Abstract
Escherichia coli O157:H7 is a Shiga toxin (stx)-producing E. coli (STEC) strain that has been classified as an adulterant in U.S. beef. However, numerous other non-O157 STEC strains are associated with diseases of various severities and have become an increasing concern to the beef industry, regulatory officials, and the public. This study reports on the prevalence and characterization of non-O157 STEC in commercial ground beef samples (n = 4,133) obtained from numerous manufacturers across the United States over a period of 24 months. All samples were screened by DNA amplification for the presence of Shiga toxin genes, which were present in 1,006 (24.3%) of the samples. Then, culture isolation of an STEC isolate from all samples that contained stx1 and/or stx2 was attempted. Of the 1,006 positive ground beef samples screened for stx, 300 (7.3% of the total of 4,133) were confirmed to have at least one strain of STEC present by culture isolation. In total, 338 unique STEC isolates were recovered from the 300 samples that yielded an STEC isolate. All unique STEC isolates were serotyped and were characterized for the presence of known virulence factors. These included Shiga toxin subtypes, intimin subtypes, and accessory virulence factors related to adherence (saa, iha, lifA), toxicity (cnf, subA, astA), iron acquisition (chuA), and the presence of the large 60-MDa virulence plasmid (espP, etpD, toxB, katP, toxB). The isolates were also characterized by use of a pathogenicity molecular risk assessment (MRA; based on the presence of various O-island nle genes). Results of this characterization identified 10 STEC isolates (0.24% of the 4,133 total) that may be considered a significant food safety threat, defined by the presence of eae, subA, and nle genes.
More than 70 different serotypes of Shiga toxin-producing Escherichia coli (STEC) that cause disease in humans have been described (13). Illnesses range from mild diarrhea to bloody diarrhea to hemorrhagic colitis (HC) and hemolytic-uremic syndrome (HUS). E. coli O157:H7 is the STEC strain most often associated with the most severe forms of disease (13, 60, 69). However, numerous non-O157 STEC isolates have also been linked to illnesses and outbreaks of disease. Six O groups (comprising 13 serotypes) have been described by the CDC to be the cause of 71% of non-O157 STEC disease (13). These serotypes have been identified as O26:H11 or nonmotile (NM); O45:H2 or NM; O103:H2, H11, H25; or NM; O111:H8 or NM; O121:H19 or H7; and O145:NM. The proportion of these non-O157 STEC serogroups breaks down as follows: 22% O26, 16% O111, 12% O103, 8% O121, 7% O45, and 5% O145. Although it is infrequently isolated and reported, it is estimated that non-O157 STEC may cause diarrhea at frequencies similar to those of other important enteric bacterial pathogens, such as Salmonella and Shigella (62), and also cause infections resulting in HUS and outbreaks (13). Due to difficulties confirming their presence in the lab, non-O157 STEC isolates are likely undercounted (31).
Cattle are recognized to be the reservoir of many STEC serotypes (46). In fact, there are ample data on the prevalence of non-O157 STEC in cattle from fed beef through processing (1) and boneless beef trim destined for grinding (10), but there is a lack of information on the prevalence of non-O157 STEC in finished ground beef. The study of beef trim destined for grinding showed that 10 to 30% of beef trim contained STEC (10); however, the results of blending the various lean and fat materials during production of ground beef on the rates of STEC present in the finished product are unknown.
An STEC seropathotype classification based upon the serotype and association with outbreaks, HUS, and diarrhea has been proposed by Karmali et al. (39). This classification system is a tool to assess the clinical and public health risks associated with different non-O157 STEC strains. The predominant enterohemorrhagic E. coli (EHEC) serotype responsible for HC and HUS, O157:H7, is classified group A. The other non-O157 STEC serotypes recognized as belonging to the EHEC group comprising the 13 serotypes listed above are considered group B. Group C in this classification system is made up of strains referred to as atypical EHEC. The atypical EHEC strains are less frequently involved in hemorrhagic diseases than typical EHEC strains but are a frequent cause of diarrhea. The principal serotypes identified in group C are O91:H21, O113:H21, and O104:H21. STEC serotypes not associated with HC, HUS, or outbreaks are numerous and belong to groups D and E. These two groups are delineated from one another by their source. Group D strains have been isolated from humans, while group E strains have been isolated only from animals (39).
Various factors and toxins contribute to the increasing virulence of STEC (72, 74). Strains that produce Shiga toxin 2 (Stx2) and, more specifically, Shiga toxin 2 subtype c (Stx2c) have been suggested to be more likely to cause HUS than those that produce Shiga toxin 1 (Stx1) alone (9, 28). Other toxins purported to increase virulence in strains are subtilase (subA) (41, 54, 75), lymphocyte-activating factor (lifA also known as efa1) (42), cytotoxic necrotizing factor (cnf) (7), and heat-stable toxin (astA) (78). EHEC strains contain a large heterologous virulence plasmid, referred to as the 60-MDa virulence plasmid, that contains a number of different genes associated with disease: espP, katP, eptD, toxB, and the EHEC hemolysin gene (ehx) (15, 16, 38, 50). Other hemolysins, such as ChuA and HylA, have also been described to influence the iron acquisition and virulence of STEC (32, 68, 76). Finally, there is a group of factors associated with adherence that influences virulence. Intimin (eae) is contained in the locus of enterocyte effacement (LEE) and along with the tir gene product is responsible for the intimate attachment and classic pedestal formation of the characteristic adherence-and-effacing (A/E) lesion in HC and HUS (46). In the absence of intimin, other factors such as saa and iha that correlate with increased adherence have been identified (36, 55, 64). The interplay between these various genes is not understood, but each has been described to provide an additional trait that increases virulence in STEC.
Finally, comparisons of the genomes of E. coli O157:H7 and E. coli K-12 have identified numerous islands of genetic material unique to each. The genomic regions specific to E. coli O157:H7 have been termed O islands [OIs]). Studies of OIs 36, 57, 71, and 122 have identified genes associated with increased virulence (19, 39, 74). In fact, a molecular risk assessment system for STEC strains based on the complement of these four OIs has been proposed (19). This project aims to determine the prevalence of non-O157 STEC strains in commercial ground beef in the United States, identify the serotypes of the STEC present, and distinguish those serotypes that may be pathogenic STEC (pSTEC). There are essentially two approaches to defining pSTEC: either epidemiological (identifying serogroups, particularly the top six) or seropathotypic (on the basis of molecular markers associated with pathogenicity). We chose the broader second option, which offers a much larger set of results and which identifies as many pSTEC serogroups as possible. Narrowly focusing on only the described top six STEC serogroups poses the problem of identifying numerous isolates of little pathogenic concern while missing other significant pSTEC serogroups, especially since nearly one-third of pSTEC strains are not within the top six serogroups. Our goal was to not allow the impact of these other pSTEC serogroups to go unappreciated in our studies.
(Part of this work was presented by L. H. Gould at the Annual Capital Area Food Protection Association Meeting, Washington, DC, 14 September 2009.)
MATERIALS AND METHODS
Design.
Four-thousand one-hundred thirty-three identity-blinded ground beef samples that had been collected previously were used (11). The samples were collected by 18 different commercial ground beef producers from unique product lots produced on different days. No more than one sample per day per supplier was used in this study. The 18 producers were located in seven of the eight Beef Industry Food Safety Council (BIFSCo) regions of the United States over a period of 24 months (11). The BIFSCo regions are 1, northwest (WA, OR, ID); 2, west (CA, NV); 3, southwest (AZ, NM, TX); 5, upper Midwest (NE, ND, SD, MN, WS); 6, central (IA, KS, MO); 7, southeast (OK, AR, LA, NC, SC, FL, AL, MS, GA, TN); and 8, northeast (IL, IN, KT, MS, ME, MD, MI, NJ, NY, NH, CN, RI, OH, WV, VA, VT, PA, DE). Region 4 (MT, CO, WY, UT) was not represented in the sample collection due to logistical problems. All samples were finished ground beef that had been screened by the manufacturer for E. coli O157:H7 before release. The presence of stx1 and stx2 genes was determined by PCR amplification. If Shiga toxin genes were present, the samples were processed using a phenotypic screening assay to isolate non-O157 STEC. STEC isolates were then characterized for serotype, the presence and subtype of virulence genes, as well as other toxin, adherence, and virulence plasmid-associated genes. Further, the presence of determinants within OIs 36, 57, 71, and 122 was determined, as was the resistance to antibiotics of each isolate.
Screening for Shiga toxin genes.
The ground beef samples (65 g) were placed in 585 ml tryptic soy broth (TSB), incubated at 42°C for 6 h, and then held for 6 to 12 h at 4°C as previously described (11). Two 1-ml portions of each enrichment were archived as frozen (−70°C) 30% glycerol stocks. Between 2 and 12 weeks after the date of freezing, 24 to 48 different glycerol stocks were thawed, vortexed to thoroughly mix, and maintained on ice. One microliter was removed from each and placed into a separate 25-μl mixture for multiplex PCR that detected stx1, stx2, eae, and ehx and that was performed as previously described (51). Products of the PCR amplifications were separated by electrophoresis, stained using ethidium bromide, and then photographed and interpreted for the presence of the four possible reaction products. Only enrichments that had stx1 and/or stx2 were considered positive for STEC and used in prevalence calculations.
Isolation of non-O157 STEC.
The sample enrichments determined by PCR to contain stx1 and/or stx2 were assayed by spiral plating of samples onto plates of washed sheep blood agar containing mitomycin (wSBAm) (63). Each enrichment was serially diluted to 1:500 and 1:5,000 in cold (4°C) buffered peptone water (BPW). Fifty microliters of each dilution was spiral plated, using an Autoplate 4000 spiral plater (Spiral Biotech, Norwood, MA), onto wSBAm plates. The plates were incubated overnight at 37°C and then viewed on a white-light box (VeriQuest 100; Photodyne Technologies, Inc., Northridge, CA) for colonies surrounded by zones of hemolysis. The suspect hemolytic phenotype was a thin zone of 1 mm or less surrounding the colony (5). However, many phenotypic variations of hemolysis not indicative of ehx were often present. In these cases, additional colonies representative of each variation were also picked for screening. A minimum of 4 colonies (if colonies were present) and a maximum of 6 colonies per sample were picked and placed into individual wells of 96-well screening plates containing 100 μl TSB per well. After suspect colonies were picked, the wSBAm plates were placed at 4°C. The 96-well screening plates were incubated overnight at 37°C and screened by PCR as described above to identify stx-containing isolates. If at least one suspect colony from a sample did not contain stx1 and/or stx2, the wSBAm plates were removed from 4°C and subjected to another round of suspect colony picking. The additional round of screening increased the number of isolate-positive samples by 15 to 20%. All stx-containing isolates were checked for purity by streaking for isolation on sorbitol MacConkey agar containing 5-bromo-4-chloro-3-indolyl-β-d-glucuronide (SMAC-BCIG; Oxoid-CM0981; Remel Inc., Lenexa, KS). If more than one colony type was observed, each one was rescreened as described above until a pure culture resulted. All purified isolates of STEC were transferred to tryptic soy agar (TSA) plates for characterization.
Characterization of STEC isolates.
All stx-containing isolates were confirmed to be E. coli by biochemical assays using Fluorocult LMX broth (Merck KGaA, Darmstadt, Germany) and SensiTiter Gram-negative ID panels (Trek Diagnostic Systems, Inc.) or API 20E strips (bioMérieux Inc., Hazelwood MO), all of which were used according to their manufacturers' recommendations. Once an isolate was established as being stx-containing E. coli, its serotype was determined by molecular and serologic identification of the O serogroup. PCR was used to molecularly identify O groups O26 (21), O45 (20), O55 (20), O103 (27), O111 (22), O113 (21), O117 (45), O121 (26), O126 (45), O145 (24), and O146 (45). A verotoxin-producing E. coli (VTEC)-specific antiserum set (Laboratorio de Referencia de E. coli, Lugo, Spain) was used to confirm the PCR results and identify O groups O1, O2, O4, O5, O6, O8, O9, O15, O16, O17, O20, O22, O25, O27, O32, O39, O41, O46, O48, O64, O74, O75, O77, O81, O82, O84, O86, O88, O91, O98, O101, O104, O105, O109, O110, O112, O116, O118, O119, O123, O128, O132, O136, O139, O141, O150, O153, O157, O162, O163, O165, O166, O168, O171, O172, O174 (OX3), O176, O177, and O178 (6, 33). The H group of each isolate was determined by molecular methods involving sequencing of the fliC gene (73) and comparison of the sequences with sequences in GenBank to detect like sequences. This method allows the identification of 44 of the 53 different H types coded for by fliC. H types H3, H17, H35, H36, H44, H47, H53, H54, and H55, which are encoded by flk, fll, and flm, were not determined by this method. In situations where the fliC gene could not be resolved from one of the other H-type-encoding genes, the fliC gene is given and the possible other H type is listed as a superscript. All other O and/or H types are referred to as untypeable, indicating that the serotype is outside the limits of the above-mentioned O- and H-typing methodologies.
Previously described PCR methods and primers were used with isolates found to be positive for stx1 to determine Shiga toxin subtype 1 or 1c (79) and with isolates found to be positive for stx2 to determine Shiga toxin subtypes 2 (referred to here as 2a), 2c, 2d, and 2e (4, 80). Isolates found to be stx2 positive but nontypeable using the PCR methods are referred to simply as stx2 positive. If an isolate contained more than one type of stx2 allele, the subtypes are given in a combined notation (e.g., stx2ac), but no attempts were made to quantitate the number of each allele present. In a similar fashion, previously described methods were used with isolates found to be positive for eae to identify intimin subtypes α1, α2, β1, β2, γ, δ, ɛ, θ, and ζ (6, 8). Since this subtyping method is not inclusive of all defined subtypes, any isolate that was eae positive but that did not subtype is referred to as untypeable and presented simply as eae positive, without the subtype indicator. The presence of four genes associated with the large 60-MDa virulence plasmid, toxB, espP, katP, and etpD, was determined by PCR using previously described methods (15, 16). Additional PCR-based assays were used to identify toxin-encoding genes (subA [53], lifA [42], cnf [7], and astA [77]), adherence-encoding genes (iha [64] and saa [52]), and hemolysin genes (hylA [43] and chuA [34]), in order to determine their distribution among the STEC isolates. Finally, the genes described for molecular risk assessment (19) associated with OIs 122 (genes nleB, nleE, and ent) 57 (genes G2-3, G5-2, and G6-2), 36 (genes nleC, H1-1, and nleB2), and 71 (genes nleG, nleG9, nleF, H1-2, nleA, and G2-1) were examined using the previously described PCR amplifications (19). The serotypes and the rates of possession of the various pathogenicity-related genes were combined to identify the seropathotypes of the STEC isolates (39).
Statistical analysis.
STEC isolates obtained by each study method were sorted according to serotype and screening PCR positive reaction pattern (stx1, stx2, eae, and ehx). Comparisons of percent isolation and mean isolation rate were examined using a one-way analysis of variance (ANOVA) and the Bonferroni multiple-comparison posttest. Comparisons of median values of the data sets were made using the Kruskal-Wallis test for nonparametric data and Dunn's multiple-comparison posttest. Comparisons of data sets with only two groups of values were made using either a two-tailed unpaired t test or the Mann-Whitney U test for nonparametric data. Interactions between study methods and isolate serotypes and/or screening PCR positive reaction patterns were examined by ANOVA factorial analysis using Prism (version 5) software (GraphPad Software, La Jolla, CA), and P values of <0.05 were considered significantly different. The DIFFER procedure of PEPI software (USD, Inc., Stone Mountain, GA) was used to calculate the pair-wise differences among the isolates in each STEC serogroup and each screening PCR-positive reaction, with significance being defined at a P value of <0.05.
RESULTS
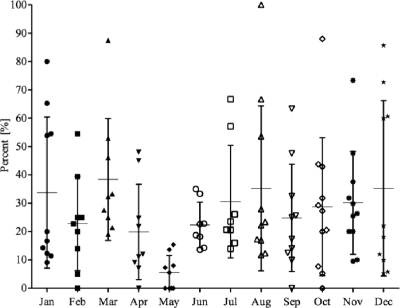
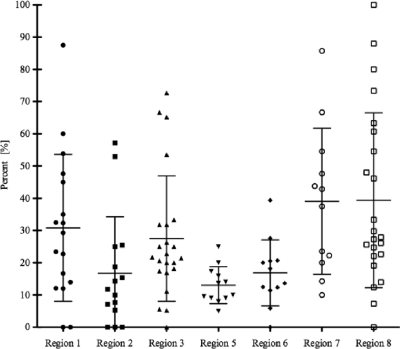
The overall suggested prevalence of STEC in ground beef on the basis of the presence of either stx1 or stx2 was 24.3% (Table 1). The samples were screened in each calendar month and were from numerous locations in 7 of the BIFSCo microbiological monitoring regions of the United States. The samples were submitted on a voluntary basis, and as such, their spatial and temporal distributions were nonuniform and the monthly and regional analyses produced wide-ranging results. The prevalence of stx in screened samples over time (Fig. 1) showed that the mean monthly prevalence ranged from a low of 5.5% in May to a high of 38.4% in March. With few exceptions, comparisons of monthly prevalence levels of stx in the screened samples were not different. Examining the prevalence of stx in screened samples by region (Fig. 2) showed that the mean prevalence across regions ranged from 13.1% to 39.4%, with regions 7 and 8 having stx prevalence rates higher (P < 0.05) than regions 2 and 5.
TABLE 1.
Summary of screening, isolation, and characterization of non-O157 STEC strains from commercial ground beef samples
| No. (%) of samples | |||
|---|---|---|---|
| Total in studya | stx positiveb | Containing STECc | Containing pathogenic STECd |
| 4,133 (100) | 1,006 (24.3) | 300 (7.3) | 10 (0.24) |
Study samples were submitted from 18 commercial ground beef producers located in 7 of the 8 BIFSCo microbiological monitoring regions for a total of 24 months.
The determination of stx positivity for a sample was based on PCR detection of stx1 and/or stx2.
STEC isolation was performed only on samples that screened positive for stx. The isolation method relied on identification of STEC on washed sheep blood agar.
The determination that an isolate was pathogenic STEC was based on detection of intimin or subtilase and genetic markers indicating the presence of at least one virulence-related OI (OI 36, 57, 71, or 122).
FIG. 1.
Distribution of stx prevalence in commercial ground beef by month. The distribution of the monthly prevalence of stx in commercial ground beef samples is based on PCR screening for the presence of stx1 and/or stx2 over a period of 24 months (n = 4,133). The mean (horizontal bars) and standard deviation (vertical bars) for each month are shown. Each point within a month represents a regional prevalence value. Note that only some regions are represented twice in each month. The STEC prevalence rate in May was significantly lower than the rates during March and November (P < 0.05). All other comparisons between months are not statistically significantly different (P > 0.05).
FIG. 2.
Distribution of stx prevalence in commercial ground beef by region. The distribution of the regional prevalence of STEC in commercial ground beef samples from seven of the eight BIFSCo microbiological monitoring regions (n = 4,133) is based on PCR screening for the presence of stx1 and/or stx2. The BIFSCo regions are defined in the text. The mean (horizontal bars) and standard deviation (vertical bars) for each region are shown. Each point within a region represents a monthly prevalence value. Note that only some months are represented twice in each region. The prevalence in samples from regions 2 and 5 was less than that of regions 7 and 8 (P < 0.05). None of the other comparisons between regions was statistically significantly different (P > 0.05).
The screening test of the samples provided the presence of intimin (eae) and the EHEC hemolysin (ehx), in addition to stx1 and stx2 (Table 2). The least common screening result (7.8% samples) was for the presence of stx1 alone, whereas just over half (53%) of the screened positive samples had only stx2. Intimin was present with stx in 41.5% of screened positive samples. Samples that screened positive for either stx1 or stx2 had similar proportions of samples that were also positive for eae: 34.6% and 37.6%, respectively. A larger proportion (48.9%) of samples (P < 0.05) that were stx1 and stx2 positive also contained eae. The presence of ehx was observed in 55.5% of the screened samples with stx, but the proportion of samples with ehx and with both stx1 and stx2 was significantly greater (P < 0.05) than that of samples with only either stx1 or stx2 alone: 73.7% compared to 37.2 and 44.7%, respectively.
TABLE 2.
STEC screening and isolation results for ground beef enrichments sorted by results of PCR for stx1, stx2, eae, and ehx
| PCR amplification result in screena |
No. screen positive (% of total) | No. STEC isolates recoveredb (% of total) | |||
|---|---|---|---|---|---|
| stx1 | stx2 | eae | ehx | ||
| + | + | + | + | 159 (3.8) | 47 (1.1) |
| + | + | + | − | 34 (0.8) | 5 (0.1) |
| + | + | − | + | 132 (3.2) | 63 (1.5) |
| + | + | − | − | 70 (1.7) | 16 (0.4) |
| + | − | + | + | 12 (0.3) | 3 (0.1) |
| + | − | + | − | 15 (0.4) | 2 (0.1) |
| + | − | − | + | 17 (0.4) | 5 (0.1) |
| + | − | − | − | 34 (0.8) | 12 (0.3) |
| − | + | + | + | 91 (2.2) | 26 (0.6) |
| − | + | + | − | 106 (2.6) | 9 (0.2) |
| − | + | − | + | 147 (3.6) | 72 (1.7) |
| − | + | − | − | 189 (4.6) | 40 (1.0) |
The screening test detected the presence of the genes for Shiga toxins 1 and 2 (stx1 and stx2), intimin (eae), and EHEC hemolysin (ehx) in 65 g of commercial ground beef.
STEC isolation was performed only on samples that screened positive for stx. The isolation method relied on identification of STEC on washed sheep blood agar.
All 1,006 samples that had screened positive for stx1 and/or stx2, regardless of the presence of eae or ehx, were subjected to the STEC isolation procedure, and 338 unique isolates resulted from 300 samples. Most samples yielded isolates of one serotype, but 32 positive samples yielded isolates of two different serotypes and 3 samples yielded isolates of three different serotypes. The rate of isolation of an STEC isolate in a screened positive sample was not different (P > 0.05) if the sample had screened positive for stx1, stx2, or stx1 and stx2. Significantly fewer (P < 0.05) STEC isolates were recovered from samples that had screened positive for the presence of eae in addition to stx (22%) than from samples that had not screened positive for eae (35%). The isolation method targeted the enterohemolytic phenotype associated with ehx on blood agar (5); therefore, nearly twice as many STEC isolates (39% versus 20%) were recovered from samples that screened positive for ehx, even though colonies that demonstrated all types of hemolytic phenotypes were examined. Samples that yielded an isolate that screened positive for stx2 and ehx made up the largest proportion of samples, while those that screened positive for stx2 and eae made up the smallest proportion.
Numerous O serogroups were present. The most frequently identified serogroups, in order of frequency, were O113, O8, O22, O117, O163, O174, O171, O116, and O20. These nine serogroups represented 53% of all that were identified, while at least 33 serogroups made up the remaining 47%. The single most frequently identified serogroup was O113, to which 11% of all the isolates belonged. Fifty-six (16.7%) of the isolates were of an untypeable O group, indicating they were either outside the range of our typing system, rough, or serologically untypeable. The H-typing protocol used identified 22 different H types in 319 isolates. Nineteen isolates were H untypeable, indicating that they may have been outside the scope of our typing scheme, or were H negative or nonmotile.
In total, 99 different serotypes were identified and are provided as follows: O1:H11, O1:H19, O5:H7, O5:H14, O8:H8, O8:H16, O8:H19, O8:H25, O8:H49, O15:H27, O17:H45, O20:H7, O20:H19, O20:Hunt, O22:H8, O22:H11, O22:H19, O22:H49, O22:Hunt, O26:H11, O26:H21, O32:H40, O41:H11, O41:H25, O41:H2(35), O48:H7, O74:H8, O74:H28, O74:H42, O82:H8, O86:H8, O88:H25, O88:Hunt, O91:H10, O91:H14, O91:H21, O101:H19, O103:H21, O103:H2(35), O104:H7, O105:H7, O105:H18, O109:H5, O109:H27, O109:H48, O110:Hunt, O112:H8, O112:H19, O112:H45, O112:H2(35), O112:Hunt, O113:H21, O113:Hunt, O116:H21, O116:Hunt, O117:H7, O117:H2, O117:Hunt, O121:H7, O121:H8, O121:H16, O132:H18, O139:H7, O139:H19, O141:H8, O141:H49, O141:Hunt, O145:H28, O146:H28, O146:H21, O147:H21, O150:H8, O157:H21, O163:H11, O163:H19, O163:H46, O168:H8, O171:H2(35), O172:H16, O174:H7, O174:H21, O174:H28, O174:H2(35), O174:Hunt, Ount:H2(35), Ount:H7, Ount:H8, Ount:H10, Ount:H11, Ount:H14, Ount:H16, Ount:H18, Ount:H19, Ount:H21, Ount:H25, Ount:H38(44), Ount:H46, Ount:H49, and Ount:Hunt. The serotypes identified most often were O113:H21 (9.5%), followed by O8:H19 and O117:H7 (4.4% and 4.7%). These serotypes were isolated from samples originating from five (O8:H19) and six (O113:H21 and O117:H7) of the regions.
The Shiga toxin of each isolate was characterized by molecular subtyping (Table 3). Shiga toxin 1 variants were present in 138 of the 338 isolates, and Shiga toxin 2 variants were present in 296. Overall, 15 different patterns of stx genes were observed. The most common stx gene pattern was the sole presence of stx2a, which was the only stx gene present in 89 isolates that were made up of 42 different serotypes. The most common serotypes in this group were O8:H19, O22:H49, O113:H21 O163:H19, and Ount:H11, each of which was identified 5 or more times. The next most common stx subtype patterns present were the possession of stx1 and stx2a and the possession of stx2a and stx2c, both of which were identified in 56 of the STEC isolates. Serotypes O8:H19, O82:H8, and O88:H25 were the most commonly observed to possess stx1 and stx2a, while O91:H21 and O113:H21 were the most commonly observed to possess stx2 and stx2c. The other frequently observed stx gene patterns were the possession of either stx1 alone or stx2c alone, present in 41 and 39 of the STEC isolates, respectively.
TABLE 3.
Summary of Shiga toxin subtyping in isolates of STECa
| No. of samples | Result for Shiga toxin subtypeb: |
|||||
|---|---|---|---|---|---|---|
| 1 | 1c | 2a | 2c | 2d | 2xc | |
| 89 | + | |||||
| 56 | + | + | ||||
| 56 | + | + | ||||
| 41 | + | |||||
| 39 | + | |||||
| 18 | + | + | + | |||
| 15 | + | + | ||||
| 10 | + | |||||
| 7 | + | + | ||||
| 3 | + | |||||
| 2 | + | + | ||||
| 1 | + | |||||
| 1 | + | + | ||||
| 1 | + | + | ||||
| 1 | + | + | ||||
A total of 338 STEC isolates from ground beef were subjected to stx subtyping using PCR-based methods. The number(s) of isolates containing each pattern of stx subtype is shown. The copy number of each allele was not determined.
stx1 was subtyped to stx1 and stx1c, and stx2 was subtyped to stx2a, stx2c, stx2d, and stx2e. No stx2e isolates were identified.
Seventeen stx2-positive isolates were untypeable by the methods used and were designated stx2x.
The most uncommon stx gene patterns present were those that included stx1c and stx2d. Three stx1 genes were subtyped stx1c. The stx1c-containing STEC isolates were serotypes O15:H27, O104:H7, and O150:H8, the first two of which also carried stx2c, while the last one, O150:H8, possessed only stx1c. Shiga toxin 2d was present in a total of 6 STEC isolates. It was the only stx present in 3 of the isolates and was present with stx2a and stx2c in 2 and 1 of the remaining isolates, respectively. Serotypes that possessed stx2d were O22:H8, O112:H2, O171:H2, and Ount:H7. There were 17 STEC isolates that possessed stx2, determined by the screening PCR, but they could not be subtyped. The nonsubtypeable stx2 strains were therefore categorized stx2x. There were 14 different serotypes in the stx2x group, 3 of which were O113:Hunt.
Genes indicating the presence of the 60-MDa large virulence plasmid (ehxA, katP, espP, and etpD) were present in 293 and absent in 45 of the STEC isolates. The most common of these genes present were ehxA and espP, detected in 266 and 246 of the isolates, respectively. The genes for katP and etpD were rarely identified and were present only 5 and 4 times, respectively. Eight different patterns of the genes of the large virulence plasmid were observed, ranging from the presence of one to the presence of three of the genes. The most common patterns present were the combination of ehxA and espP, found in 218 isolates, and each of these markers alone: 41 isolates with ehxA and 25 with espP. No isolates contained all four markers, but 3 isolates did possess 3 of the genes. A serotype O163:H19 isolate contained the combination of ehxA-katP-etpD, while isolates of serotypes O145:H28 and O26:H11 contained the combination of ehxA-katP-espP. Twenty-three serotypes were identified among the STEC isolates that lacked any markers for the large virulence plasmid. A third of the isolates that appeared to lack the large virulence plasmid were serotypes O171:H2, O22:H8, and O121:H7.
The intimin gene was present in nine of the isolates (see the table in the supplemental material). Subtyping of this gene showed that five variants could be identified. The intimin gene subtypes were specific within certain serotypes. The β1 subtype was found in a serotype O26:H11 isolate, the γ subtype was found in serotypes O145:H28 and O117:Hunt, and the ɛ subtype was present in 4 isolates of serotype O103:H2, while the ζ subtype was present in serotype Ount:H25 and an untypeable intimin gene subtype was present in a serotype Ount:H8 isolate. These STEC isolates were recovered from samples originating from 4 different regions and between the months of October through May.
The intimin gene-containing isolates and an additional isolate of serotype O26:H21 were the only isolates to also possess any of the markers indicating the presence of virulence-associated OIs, OIs 122, 57, 36, and 71 (Table 4). Three genes were screened for in OIs 122, 57 and 36, while six genes were targeted in OI-71. In this screening, between 2 and 10 of the OI genes were identified, indicating that 10 STEC isolates contained 2 to 4 of these OIs. One isolate of serotype O26:H11, four isolates of serotype O103:H2, and one isolate of serotype Ount:H8 had between 6 and 10 of these markers, indicating the presence of all 4 OIs. Only the O26:H11 STEC isolate had markers suggesting that complete OIs 122 and 57 were present. Two of the O103:H2 isolates contained markers suggesting that a complete OI-36 was present. All other OIs, when they were present in any of the isolates, were incomplete. Two isolates did not contain any markers indicating the presence of OI-36 but did contain portions of OIs 122, 57, and 71. These were serotypes O145:H28 and O26:H11. Out of all the genetic markers screened, nleB and nleE in OI-122, nleB2 in OI-36, G2-3 in OI-57, and nleF in OI-71 were the most frequently observed. Two markers in OI-71, H1-2 and G2-1, were not found in any of the STEC isolates and were found only in the E. coli O157:H7 control.
TABLE 4.
Distribution of genetic markers within virulence-associated OIs of STEC isolatesa positive for one or more of the indicated genesa
| STEC isolate | Detection of indicated gene in OI: |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 122 |
57 |
36 |
71b |
||||||||||
| nleB | nleE | ent | G2-3 | G5-2 | G6-2 | nleC | H1-1 | nleB2 | nleG | nleG9 | nleF | nleA | |
| O26:H11 | + | + | + | + | + | + | + | + | + | + | |||
| O103:H2 | + | + | + | + | + | + | + | + | |||||
| O103:H2 | + | + | + | + | + | + | + | ||||||
| O103:H2 | + | + | + | + | + | + | + | ||||||
| O103:H2 | + | + | + | + | + | + | |||||||
| O26:H21c | + | + | + | + | + | + | |||||||
| Ount:H8 | + | + | + | + | + | + | |||||||
| O145:H28 | + | + | + | + | |||||||||
| O117:Hunt | + | + | |||||||||||
| Ount:H25 | + | + | |||||||||||
A total of 338 STEC isolates from ground beef were analyzed for the presence of the four OIs shown. Any isolates containing a genetic marker indicating the presence of an OI are listed.
Within OI-71, six genes were targeted (nleG, nleG9, nleF, H1-2, nleA, and G2-1), but H1-2 and G2-1 were not detected in any isolate; therefore, these markers are not listed.
All other STEC isolates contained intimin (eae), whereas this isolate contained subtilase (subA).
The STEC isolates were also screened for the presence of additional putative virulence factors (subA, lifA, cnf, and astA), adherence factors (iha and saa), and iron acquisition factors (chuA and hlyA) (see the table in the supplemental material). The subA gene was present in 49% of the STEC isolates, and no isolate that contained subA contained eae. The presence of subA was observed in 52 different serotypes, and in some cases the presence or absence of subA appeared to be a defining characteristic of a serotype. For instance, 88% of STEC O8:H19 isolates, 97% of STEC O113:H21 isolates, 100% of STEC O116:H21 isolates, and 100% of STEC isolates of serogroups O163 (H11, H19, and H46) contained subA, while 100% of STEC O91:H21 and O22:H8 isolates did not.
A small number of STEC isolates contained lifA, cnf, or astA. Six isolates of serotypes O26:H8, O26:H11, and O103:H21 contained lifA. Five of these six isolates also contained eae and nle gene markers for OIs. Nine STEC isolates of 6 different serotypes contained cnf. Half of this group was made up of serotypes O8:H16 and Ount:H21. STEC isolates that contained cnf did not contain subA or astA and more commonly (7 of 9) contained only stx1. Seventeen STEC isolates contained allele I and/or allele II of astA. Two of the astA isolates containing STEC possessed allele I and were of serotypes O145:H28 and Ount:H16, while 14 STEC isolates contained allele I and were of 10 serotypes. One STEC isolate contained both alleles of astA, and this was the STEC Ount:H25 strain that contained eae subtype ζ. The group of STEC isolates containing astA possessed varied types of stx genes. Another uncommon factor was chuA, which was present in 29 of the STEC isolates. STEC isolates of 21 different serotypes contained chuA, and there was no remarkable association with any of the other virulence factors examined.
A large portion of the STEC isolates contained iha (88%) and saa (73%). STEC isolates that did not contain iha were mostly nonremarkable, with the exception of STEC O103:H2 isolates, all of which contained eae and lacked iha. The saa gene was always present in the absence of eae and was more often found in isolates that also carried some portion of the large virulence plasmid (97%) compared to the incidence in those that lacked the large virulence plasmid (29%).
DISCUSSION
These studies were performed to determine the prevalence of STEC in commercial ground beef in the United States and to attempt to determine what proportion of such isolates may be significant pathogens. Commercial ground beef was the selected product because of its wide distribution to consumers and because it is formulated from varied beef source materials, including finely textured beef (FTB) and imported frozen lean boneless beef trim. Numerous providers from across the United States supplied samples for 24 months to generate as extensive and as representative of a sample pool as possible.
Screening of samples for stx genes showed regional and seasonal differences in their prevalence. However, we must warn against attempts to overinterpret these data. For instance, although the prevalence in May was statistically lower than the prevalence in March and November, the samples submitted for this month were more limited in number and location.
Analysis of the isolates by region showed that the most common serotypes of STEC could be found in all regions. The rate of STEC isolation by month in a region ranged from as low as 10% in regions 2 and 5 to as high as 60% in region 7. This variability in the rates of STEC isolation may be due to the load of STEC present in the samples. However, the use of methods to enumerate or measure the concentration of STEC before enrichment is not feasible in a large-scale study such as this. In future experiments, use of direct measurements or a most-probable-number (MPN) method should be considered to determine if such information can provide a reliable estimate of the STEC concentration in a sample.
Screening for the presence of stx1 and stx2 by PCR may not have been the best method for the initial determination of the prevalence of STEC, since other bacteria can carry the stx genes. Our results were similar to those that reported a 23% stx gene prevalence in samples of beef collected from grocery stores in Seattle, WA (61). In studies of minced meat and beef samples from France, the prevalence was found to be 15% in one study (57) and only 11% in another (59). Sixteen percent of ground beef samples examined in an Australian study (3) were reported to contain stx genes. The studies listed for comparison used geographically limited sampling areas. If the results are limited to only one sample set from one supplier, we observed stx prevalence rates that ranged from 0% to 100%. The prevalence of STEC in ground beef samples appears to be highly variable, and the factors contributing to this are an area for continued study.
All samples that screened positive for stx1 and/or stx2 were taken forward for the isolation of STEC. The rates of isolation of STEC from eae- and ehx-positive samples varied (Table 2). These values are higher than those previously reported by our group when an alternative STEC isolation method (colony hybridization) was used (1). There are several potential reasons that not every PCR-positive sample produced an STEC isolate. (i) Other species of bacteria besides E. coli can possess stx genes; therefore, the positive PCR signal may not have been from an STEC isolate. (ii) PCR can detect one copy of the stx genes; therefore, the concentration of STEC in the samples may have been too low to recover an isolate while still producing a positive PCR signal. (iii) The isolation method relied upon hemolytic phenotypes, but some STEC isolates do not posses either ehx or other hemolysins (13).
There are admitted limitations to the methods used in this study. The serotyping scheme was limited to the 70 VTEC O antisera available, and the H typing was based on sequencing of only fliC. Therefore, the numbers of O-untypeable and H-untypeable results were likely greater than would be reported if alternative serotyping methods had been used. Our other characterization methods relied on PCR amplification of genes and did not follow up on PCR-negative strains by testing with a secondary method such as hybridization to determine if a factor was indeed absent due to the large number of STEC isolates that were recovered for characterization.
The most frequent serotype identified in ground beef was O113:H21. This serotype was isolated from multiple independent sources over multiple months of sample collection. STEC isolates of this serotype were associated with a cluster of three HUS cases in Australia in 1998 (56), but they are rarely implicated in human disease in the United States (13). Without a direct comparison between our O113:H21 isolates and the Australian strains, it is difficult to determine why such a prevalent STEC strain is not more frequently reported in the United States. One isolate each of O145:H28 and O26:H11 was found in the ground beef samples. STEC isolates of these serotypes are considered frequent and significant sources of human disease (13), suggesting that those infections may not necessarily be from beef.
The virulence of STEC is a multifactorial trait that is not fully understood, and the genetic markers that define a pSTEC strain need to be determined. We attempted to use known and suggested virulence factors to characterize and identify those STEC isolates most likely to be pSTEC or defined as a seropathotype B or C isolate (39).
The first trait examined was the stx subtype. A number of studies have documented that Shiga toxin subtypes stx2a and stx2c are more often associated with HUS (17, 23), while stx2d and stx2e are less often associated with HUS (35, 37). The subtyping method used was based on PCR amplification of unique regions of the stx2 and stx1 (stx1c) genes. Although the use of PCR-restriction fragment length polymorphism (PCR-RFLP) has often been described for subtyping of stx variants (4), we prefer the PCR amplification methods used here because PCR-RFLP is vulnerable to single-nucleotide changes and the results can be difficult to interpret if an isolate contains more than one subtype of stx, as was often seen in the ground beef STEC isolates.
The most frequently observed stx2 variant in our isolates was stx2a, seen in 222 of the isolates, followed by stx2c in 130 isolates. These two stx variants were present together in 74 isolates of numerous serotypes. The presence of multiple alleles of stx is not an uncommon feature of STEC (58) but has been described to be significantly less frequent in non-O157 STEC than O157:H7 isolates (23, 29). The least common stx2 subtype was stx2d. The mucus-activatable stx, stx2d, has been associated with severe human disease and has been described to have limited occurrence in cattle reservoirs (30, 65, 80). Its rate of occurrence in isolates originating from cows and beef was shown to be 6 of 153 isolates, or 3.9% (80). Our study showed stx2d to be present at half this rate, 1.8% of all STEC isolates recovered. This is most likely due to the fact that all of our isolates were from ground beef, whereas the previous study also included isolates from humans, which may have skewed the prevalence upward. Two of the stx2d STEC isolates in our study were serogroup O22, a serogroup that has previously been described to carry this stx variant (80).
Unlike stx2, stx1 has few easily identifiable subtypes. Subtype 1c has been described to be commonly found among STEC isolates from sheep but seldom among those from cattle (12) and has also been isolated from asymptomatic or mildly ill humans (48). There were only 3 stx1c-containing isolates found in our study (a low percentage of the total), and they were found in serotypes O15:H27, O104:H7, and O150:H8. To the best of our knowledge, these serotypes have not previously been reported to contain stx1c.
Most of the primary virulence determinants of STEC are chromosomally encoded. These include the Shiga toxin variants and the LEE, as well as other OIs. However, plasmids have been described as playing role in the pathogenesis of STEC-related disease (38). The 60-MDa plasmid, also termed pO157, because it is found in nearly all clinical O157:H7 isolates from humans (44, 50), contains numerous genes associated with STEC pathogenesis. Reports correlate this large virulence plasmid with the hemolytic activity of ehx and adherence to intestinal epithelial cells through other genes (66). STEC isolates of different serotypes are known to harbor large plasmids, and the large plasmids of STEC are not uniform genetic elements but heterogeneous in both their gene composition and arrangement (15, 38). However, their necessity to conferring virulence may be arguable because the pO157 plasmid from sorbitol-fermenting E. coli O157:H7 lacks katP, espP, and toxB (14). The heterogeneous nature of the large virulence plasmid was borne out by our results. Our analysis of the large virulence plasmid in the STEC isolates from ground beef showed that there were at least eight variations present on the basis of the presence of the five genetic markers examined. None of the STEC isolates that contained the large virulence plasmid possessed the toxB gene. Other reports have described toxB to be the least frequent of these genes in non-O157 STEC isolates (18).
Other toxins reported to contribute to the virulence of STEC and the severity of STEC disease have been described. These include subA (53, 54), astA (25, 40, 71, 78), and cnf (7). subA is the prototype subtilase cytotoxin that was detected in a LEE-negative O113:H21 STEC strain that was responsible for a 1998 outbreak of HUS in Australia (56). subA is a highly potent and lethal subtilase cytotoxin that is unrelated to any bacterial toxin. Khaitan et al. (41) found a significant number of subA-positive STEC isolates among North American STEC isolates, whereas Wolfson et al. (75) did not. Nearly one-half of our STEC isolates (49%) contained subA, which is greater than the proportion reported by Khaitan et al. (41), who found that 6 of 24 (25%) cattle isolates contained subA. Heat-stable enterotoxin 1 (astA) is a genetically distinct toxin structurally related to heat-stable enterotoxin I (ST I) of enterotoxigenic E. coli (ETEC) (78). The astA gene was found in 31% (40) and 43% (25) of STEC isolates of swine origin and identified in serogroups O8:H−, O100:H−, O121:H−, O121:H10, and O159:H−. The prevalence of astA in ground beef STEC isolates was infrequent, being found in 17 isolates (5% of STEC) which were of 13 different serotypes. The serotypes of STEC isolates containing astA recovered from ground beef were not the same as any of those described for swine. cnf encodes a pair of toxins produced by uropathogenic E. coli strains that mediate its effects via the activation of small GTP-binding proteins. We used a global primer set that did not distinguish between the two (cnf1 and cnf2). Nine (2.7%) of the STEC isolates possessed cnf, and this low rate of prevalence in STEC is not uncommon (7, 49).
Several other factors that increase the ability of STEC to adhere to hosts and establish an infection have been described in STEC (67). These include ToxB (66), Saa (55), Iha (64), and Efa1, also termed LifA (47). These adhesins are encoded either in the large plasmid harbored by STEC strains or in chromosomal OIs. ToxB and Saa are plasmid encoded, and an association between the presence of saa and enterohemolysin gene ehx has been reported (55). Iha is encoded in OIs 43 and 48, while Efa1 is encoded in OI-122 (39). Therefore, our observations that the occurrence of these genes correlates with the presence of the large virulence plasmid or with the presence of some of the OI genetic markers are not surprising.
In E. coli O157:H7, the gene chuA codes for a 69-kDa outer membrane protein responsible for heme uptake (68). ChuA is thought to contribute to the colonization of human hosts and to the pathogenicity of E. coli strains causing extraintestinal infections. Hoffmann et al. (34) reported that 30% of E. coli strains isolated from the environment and about 70% of E. coli strains isolated from human sources carried chuA. The chuA gene was present in 29 (8.6%) of the STEC isolates in this study, and many were of serotypes associated with HUS or severe human disease, including an O145:H28 isolate and an O103:H2 isolate. Therefore, chuA may serve as a useful marker to identify pSTEC.
After the molecular risk assessment profile described by Coombes et al. (19) was applied, only 10 STEC isolates that contained some genetic determinants were identified, indicating the presence of at least a portion of OI-122, OI-57, OI-36, and/or OI-71 as well as other significant virulence factors (Table 5). As expected, isolates that were O26:H11, O103:H2, and O145:H28 were identified to possess 4 to 10 of the genetic determinants and all 4 OIs, except for O145, in which OI-36 was absent. Additionally, this screening indicated that STEC O26:H21, O117:Hunt, Ount:H8, and Ount:H25 contained some of these genetic determinants as well. Efforts to focus primarily on the six most frequent serogroups of STEC may not identify these additional potentially pathogenic STEC isolates. The analysis which assigned STEC isolates to seropathotypes showed that only 3% contained any portion of an OI associated with disease, suggesting that they belong to seropathotype group B or C (severe disease potential). This implies that 97% of STEC isolates from beef belong in low-incidence and no-severe-disease-seropathotype groups D and E.
TABLE 5.
Summary of all potential pathogenic STEC isolates of seropathotype B or C isolated from 4,133 samples of ground beef
| STEC isolate characterizationa |
MRAg |
||||||
|---|---|---|---|---|---|---|---|
| Isolate | Serotypeb | stx1c | stx2d | eaee | subAf | OI | Marker |
| 1 | O26:H11 | 1 | − | B1 | − | 4 | 10 |
| 2 | O103:H2 | 1 | − | E | − | 4 | 8 |
| 3 | O103:H2 | 1 | c | E | − | 4 | 7 |
| 4 | O103:H2 | 1 | − | E | − | 4 | 6 |
| 5 | O103:H2 | 1 | − | E | − | 4 | 7 |
| 6 | O145:H28 | 1 | − | G | − | 3 | 4 |
| 7 | O26:H21 | − | a | − | + | 3 | 6 |
| 8 | O117:Hunt | 1 | c | G | − | 2 | 2 |
| 9 | Ount:H8 | 1 | a | + | − | 4 | 6 |
| 10 | Ount:H25 | 1 | − | Z | − | 2 | 2 |
Ten STEC isolates meet the criteria of being considered a pathogenic STEC.
Untypeable O group or H type, indicated by “unt.”
stx1 subtype 1 or 1c is indicated.
stx2 subtype 2a (a), 2c (c), or 2d (d) is indicated.
Intimin subtype β1 (B1), ɛ (E), γ (G), or ζ (Z) is indicated; +, intimin positive but the subtype was not reactive with any of the preceding subtypes.
subA represents the subtilase cytotoxin.
MRA, molecular risk assessment, based on the presence of OIs 36, 57, 71, and 122 and genetic markers within those OIs (up to 16 possible). Values indicate the number of OIs or partial OIs present and the number of genetic markers present for the indicated OIs.
Seropathotype groups B and C were found in all regions at least once. However, these categories of isolates were not found in July, August, or September, which are the high-prevalence months for E. coli O157:H7. Since some non-O157 STEC isolates have been described to cross-react with commonly used E. coli O157:H7 molecular detection tests (2), it is possible that the industry practice of E. coli O157:H7 test and hold reduced the prevalence of these pathogens during that time period as well. The most frequently isolated serotype of pSTEC was O103:H2. This serotype was isolated from samples originating in 3 different regions (regions 5, 7, and 8). Only one of these O103:H2 isolates was positive for stx2c; all others contained only stx1. All contained intimin of subtype ɛ. This is the most prevalent of the top six serogroups found in this study.
Much attention is currently being placed on the development of tests for detection of the most common human disease-related STEC isolates. The most recently proposed methods released by the USDA Food Safety Inspection Service (70) state that enrichments should be screened for stx and eae. In this study, four of the STEC isolates of the greatest pathogenic potential were isolated from enrichments that had initially screened negative for eae. Had a positive screen for eae been a selection criterion, one STEC O26:H11 isolate and two O103:H2 isolates would have gone undetected.
Of the six most frequent STEC serogroups found in human disease, only isolates of serogroups O26, O103, O121, and O145 were identified in ground beef. Further, a number of these isolated STEC isolates lacked virulence factors associated with severe disease. An additional O26:H21 isolate and O103:H21 isolate that contained Shiga toxins but few of the other virulence factors were isolated in our evaluation. Nine STEC O121 isolates of four H types (H7, H8, H16, and H19) were isolated. Only two contained subA and four possessed saa and genes of the large virulence plasmid, which are not enough to raise them to the category of pSTEC or seropathotype group B or C. Narrowly focusing on only the described top six STEC serogroups (13) will identify numerous isolates of little pathogenic concern, while it will miss others that should not go unnoticed.
In conclusion, from 4,133 samples of U.S. commercial ground beef, we have determined that the prevalence of STEC may be as high as 24.3% and was confirmed in 7.3% of the samples. When the 338 resulting isolates were narrowed down to those most likely to be pSTEC or in seropathotype group B or C, the prevalence rate was reduced to 0.24%. On the basis of the virulence factor analysis and our best judgment of the data analysis, we believe that screening of ground beef enrichments for some combination of stx1, stx2, ehx, eae, subA, chuA, nleB, and nleF may be a good approach to identifying samples that might harbor pSTEC. However, further work is needed to validate this approach.
Supplementary Material
Acknowledgments
We gratefully thank the participating ground beef producers for their role in supplying, collecting, and submitting samples for this work. We also thank Dennis Johnson for assistance organizing the receipt and maintenance of confidentiality of the suppliers. We thank Greg Smith, Sydney Bidleman, and Scott Schroetlin for technical support; Cheryl Yates for secretarial support; and Terrance Arthur and Norasak Kalchayanand for scientific support.
This project was funded in part by The Beef Checkoff.
Names are necessary to report factually on available data; however, the USDA neither guarantees nor warrants the standard of the product, and the use of the name by USDA implies no approval of the product to the exclusion of others that may also be suitable.
Footnotes
Published ahead of print on 21 January 2011.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Arthur, T. M., G. A. Barkocy-Gallagher, M. Rivera-Betancourt, and M. Koohmaraie. 2002. Prevalence and characterization of non-O157 Shiga toxin-producing Escherichia coli on carcasses in commercial beef cattle processing plants. Appl. Environ. Microbiol. 68:4847-4852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arthur, T. M., J. M. Bosilevac, X. Nou, and M. Koohmaraie. 2005. Evaluation of culture- and PCR-based detection methods for Escherichia coli O157:H7 in inoculated ground beef. J. Food Prot. 68:1566-1574. [DOI] [PubMed] [Google Scholar]
- 3.Barlow, R. S., K. S. Gobius, and P. M. Desmarchelier. 2006. Shiga toxin-producing Escherichia coli in ground beef and lamb cuts: results of a one-year study. Int. J. Food Microbiol. 111:1-5. [DOI] [PubMed] [Google Scholar]
- 4.Bastian, S. N., I. Carle, and F. Grimont. 1998. Comparison of 14 PCR systems for the detection and subtyping of stx genes in Shiga-toxin-producing Escherichia coli. Res. Microbiol. 149:457-472. [DOI] [PubMed] [Google Scholar]
- 5.Bettelheim, K. A. 1995. Identification of enterohaemorrhagic Escherichia coli by means of their production of enterohaemolysin. J. Appl. Bacteriol. 79:178-180. [DOI] [PubMed] [Google Scholar]
- 6.Blanco, J. E., et al. 1996. O serogroups, biotypes and eae genes in Escherichia coli isolated from diarrheic and healthy rabbits. J. Clin. Microbiol. 34:3101-3107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blanco, J., et al. 1992. Serogroups of Escherichia coli strains producing cytotoxic necrotizing factors CNF1 and CNF2. FEMS Microbiol. Lett. 96:155-160. [DOI] [PubMed] [Google Scholar]
- 8.Blanco, M., et al. 2004. Serotypes, virulence genes, and intimin types of Shiga toxin (verotoxin)-producing Escherichia coli isolates from cattle in Spain and identification of a new intimin variant gene (eae-xi). J. Clin. Microbiol. 42:645-651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boerlin, P., et al. 1999. Associations between virulence factors of Shiga toxin-producing Escherichia coli and disease in humans. J. Clin. Microbiol. 37:497-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bosilevac, J. M., M. N. Guerini, D. M. Brichta-Harhay, T. M. Arthur, and M. Koohmaraie. 2007. Microbiological characterization of imported and domestic boneless beef trim used for ground beef. J. Food Prot. 70:440-449. [DOI] [PubMed] [Google Scholar]
- 11.Bosilevac, J. M., M. N. Guerini, N. Kalchayanand, and M. Koohmaraie. 2009. Prevalence and characterization of salmonellae in commercial ground beef in the United States. Appl. Environ. Microbiol. 75:1892-1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brett, K. N., et al. 2003. stx1c is the most common Shiga toxin 1 subtype among Shiga toxin-producing Escherichia coli isolates from sheep but not among isolates from cattle. J. Clin. Microbiol. 41:926-936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brooks, J. T., et al. 2005. Non-O157 Shiga toxin-producing Escherichia coli infections in the United States, 1983-2002. J. Infect. Dis. 192:1422-1429. [DOI] [PubMed] [Google Scholar]
- 14.Brunder, W., H. Karch, and H. Schmidt. 2006. Complete sequence of the large virulence plasmid pSFO157 of the sorbitol-fermenting enterohemorrhagic Escherichia coli O157:H− strain 3072/96. Int. J. Med. Microbiol. 296:467-474. [DOI] [PubMed] [Google Scholar]
- 15.Brunder, W., H. Schmidt, M. Frosch, and H. Karch. 1999. The large plasmids of Shiga-toxin-producing Escherichia coli (STEC) are highly variable genetic elements. Microbiology 145:1005-1014. [DOI] [PubMed] [Google Scholar]
- 16.Burland, V., et al. 1998. The complete DNA sequence and analysis of the large virulence plasmid of Escherichia coli O157:H7. Nucleic Acids Res. 26:4196-4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Caprioli, A., I. Luzzi, A. Gianviti, H. Russmann, and H. Karch. 1995. Pheno-genotyping of verotoxin 2 (VT2)-producing Escherichia coli causing haemorrhagic colitis and haemolytic uraemic syndrome by direct analysis of patients' stools. J. Med. Microbiol. 43:348-353. [DOI] [PubMed] [Google Scholar]
- 18.Cergole-Novella, M. C., et al. 2007. Distribution of virulence profiles related to new toxins and putative adhesins in Shiga toxin-producing Escherichia coli isolated from diverse sources in Brazil. FEMS Microbiol. Lett. 274:329-334. [DOI] [PubMed] [Google Scholar]
- 19.Coombes, B. K., et al. 2008. Molecular analysis as an aid to assess the public health risk of non-O157 Shiga toxin-producing Escherichia coli strains. Appl. Environ. Microbiol. 74:2153-2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.DebRoy, C., P. M. Fratamico, E. Roberts, M. A. Davis, and Y. Liu. 2005. Development of PCR assays targeting genes in O-antigen gene clusters for detection and identification of Escherichia coli O45 and O55 serogroups. Appl. Environ. Microbiol. 71:4919-4924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.DebRoy, C., et al. 2004. Detection of Escherichia coli serogroups O26 and O113 by PCR amplification of the wzx and wzy genes. Appl. Environ. Microbiol. 70:1830-1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Durso, L. M., J. L. Bono, and J. E. Keen. 2007. Molecular serotyping of Escherichia coli O111:H8. J. Microbiol. Methods 69:381-383. [DOI] [PubMed] [Google Scholar]
- 23.Eklund, M., K. Leino, and A. Siitonen. 2002. Clinical Escherichia coli strains carrying stx genes: stx variants and stx-positive virulence profiles. J. Clin. Microbiol. 40:4585-4593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Feng, L., et al. 2005. Structural and genetic characterization of enterohemorrhagic Escherichia coli O145 O antigen and development of an O145 serogroup-specific PCR assay. J. Bacteriol. 187:758-764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fratamico, P. M., A. A. Bhagwat, L. Injaian, and P. J. Fedorka-Cray. 2008. Characterization of Shiga toxin-producing Escherichia coli strains isolated from swine feces. Foodborne Pathog. Dis. 5:827-838. [DOI] [PubMed] [Google Scholar]
- 26.Fratamico, P. M., C. E. Briggs, D. Needle, C.-Y. Chen, and C. DebRoy. 2003. Sequence of the Escherichia coli O121 O-antigen gene cluster and detection of enterohemorrhagic E. coli O121 by PCR amplification of the wzx and wzy genes. J. Clin. Microbiol. 41:3379-3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fratamico, P. M., C. DebRoy, T. P. Strobaugh, Jr., and C.-Y. Chen. 2005. DNA sequence of the Escherichia coli O103 O antigen gene cluster and detection of enterohemorrhagic E. coli O103 by PCR amplification of the wzx and wzy genes. Can. J. Microbiol. 51:515-522. [DOI] [PubMed] [Google Scholar]
- 28.Friedrich, A. W., et al. 2002. Escherichia coli harboring Shiga toxin 2 gene variants: frequency and association with clinical symptoms. J. Infect. Dis. 185:74-84. [DOI] [PubMed] [Google Scholar]
- 29.Fürst, S., et al. 2000. Identification and characterization of Escherichia coli strains of O157 and non-O157 serogroups containing three distinct Shiga toxin genes. J. Med. Microbiol. 49:383-386. [DOI] [PubMed] [Google Scholar]
- 30.Gobius, K. S., G. M. Higgs, and P. M. Desmarchelier. 2003. Presence of activatable Shiga toxin genotype (stx2d) in Shiga-toxigenic Escherichia coli from livestock sources. J. Clin. Microbiol. 41:3777-3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reference deleted.
- 32.Guerinot, M. L. 1994. Microbial iron transport. Annu. Rev. Microbiol. 48:743-772. [DOI] [PubMed] [Google Scholar]
- 33.Guinee, P. P. M., C. M. Agterberg, and W. H. Jansen. 1972. Escherichia coli O antigen typing by means of a mechanized microtechnique. Appl. Microbiol. 24:127-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hoffmann, H., M. W. Hornef, S. Schubert, and A. Roggenkamp. 2001. Distribution of the outer membrane haem receptor protein ChuA in environmental and human isolates of Escherichia coli. Int. J. Med. Microbiol. 291:227-230. [DOI] [PubMed] [Google Scholar]
- 35.Ito, H., A. Terai, H. Kurazono, Y. Takeda, and M. Nishibuchi. 1990. Cloning and nucleotide sequencing of Vero toxin 2 variant genes from Escherichia coli O91:H21 isolated from a patient with the hemolytic uremic syndrome. Microb. Pathog. 8:47-60. [DOI] [PubMed] [Google Scholar]
- 36.Jenkins, C., et al. 2003. Distribution of the saa gene in strains of Shiga toxin-producing Escherichia coli of human and bovine origins. J. Clin. Microbiol. 41:1775-1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jenkins, C., et al. 2003. Subtyping of virulence genes in verocytotoxin-producing Escherichia coli (VTEC) other than serogroup O157 associated with disease in the United Kingdom. J. Med. Microbiol. 52:941-947. [DOI] [PubMed] [Google Scholar]
- 38.Johnson, T. J., and L. K. Nolan. 2009. Pathogenomics of the virulence plasmids of Escherichia coli. Microbiol. Mol. Biol. Rev. 73:750-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Karmali, M. A., et al. 2003. Association of genomic O island 122 of Escherichia coli EDL 933 with verocytotoxin-producing Escherichia coli seropathotypes that are linked to epidemic and/or serious disease. J. Clin. Microbiol. 41:4930-4940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kaufmann, M., et al. 2006. Escherichia coli O157 and non-O157 Shiga toxin-producing Escherichia coli in fecal samples of finished pigs at slaughter in Switzerland. J. Food Prot. 69:260-266. [DOI] [PubMed] [Google Scholar]
- 41.Khaitan, A., D. M. Jandhyala, C. M. Thorpe, J. M. Ritchie, and A. W. Paton. 2007. The operon encoding SubAB, a novel cytotoxin, is present in Shiga toxin-producing Escherichia coli isolates from the United States. J. Clin. Microbiol. 45:1374-1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Klapproth, J.-M. A. 2010. The role of lymphostatin/EHEC factor for adherence-1 in the pathogenesis of gram negative infection. Toxins 2:954-962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lehmacher, A., H. Meier, S. Aleksic, and J. Bockemühl. 1998. Detection of hemolysin variants of Shiga toxin-producing Escherichia coli by PCR and culture on vancomycin-cefixime-cefsulodin blood agar. Appl. Environ. Microbiol. 64:2449-2453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Levine, M. M., et al. 1987. A DNA probe to identify enterohemorrhagic Escherichia coli of O157:H7 and other serotypes that cause hemorrhagic colitis and hemolytic uremic syndrome. J. Infect. Dis. 156:175-182. [DOI] [PubMed] [Google Scholar]
- 45.Liu, Y., C. DebRoy, and P. Fratamico. 2007. Sequencing and analysis of the Escherichia coli serogroup O117, O126, and O146 O-antigen gene clusters and development of PCR assays targeting serogroup O117-, O126-, and O146-specific DNA sequences. Mol. Cell. Probes 21:295-302. [DOI] [PubMed] [Google Scholar]
- 46.Nataro, J. P., and J. B. Kaper. 1998. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 11:142-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nicholls, L., T. H. Grant, and R. M. Robins-Browne. 2000. Identification of a novel genetic locus that is required for in vitro adhesion of a clinical isolate of enterohaemorrhagic Escherichia coli to epithelial cells. Mol. Microbiol. 35:275-288. [DOI] [PubMed] [Google Scholar]
- 48.Ochoa, T. J., and T. G. Cleary. 2003. Epidemiology and spectrum of disease of Escherichia coli O157. Curr. Opin. Infect. Dis. 16:259-263. [DOI] [PubMed] [Google Scholar]
- 49.Osek, J., P. Gallien, and D. Protz. 2000. Characterization of Shiga toxin-producing Escherichia coli strains isolated from calves in Poland. Comp. Immunol. Microbiol. Infect. Dis. 23:267-276. [DOI] [PubMed] [Google Scholar]
- 50.Ostroff, S. M., et al. 1989. Toxin genotypes and plasmid profiles as determinants of systemic sequelae in Escherichia coli O157:H7 infections. J. Infect. Dis. 160:994-998. [DOI] [PubMed] [Google Scholar]
- 51.Paton, A. W., and J. C. Paton. 1998. Detection and characterization of Shiga toxigenic Escherichia coli by using multiplex PCR assays for stx1, stx2, eaeA, enterohemorrhagic E. coli hlyA, rfbO111, and rfbO157. J. Clin. Microbiol. 36:598-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Paton, A. W., and J. C. Paton. 2002. Direct detection and characterization of Shiga toxigenic Escherichia coli by multiplex PCR for stx1, stx2, eae, ehxA, and saa. J. Clin. Microbiol. 40:271-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Paton, A. W., and J. C. Paton. 2005. Multiplex PCR for direct detection of Shiga toxigenic Escherichia coli strains producing the novel subtilase cytotoxin. J. Clin. Microbiol. 43:2944-2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Paton, A. W., P. Srimanote, U. M. Talbot, H. Wang, and J. C. Paton. 2004. A new family of potent AB(5) cytotoxins produced by Shiga toxigenic Escherichia coli. J. Exp. Med. 200:35-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Paton, A. W., P. Srimanote, M. C. Woodrow, and J. C. Paton. 2001. Characterization of Saa, a novel auto agglutinating adhesin produced by locus of enterocyte effacement-negative Shiga-toxigenic Escherichia coli strains that are virulent for humans. Infect. Immun. 69:6999-7009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Paton, A. W., M. C. Woodrow, R. M. Doyle, J. A. Lanser, and J. C. Paton. 1999. Molecular characterization of a Shiga toxigenic Escherichia coli O113:H21 strain lacking eae responsible for a cluster of cases of hemolytic-uremic syndrome. J. Clin. Microbiol. 37:3357-3361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Perelle, S., F. Dilasser, J. Grout, and P. Fach. 2006. Screening food raw materials for the presence of the world's most frequent clinical cases of Shiga toxin-encoding Escherichia coli O26, O103, O111, O145, and O157. Int. J. Food Microbiol. 113:284-288. [DOI] [PubMed] [Google Scholar]
- 58.Persson, S., K. E. P. Olsen, S. Ethelberg, and F. Scheutz. 2007. Subtyping method for Escherichia coli Shiga toxin (verocytotoxin) 2 variants and correlations to clinical manifestations. J. Clin. Microbiol. 45:2020-2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pradel, N., et al. 2000. Prevalence and characterization of Shiga toxin-producing Escherichia coli isolated from cattle, food, and children during a one-year prospective study in France. J. Clin. Microbiol. 38:1023-1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rivero, M. A., J. A. Passucci, E. M. Rodriguez, and A. E. Parma. 2010. Role and clinical course of verotoxigenic Escherichia coli infections in childhood acute diarrhoea in Argentina. J. Med. Microbiol. 59:345-352. [DOI] [PubMed] [Google Scholar]
- 61.Samadpour, M., et al. 1994. Occurrence of Shiga-like toxin-producing Escherichia coli in retail fresh seafood, beef, lamb, pork, and poultry from grocery stores in Seattle, Washington. Appl. Environ. Microbiol. 60:1038-1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Slutsker, L., et al. 1997. Escherichia coli O157:H7 diarrhea in the United States: clinical and epidemiologic features. Ann. Intern. Med. 126:505-513. [DOI] [PubMed] [Google Scholar]
- 63.Sugiyama, K., K. Inoue, and R. Sakazaki. 2001. Mitomycin-supplemented washed blood agar for the isolation of Shiga toxin-producing Escherichia coli other than O157:H7. Lett. Appl. Microbiol. 33:193-195. [DOI] [PubMed] [Google Scholar]
- 64.Tarr, P. I., et al. 2000. Iha: a novel Escherichia coli O157:H7 adherence-conferring molecule encoded on a recently acquired chromosomal island of conserved structure. Infect. Immun. 68:1400-1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tasara, T., et al. 2008. Activatable Shiga toxin 2d (Stx2d) in STEC strains isolated from cattle and sheep at slaughter. Vet. Microbiol. 131:199-204. [DOI] [PubMed] [Google Scholar]
- 66.Tatsuno, I., et al. 2001. toxB gene on pO157 of enterohemorrhagic Escherichia coli O157:H7 is required for full epithelial cell adherence phenotype. Infect. Immun. 69:6660-6669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Toma, C., et al. 2004. Distribution of putative adhesins in different seropathotypes of Shiga toxin-producing Escherichia coli. J. Clin. Microbiol. 42:4937-4946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Torres, A. G., and S. M. Payne. 1997. Haem iron-transport system in enterohaemorrhagic Escherichia coli O157:H7. Mol. Microbiol. 23:825-833. [DOI] [PubMed] [Google Scholar]
- 69.Tozzi, A. E., et al. 2003. Shiga toxin-producing Escherichia coli infections associated with hemolytic uremic syndrome, Italy, 1988-2000. Emerg. Infect. Dis. 9:106-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.USDA, Food Safety Inspection Service. 2010. Detection and ISolation of non-O157 Shiga-toxin producing Escherichia coli strains (STEC) from meat products. In Microbiological laboratory guidebook, version 5B.00. USDA, Food Safety Inspection Service, Washington, DC. www.fsis.usda.gov/PDF/Mlg_5B_00.pdf. Accessed 30 November 2010.
- 71.Vial, P. A., et al. 1988. Characterization of enteroadherent-aggregative Escherichia coli, a putative agent of diarrheal disease. J. Infect. Dis. 158:70-79. [DOI] [PubMed] [Google Scholar]
- 72.Wang, G., C. G. Clark, and F. G. Rodgers. 2002. Detection in Escherichia coli of the genes encoding the major virulence factors, the genes defining the O157:H7 serotype, and components of the type 2 Shiga toxin family by multiplex PCR. J. Clin. Microbiol. 40:3613-3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang, L., D. Rothemund, H. Curd, and P. R. Reeves. 2003. Species-wide variation in the Escherichia coli flagellin (H-antigen) gene. J. Bacteriol. 185:2936-2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wickham, M. E., et al. 2006. Bacterial genetic determinants of non-O157 STEC outbreaks and hemolytic-uremic syndrome after infection. J. Infect. Dis. 194:819-827. [DOI] [PubMed] [Google Scholar]
- 75.Wolfson, J. J., et al. 2009. Prevalence of the operon encoding subtilase cytotoxin in non-O157 Shiga toxin-producing Escherichia coli isolated from humans in the United States. J. Clin. Microbiol. 47:3058-3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wyckoff, E. E., et al. 1998. Structure of the Shigella dysenteriae haem transport locus and its phylogenetic distribution in enteric bacteria. Mol. Microbiol. 28:1139-1152. [DOI] [PubMed] [Google Scholar]
- 77.Yamamoto, T., and P. Echeverria. 1996. Detection of the enteroaggregative Escherichia coli heat-stable enterotoxin 1 gene sequences in enterotoxigenic E. coli strains pathogenic for humans. Infect. Immun. 64:1441-1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yatsuyanagi, J., S. Saito, Y. Miyajima, K. Amano, and K. Enomoto. 2003. Characterization of atypical enteropathogenic Escherichia coli strains harboring the astA gene that were associated with a waterborne outbreak of diarrhea in Japan. J. Clin. Microbiol. 41:2033-2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhang, W., M. Bielaszewska, T. Kuczius, and H. Karch. 2002. Identification, characterization, and distribution of a Shiga toxin 1 gene variant (stx1c) in Escherichia coli strains isolated from humans. J. Clin. Microbiol. 40:1441-1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zheng, J., et al. 2008. Identification and characterization of Shiga toxin type 2 variants in Escherichia coli isolates from animals, food, and humans. Appl. Environ. Microbiol. 74:5645-5652. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.