, is depicted in Fig. 1A. As expected, two groups were clearly distinguishable.
FIG. 1.
Representative allelic discrimination plots. (A) SNP 2287 located on a single-copy gene. Blue circle, presence of adenine (white and gray squares in Tables 2 and 3); red circle, presence of guanine (gray and white squares in Tables 2 and 3). (B) SNP 7726 located on the multicopy IS1111 element. Blue circle, presence of guanine only (white and green squares in Tables 2 and 3); red circle, presence of cytosine only (green and white squares in Tables 2 and 3); green circle and single pink dot, presence of both guanine and cytosine (dark green and dark green and light green and dark green squares in Tables 2 and 3, respectively) in distinct quantities.
SNPs 7087, 7726, and 7974 are located within multicopy insertion sequence IS1111 (positions within the first IS1111 encountered, as indicated in the strain RSA493 reference sequence, GenBank accession no. AE016828.2), which is distributed throughout the C. burnetii genome. Because of the genetic variation in the up to 100 (13) different IS1111 elements within one strain, in theory more than 2 real-time PCR profiles can be expected. Two fluorescent profiles were encountered for SNP 7087, 4 for SNP 7726, and 3 for SNP 7974. The representative allelic discrimination of SNP 7726 is depicted in Fig. 1B. All fluorescent profiles observed from the 30 strains in the validation panel and the 24 outbreak samples could be allocated to one of the depicted profiles. A total of 9 distinct SNP genotypes were distinguished (Table 2).
The experimental SNP data obtained for strains RSA493, RSA331, and CbuG_Q212 were in agreement with the expectation based on the sequence information. The SNP profile of the Nine Mile strains after passage 3 in embryonated hen eggs (EP3; RSA493, phase I) was identical to the profile of the strains after passage 165 (EP165; RSA439, phase II). For strain Priscilla, EP3 and EP50 yielded results identical to those for a Priscilla strain from another source. Two different batches of strains Henzerling and Luga from two different sources also yielded identical results (data not shown). Thus, the fluorescent profiles appear to be highly reproducible and provide reliable SNP profiles. To assess the discriminatory power of the SNP genotyping test, we included the MLVA types in Table 2. Among the 28 reference strains representing 14 MLVA types, 9 SNP types were distinguished. The concordance between the SNP genotyping test and MLVA typing was 93%.
(ii) Dutch outbreak-related samples.
We subsequently determined the SNP types of the strains in the Dutch human clinical and veterinary materials (Table 3). The strains from all 7 human vascular tissue samples, 2 serum samples, and the semen sample clustered in SNP genotype group 1. The 2 serum samples were from one patient who had not progressed to chronic Q fever and from a patient still awaiting analysis of the 12-month sample. The residences of the 10 patients clustering in SNP genotype group 1 were distributed throughout the whole outbreak area in Noord-Brabant. SNP genotype group 1 was also present in cow colostrum sample 1; goat placenta; sheep placenta; bulk goat milk samples from a tank; individual goat milk samples from farms 5, 6, 8, and 9; a bulk cow milk sample from farm 10; and the fur swab at farm 12. The majority of the goat farms with SNP genogroup 1 were located in the center of the Q fever epidemic. Goat farm 9 was located well outside this area. From the reference panel, 3 caprine strains from France and 1 ovine strain from Morocco clustered in SNP genotype group 1. Three other human serum samples clustered in SNP genotype group 6. One sample was derived from a patient who had cleared the infection, while the other 2 samples were from patients who had progressed to chronic Q fever. These patients lived within a 3-mile radius of each other. Bulk goat milk and two swabs from farm 13, located near the residences of these three patients, contained the same SNP genotype. From the reference panel, the goat-derived Priscilla strain from the United States and two ovine strains from France clustered in SNP genotype group 6.
Strains from bulk goat milk samples derived from farms 1 and 4, both located outside the epidemic center of the Q fever outbreak, clustered in SNP genotype group 2. To this SNP genotype also belonged the strains from bulk cow milk samples from farms 4, 7, and 10; cow colostrum sample 2; and the commercial cow milk. Of the reference strains, French caprine strain CbC1 and all bovine strains from France except CbB10 clustered in SNP genotype group 2. The strain from the bulk goat milk sample from a tank from farm 2 was the only one of SNP genotype group 4. Of the reference panel, strain L35 derived from a Slovakian human blood sample clustered in this group. Serum sample 6 from Belgium was the only sample that contained SNP genotype 8. A number of tick-derived reference strains from Slovakia and Russia and human-derived strains Henzerling from Italy and Florian from Slovakia also displayed this genotype. None of the Dutch human clinical materials clustered in SNP genotype group 2 or 4.
DISCUSSION
SNP genotyping of C. burnetii provides informative epidemiological insight and is particularly suitable for direct typing of strains from clinical and veterinary materials. Because only small stretches of target DNA are amplified, the SNP assays are especially appropriate for use with serum samples in which fragmented DNA is supposedly present (24). In addition, the SNP genotyping method has a short turnaround time (approximately 2 h from the time of DNA extraction to a result); can be performed in any real-time PCR machine, which is standard equipment in many molecular laboratories; and does not require expensive DNA sequencing equipment, which is needed for, e.g., MLVA. Another advantage of the SNP profiling test is that processing after PCR is not required in real-time PCR analysis. This substantially reduces the risk of contamination with PCR products (10).
Employing a panel of 10 carefully selected SNPs, 7 located in single-copy genes and 3 located in multicopy IS1111, a total of 9 SNP genotypes could be distinguished among 28 C. burnetii reference strains representing 14 MLVA types. On the basis of the concordance rate, the discriminatory power of SNP genotyping may be somewhat lower than that of MLVA typing. Nevertheless, the differences observed between many of the MLVA types which the SNP genotyping could not distinguish were based on a single repeat difference in all strains except strain CbC4 (1, 22). Intracellular bacteria, of which C. burnetii is a recent associate, are known for their genome reduction by shedding of noncrucial genomic elements (27). Hence, the question that remains is whether the repeat regions selected for MLVA genotyping show adequate stability, an issue previously raised and discussed (1, 17, 25). The same might be true for the markers chosen for the SNP genotyping method. With regard to marker stability, however, we analyzed and compared the SNP profiles generated from the DNA derived from the Priscilla and the Nine Mile strains after EP3 to those observed after EP50 and EP165, respectively. The SNP profiles before and after passage proved to be identical. This indicates that the selected SNPs show sufficient stability to not be influenced by a large number of passages.
Caprine strains CbC2, CbC4, and CbC5 from France and ovine strain CbO4 from Morocco grouped together in SNP genotype group 1. Strains CbB1, CbB2, CbB3, CbB4, CbB5, and CbB7, all of bovine origin, and caprine strain CbC1 were also isolated in France and clustered in SNP genotype group 2. Strains L35, CbC7, and CbB10 were designated SNP genotypes 4, 5, and 7, respectively. Strain CbB10 displayed an SNP profile indicative of a duplication of (part) of the DNA gyrase B subunit-encoding gene (data not shown). The three Priscilla strains clustered together, as expected, in SNP genotype 6, which was shared by ovine strains CbO1 and CbO2, both isolated in France. SNP genotype group 8 consisted of strains isolated from humans and ticks in Italy, Slovakia, and Russia. The clustering of strains CbuG_Q212, Q229, and S in SNP genotype 9 could be supported by a geographical link (Nova Scotia and the United States) between the strains, and in addition, all three were isolated in the early 1980s.
SNP genotyping of the Dutch outbreak panel revealed 3 distinct genotypes among human samples and 4 distinct genotypes among livestock. Thus, multiple C. burnetii strains have infected humans in the Dutch Q fever outbreak. The human tissues clustered in SNP genotype 1, together with two human serum samples, a human semen sample, a cow colostrum sample, a goat placenta sample, a sheep placenta sample, goat milk from seven farms, bulk cow milk from one of these farms, and a fur swab from another farm. It is interesting to note that some farmers use raw cow colostrum to feed the newborn goats. Three human serum samples clustered in SNP group 6, together with several materials from farm 13, the Priscilla strain, and ovine strains CbO1 and CbO2 from France. Although SNP genotype 6 was detected only in the serum samples taken during the acute phase of disease, one of these patients later developed chronic disease. Assuming that this patient was not reinfected with a second C. burnetii strain, both SNP genotype 1 and genotype 6 may cause chronic disease in humans. If the C. burnetii strain from genotype 6 found in the patient samples shares not only the SNP profile but also the antibiotic resistance properties of the Priscilla strains, this may have consequences for the antibiotic regimen used for patients infected with this strain (28). Almost all the sources of the samples in which SNP genotype 1 was identified were located within a surface area of approximately 150 square miles and could therefore be epidemiologically linked. One goat milk sample from a farm located outside the outbreak area also yielded SNP genotype 1. This indicates the further spread of this C. burnetii strain in the Netherlands. The patients infected with C. burnetii SNP genotype 6 lived at the edge of the main outbreak area within a 3-mile radius from each other and the goat farm with this genotype.
The direct SNP typing of bulk milk samples raises questions about clonality, since multiple genotypes could theoretically be present at one farm. At farm 4, two different groups of goats and the cows displayed one identical SNP genotype. The SNP profiles from bulk milk samples showed the presence of 1 allele in the SNPs located on single-copy genes. These findings suggest the presence of a single strain at each farm. At farm 10, however, 2 SNP genotypes were found. Bulk milk from goats as well as bulk milk from one group of cows contained SNP genotype 1, while bulk milk from a distinct group of cows yielded SNP genotype 2. Apparently, two C. burnetii strains have infected separate groups of animals on that farm.
Two of the veterinary genotypes have thus far not been linked to human cases in the Netherlands. Two of the farms where one of these genotypes was found were located outside the major outbreak area, in the northwest and center of the Netherlands, where only a small number of people fell ill.
One human serum sample from the Dutch-Belgian border area contained SNP genotype 8. This SNP genotype has not been linked to any animal source thus far.
Strains Priscilla, RSA493, Henzerling, and Luga were analyzed multiple times. All analyses yielded identical results indicating that the SNP genotyping assay provided good reproducibility. On the basis of these findings, we expect that the technique is suitable for interlaboratory standardization and database building. Gel-based techniques have proven to be more difficult to standardize and are therefore less suitable for these purposes (1).
With regard to the analysis of weakly positive samples, caution is advised for the interpretation of the signals. The sensitivity of SNP genotyping was estimated to be about 100 genome copies per reaction for the SNPs located on single-copy genes and about 10 genome copies per reaction for the SNPs located on IS1111. Although the 3 IS1111 SNPs do not provide the full resolution of the total SNP panel, typing of materials containing small amounts of DNA with only these SNPs may provide crucial information regarding the C. burnetii type.
In conclusion, we have developed a novel SNP genotyping method suitable for direct typing of C. burnetii from a wide variety of clinical and veterinary samples. With this typing method, we have shown that at least five distinct genotypes (three found in humans and four found in livestock) are present in the Dutch outbreak. This implies that environmental circumstances (such as the high density of farms and people, dry periods during spring) rather than one highly virulent C. burnetii strain favored the Dutch Q fever spread. Further research will have to resolve the question of whether all genotypes present in livestock are able to cause human disease or whether some strains are more virulent or infectious than others.
Acknowledgments
We thank Annie Rodolakis for kindly providing part of the panel of C. burnetii strains typed by MLVA. We cordially thank the goat and cow farmers who provided the samples. We thank Bas Beerens for technical assistance and Kathelijn Geraats-Peters for critical reading of the manuscript.
The contribution of Rudolf Toman to this work was supported by grant no. 2/0127/10 from the Scientific Grant Agency of Ministry of Education of the Slovak Republic and the Slovak Academy of Sciences.
Footnotes
Published ahead of print on 21 January 2011.
REFERENCES
- 1.Arricau-Bouvery, N., et al. 2006. Molecular characterization of Coxiella burnetii isolates by infrequent restriction site-PCR and MLVA typing. BMC Microbiol. 26:6-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benenson, A. S., and W. D. Tigertt. 1956. Studies on Q. fever in man. Trans. Assoc. Am. Physicians 69:98-104. [PubMed] [Google Scholar]
- 3.Dutch Central Bureau of Statistics. 2010. STATLINE: landbouw; gewassen, dieren, grondgebruik naar region. Dutch Central Bureau of Statistics, The Hague, Netherlands. http://www.cbs.nl/nl-NL/menu/home/default.htm. Accessed 21 August 2010.
- 4.Enserink, M. 2010. Questions abound in Q-fever explosion in the Netherlands. Science 327:266-267. [DOI] [PubMed] [Google Scholar]
- 5.Glazunova, O., et al. 2005. Coxiella burnetii genotyping. Emerg. Infect. Dis. 11:1211-1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gopaul, K. K., M. S. Koylass, C. J. Smith, and A. M. Whatmore. 2008. Rapid identification of Brucella isolates to the species level by real time PCR based single nucleotide polymorphism (SNP) analysis. BMC Microbiol. 2:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gopaul, K. K., J. Sells, B. J. Bricker, O. R. Crasta, and A. M. Whatmore. 2010. Rapid and reliable single nucleotide polymorphism-based differentiation of Brucella live vaccine strains from field strains. J. Clin. Microbiol. 48:1461-1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hendrix, L. R., J. E. Samuel, and L. P. Mallavia. 1991. Differentiation of Coxiella burnetii isolates by analysis of restriction-endonuclease-digested DNA separated by SDS-PAGE. J. Gen. Microbiol. 137:269-276. [DOI] [PubMed] [Google Scholar]
- 9.Hernychova, L., et al. 2008. Detection and identification of Coxiella burnetii based on the mass spectrometric analyses of the extracted proteins. Anal. Chem. 80:7097-7104. [DOI] [PubMed] [Google Scholar]
- 10.Huijsmans, R., J. Damen, H. van der Linden, and M. Hermans. 2007. Single nucleotide polymorphism profiling assay to confirm the identity of human tissues. J. Mol. Diagn. 9:205-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kazar, J. 2005. Coxiella burnetii infection. Ann. N. Y. Acad. Sci. 1063:105-114. [DOI] [PubMed] [Google Scholar]
- 12.Klaassen, C. H., M. H. Nabuurs-Franssen, J. J. Tilburg, M. A. Hamans, and A. M. Horrevorts. 2009. Multigenotype Q fever outbreak, the Netherlands. Emerg. Infect. Dis. 15:613-614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Klee, S. R., H. Ellerbrok, J. Tyczka, T. Franz, and B. Appel. 2006. Evaluation of a real-time PCR assay to detect Coxiella burnetii. Ann. N. Y. Acad. Sci. 1078:563-565. [DOI] [PubMed] [Google Scholar]
- 14.Koylass, M. S., et al. 2010. Comparative performance of SNP typing and ‘Bruce-ladder’ in the discrimination of Brucella suis and Brucella canis. Vet. Microbiol. 142:450-454. [DOI] [PubMed] [Google Scholar]
- 15.Madariaga, M. G., K. Rezai, G. M. Trenholme, and R. A. Weinstein. 2003. Q fever: a biological weapon in your backyard. Lancet Infect. Dis. 3:709-721. [DOI] [PubMed] [Google Scholar]
- 16.Nguyen, S. V., and K. Hirai. 1999. Differentiation of Coxiella burnetii isolates by sequence determination and PCR-restriction fragment length polymorphism analysis of isocitrate dehydrogenase gene. FEMS Microbiol. Lett. 15:249-254. [DOI] [PubMed] [Google Scholar]
- 17.Noller, A. C., M. C. McEllistrem, K. A. Shutt, and L. H. Harrison. 2006. Locus-specific mutational events in a multilocus variable-number tandem repeat analysis of Escherichia coli O157:H7. J. Clin. Microbiol. 44:374-377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pickens, E. G., and J. A. Gaon. 1961. Growth of Coxiella burnetii in agar tissue culture. Am. J. Trop. Med. Hyg. 10:49-52. [DOI] [PubMed] [Google Scholar]
- 19.Raoult, D., T. J. Marrie, and J. L. Mege. 2005. Natural history and pathophysiology of Q fever. Lancet Infect. Dis. 5:219-226. [DOI] [PubMed] [Google Scholar]
- 20.Schimmer, B., et al. 2009. Sustained intensive transmission of Q fever in the south of the Netherlands. Euro Surveill. 14(19):pii=19210. [DOI] [PubMed] [Google Scholar]
- 21.Seshadri, R., et al. 2003. Complete genome sequence of the Q-fever pathogen Coxiella burnetii. Proc. Natl. Acad. Sci. U. S. A. 100:5455-5460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Svraka, S., R. Toman, L. Skultety, K. Slaba, and W. L. Homan. 2006. Establishment of a genotyping scheme for Coxiella burnetii. FEMS Microbiol. Lett. 254:268-274. [DOI] [PubMed] [Google Scholar]
- 23.Thiele, D., H. Willems, G. Köpf, and H. Krauss. 1993. Polymorphism in DNA restriction patterns of Coxiella burnetii isolates investigated by pulsed field gel electrophoresis and image analysis. Eur. J. Epidemiol. 9:419-425. [DOI] [PubMed] [Google Scholar]
- 24.Tilburg, J. J., et al. 2010. Interlaboratory evaluation of different extraction and real-time PCR methods for the detection of Coxiella burnetii DNA in serum. J. Clin. Microbiol. 48:3923-3927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van Belkum, A. 2007. Tracing isolates of bacterial species by multilocus variable number of tandem repeat analysis (MLVA). FEMS Immunol. Med. Microbiol. 49:22-27. [DOI] [PubMed] [Google Scholar]
- 26.van der Hoek, W., et al. 2010. Q fever in the Netherlands: an update on the epidemiology and control measures. Euro Surveill. 15(12):pii=19520. [PubMed] [Google Scholar]
- 27.Wernegreen, J. J. 2005. For better or worse: genomic consequences of intracellular mutualism and parasitism. Curr. Opin. Genet. Dev. 15:572-583. [DOI] [PubMed] [Google Scholar]
- 28.Yeaman, M. R., and O. G. Baca. 1990. Antibiotic susceptibility of Coxiella burnetii, p. 213-221. In T. J. Marrie, Q-fever the disease, vol. 1, CRC Press, Boca Raton, FL. [Google Scholar]
- 29.Zhang, G. Q., et al. 1998. Direct identification of Coxiella burnetii plasmids in human sera by nested PCR. J. Clin. Microbiol. 36:2210-2213. [DOI] [PMC free article] [PubMed] [Google Scholar]