Abstract
The diversities leaf-associated bacteria on nonnodulated (Nod−), wild-type nodulated (Nod+), and hypernodulated (Nod++) soybeans were evaluated by clone library analyses of the 16S rRNA gene. To analyze the impact of nitrogen fertilization on the bacterial leaf community, soybeans were treated with standard nitrogen (SN) (15 kg N ha−1) or heavy nitrogen (HN) (615 kg N ha−1) fertilization. Under SN fertilization, the relative abundance of Alphaproteobacteria was significantly higher in Nod− and Nod++ soybeans (82% to 96%) than in Nod+ soybeans (54%). The community structure of leaf-associated bacteria in Nod+ soybeans was almost unaffected by the levels of nitrogen fertilization. However, differences were visible in Nod− and Nod++ soybeans. HN fertilization drastically decreased the relative abundance of Alphaproteobacteria in Nod− and Nod++ soybeans (46% to 76%) and, conversely, increased those of Gammaproteobacteria and Firmicutes in these mutant soybeans. In the Alphaproteobacteria, cluster analyses identified two operational taxonomic units (OTUs) (Aurantimonas sp. and Methylobacterium sp.) that were especially sensitive to nodulation phenotypes under SN fertilization and to nitrogen fertilization levels. Arbuscular mycorrhizal infection was not observed on the root tissues examined, presumably due to the rotation of paddy and upland fields. These results suggest that a subpopulation of leaf-associated bacteria in wild-type Nod+ soybeans is controlled in similar ways through the systemic regulation of autoregulation of nodulation, which interferes with the impacts of N levels on the bacterial community of soybean leaves.
Although diverse microorganisms reside in the phytosphere as endophytes, epiphytes, and rhizosphere bacteria, many questions about the driving forces and ecological rules underlying the relationships between these microbes and plants remain unanswered (18, 39). During their evolution, legumes have developed two systems for attaining mutual symbioses with rhizobia and mycorrhizae. One of the systems genetically required for rhizobial and arbuscular mycorrhizal interactions in plants overlaps in a common signaling pathway (CSP) leading to successful symbioses (24). Plants also have a control system for regulating the degree of nodulation and mycorrhization on roots by rhizobia and mycorrhizae, respectively. This autoregulatory system occurs through long-distance signaling between shoots and roots (33). Leguminous plants deficient in the CSP and autoregulation systems develop nonnodulated (Nod−) and hypernodulated (Nod++) roots, respectively. However, the degree to which plants use similar or identical systems, such as CSP and autoregulation, for interactions with other microorganisms in the phytosphere remains unclear.
Recently, it was shown that the bacterial and fungal community structures in the roots of symbiosis-defective mutants of Medicago truncatula (32) and soybean (22) differ from those in the roots of wild-type host plants; it was also shown that certain microbes preferentially associate with arbuscular mycorrhizal roots (41) and nodulated (Nod+) roots (22). However, unexpectedly, analyses of the rhizosphere community in soybeans have revealed that the bacterial community in nonnodulated soybeans is more similar to that in hypernodulated soybeans than to that in wild-type soybeans (22).
The autoregulation of nodulation occurs through long-distance signaling between shoots and roots (33), and a heavy supply of nitrate to the roots of leguminous plants inhibits nodulation through autoregulation (6, 34). Thus, it is possible that the nodulation phenotype and host nitrogen status affect the structure of the microbial community in aboveground tissues, such as stems and leaves, as has been observed in roots (22). Indeed, the results of our previous study of stem-associated bacteria suggested that a subpopulation of Proteobacteria in the stems of soybeans was controlled in similar ways through both the system regulating plant-rhizobium symbiosis and the system sensing exogenous nitrogen fertilization in plants (21).
These findings led to the question of whether soybean-associated bacteria are more tightly regulated in leaves than in stems, since the shoot-derived factor (SDF) for the autoregulation of nodulation is thought to be produced there (26, 47). If this is the case, then the observation of more direct and clear effects of autoregulation systems on soybean-associated bacteria, including endophytes and epiphytes, could be expected in the leaves than in the stems. Here, the impacts of the nodulation phenotype and the nitrogen fertilization level on the community structures of soybean leaf-associated bacteria were examined by a clone library analysis of the 16S rRNA gene sequence.
MATERIALS AND METHODS
Plant materials and field experimental design.
The plants included the soybean cultivar Enrei (wild-type nodulating cultivar; Nod+), the lines En 1314 and En 1282 (nonnodulating mutants derived from Enrei; Nod−) (13), and the lines En 6500 and Sakukei 4 (hypernodulating mutants derived from Enrei; Nod++) (1). Mutations in the NFR1 gene, which is responsible for a LysM-type receptor kinase required for Nod factor recognition, have been found in En 1314 and En 1282 soybeans (38; M. Hayashi, National Institute of Agrobiological Sciences, Tsukuba, Japan, personal communication). A mutation in En 6500 was found in the GmNARK (NTS1) gene (31), which mediates systemic autoregulation of nodulation (44), and Sakukei 4 is a descendant of En 6500 with the same hypernodulating phenotype due to the lack of a functional GmNARK gene (29).
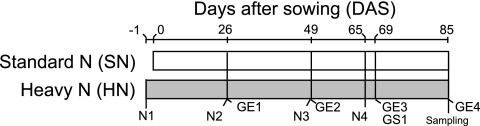
The seeds were planted on 3 June 2009 in an experimental field at Tohoku University (Kashimadai, Miyagi, Japan), which has been cultivated with a rotation of rice (under paddy field conditions) and soybean (upland field conditions) every year since 1997. The field design is outlined in Fig. 1. Soybeans of all nodulation genotypes were grown in two neighboring fields (standard nitrogen [SN] and heavy nitrogen [HN]). The two fields were dressed with SN fertilization (15 kg N ha−1) as a basal fertilization (N1 in Fig. 1), and the HN field was additionally dressed four times with HN fertilization (N1 to N4 in Fig. 1) (each application contained 150 kg N ha−1 as urea; total, 600 kg N ha−1). The field soil was classified as a Gray Lowland (pH [H2O], 5.9; pH [KCl], 4.3; total carbon content, 1.21%; total nitrogen content, 0.11%; Truog phosphorus content, 69 mg P2O5 kg−1).
FIG. 1.
Timetable and design of the field experiment. Basal fertilization was supplied to both the SN and HN fields as commercial fertilizer at 15 kg N ha−1. The HN field was also supplied with nitrogen fertilization as urea four times (N1 to N4; 150 kg N ha−1 for each fertilization). GE1 to GE4 represent the days on which soybean growth indexes (see Fig. S1 in the supplemental material) were measured. GS1 is the growth stage at which the dry weights and nitrogen contents of the soybean leaves were measured (see Fig. S2 and S3 in the supplemental material).
Growth evaluation and sampling.
To define the environmental factors relevant to changes in bacterial community structure, the plant growth stage was monitored by measuring the vegetative and reproductive indexes, plant dry weights, and nitrogen contents in plant tissues and field soils. The growth stage examination (GE) was carried out four times throughout the growing period (GE1 to GE4 in Fig. 1), in accordance with the criteria of Fehr et al. (11). The dry weights of shoots, roots, and nodules, as well as the number of nodules, were measured 69 days after sowing (at growth stage 1 [GS1] [Fig. 1]). Total nitrogen in soybean shoots at GS1 was analyzed by the Kjeldahl method. Two plants were processed as a composite sample to measure the nitrogen content of the soybean shoots. The nitrogen content in soils under SN or HN fertilization was also measured three times, before each of the additional nitrogen fertilizations of the HN field (Fig. 1, N2, N3, and N4). Soils were air dried and sieved through a 2-mm mesh. The contents of NH4+ and NO3− were determined with the autoanalyzers Quaatro and AACS-II (Bran+Luebbe, Hamburg, Germany), respectively. Leaflets were collected manually and stored at −80°C until they were used (Fig. 1, Sampling). Four plants were processed as a composite sample for bacterial cell enrichment. Three composite samples were used for DNA preparation and PCR amplification of each genotype in triplicate.
Clone library construction and sequencing.
Leaves (approximately 50 g) of a composite sample were homogenized without surface sterilization to prepare leaf-associated bacterial cells (including both epiphytes and endophytes), and the bacterial cells were extracted from leaf tissues and purified by an enrichment method (19), with slight modification (namely, two additional washing steps with high-speed centrifugation to eliminate contamination by chloroplast DNA in the final bacterial fraction). Total bacterial DNA was extracted from each bacterial cell fraction by the DNA extraction method described by Ikeda et al. (22). The final DNA samples were resuspended in 100 μl of sterilized water. The quality and quantity of DNA were assessed spectrophotometrically by calculating absorbance at a wavelength of 260 nm (A260) and the A260/A230 and A260/A280 ratios. For each soybean genotype grown under SN or HN fertilization conditions, triplicate DNA samples were prepared from composite samples. PCR clone libraries for 16S rRNA genes were constructed as follows. Briefly, 25 ng of total bacterial DNA was used as a template in a final reaction volume of 12.5 μl, including 25 pmol of each primer and 1 U of Ex Taq DNA polymerase (Takara Bio, Otsu, Japan). The universal primers 27F (50-AGAGTTTGATCMTGGCTCAG-30) and 1525R (50-AAGGAGGTGWTCCARCC-30) were used (25). Cycling conditions were as follows: initial denaturation for 2 min at 94°C; then, 25 cycles consisting of 30 s at 94°C, 30 s at 55°C, and 2 min at 72°C; and a final extension for 10 min at 72°C. Three PCR products derived from triplicate DNA samples were combined into a sample, and the PCR products were resolved by 1% agarose gel electrophoresis in 1× TBE (89 mM Tris-borate, 0.2 mM EDTA) buffer. PCR products of the predicted size (approximately 1,500 bp) were extracted from the gels by using NucleoSpin Extract II (Macherey-Nagel GmbH & Co. KG, Düren, Germany) and ligated into the pGEM-T Easy plasmid vector (Promega Japan, Tokyo, Japan) at 25°C for 1 h. ElectroTen-Blue electroporation-competent cells (Stratagene, La Jolla, CA) were then electroporated with the ligated DNA by using a Gene Pulser electroporator (Bio-Rad Laboratories, Tokyo, Japan). After the transformants had been cultured overnight at 37°C on Luria-Bertani agar plates containing ampicillin (50 mg ml−1), 384 colonies were randomly selected from each library for sequencing. Sequencing analysis was conducted with a type 3730×l DNA Analyzer (Applied Biosystems, Foster City, CA) using a BigDye Terminator Cycle Sequencing Reaction Kit (Applied Biosystems). Template DNAs were prepared by using an Illustra TempliPhi DNA Amplification Kit (GE Healthcare, Uppsala, Sweden). A partial sequence of the 16S rRNA gene was obtained using the 27F primer. Sequences were processed with Phred (9, 10) at a cutoff value of 0.001 to eliminate low-quality regions. Approximately 500 bases of the 16S rRNA gene (corresponding to 109 to 665 bases of the Escherichia coli 16S rRNA gene) were then used for the sequence analyses.
Sequence analysis.
Sequences were analyzed for orientation and detection of non-16S rRNA gene sequences by using OrientationChecker (3). The presence of chimeras was assessed by MALLARD (3). A sequence identified at the 99.9% threshold was discarded as chimeric. The remaining sequences were aligned by using CLUSTAL W (45). On the basis of the alignment, a distance matrix was constructed by using the DNADIST program from PHYLIP ver. 3.66 (http://evolution.genetics.washington.edu/phylip.html) with the default parameters. The resulting matrices were run in DOTUR (42) to generate diversity indexes. The default DOTUR settings were used with threshold values of 97% sequence identity for the definition of operational taxonomic units (OTUs). Library coverage was calculated with the nonparametric estimator C (15), as described by Kemp and Aller (23). The reciprocal of Simpson's index (1/D) was used as a measure of diversity to evaluate the level of dominance in a community (49). The number of OTUs shared between libraries was calculated by using SONS (43). UniFrac (17, 27) was applied to examine the similarities between clone libraries. A tree file generated by CLUSTAL W (45) and an environment file, which links a file to a library, were uploaded to UniFrac (27). Principal coordinates analysis (PCoA) was performed by using UniFrac with the abundance-weighted option (27).
Phylogenetic analysis.
The phylogenetic composition of the sequences in each library was evaluated by using the Classifier program of RDP-II release 10 (46), with confidence levels of 80%. BLASTN (2) was also used to classify the clones and to identify the closest relatives in the GenBank database. For the phylogenetic analysis, sequences were aligned by using the CLUSTALW program (45). The neighbor-joining method was used to build the trees (40). The PHYLIP format tree output was obtained by using the bootstrapping procedure (12); 1,000 bootstrap trials were used. The trees were constructed by using TreeView software (36).
Nucleotide sequence accession numbers.
Nucleotide sequences of 16S rRNA genes for the clone libraries have been deposited in the DDBJ database under the accession numbers shown in Table 1.
TABLE 1.
Accession numbers of sequences deposited in the DDBJ
| Genotype | N fertilization level |
|
|---|---|---|
| SN | HN | |
| Enrei | AB581722-AB581903 | AB582442-AB582565 |
| En 1282 | AB581904-AB582062 | AB582566-AB582728 |
| En 1314 | AB582063-AB582223 | AB582729-AB582888 |
| En 6500 | AB582224-AB582313 | AB582889-AB583026 |
| Sakukei 4 | AB582314-AB582441 | AB583027-AB583138 |
RESULTS
Soybean growth.
The soybean leaf-associated bacteria, including endophytes and epiphytes sampled at the beginning of the soybean reproductive stage (GE4 in Fig. 1), were subjected to clone library analyses. Measurement of growth indexes at the reproductive growth stage revealed no differences among soybean genotypes or nitrogen fertilization treatments at the time of sampling, although during the vegetative growth stage, slight variations were observed between soybean genotypes under HN fertilization (V13 to V15) and those under SN fertilization (V13 to V14) (see Fig. S1 in the supplemental material). Thus, at the time of sampling, the reproductive growth index was R5 (pod-maturing stage) (11) for all samples in both the SN and the HN fertilization fields, and the vegetative growth indexes ranged approximately from V13 to V14 for most samples. Hence, we concluded that the effects of the soybean growth stage on the community analysis would not be significant among the genotypes examined under SN or HN fertilization.
HN fertilization was performed to eliminate the effects of nitrogen deficiency in Nod− soybeans on the microbial community and to inhibit nodulation in the wild-type soybean (Nod+). HN fertilization increased the nitrate content in the soil (6 to 16 mmol/kg dry soil) during soybean cultivation (see Table S1 in the supplemental material). Plant dry weights and leaf nitrogen contents showed no significant differences between nodulated Enrei and the Nod− mutants under HN fertilization (see Fig. S2A and B and S3 in the supplemental material). Under HN fertilization, the number of nodules and the nodule dry weight on wild-type soybeans were markedly inhibited (see Fig. S2C and D in the supplemental material).
Statistics on clone libraries.
The statistics on the clone libraries are summarized in Table 2 . Although the number of sequences analyzed was as low as 90 for En 6500 under SN fertilization because of the high ratio of chimeric sequences, the library coverage among all libraries ranged from 91.4% to 98.1%. Under SN fertilization, Nod+ soybeans (Enrei) had the highest number of OTUs and greater diversity than the mutants in terms of the Shannon and Simpson indexes. No clear effect of the nodulation phenotype on the number of OTUs or on diversity index values was observed under HN fertilization.
TABLE 2.
Statistical analysis of 16S rRNA gene clone libraries derived from soybean leaves
| Parameter | Value for indicated library of specified nodulation phenotype at N fertilization level: |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| SN |
HN |
|||||||||
| Enrei (Nod+) | En 1282 (Nod−) | En 1314 (Nod−) | En 6500 (Nod++) | Sakukei 4 (Nod++) | Enrei (Nod+) | En 1282 (Nod−) | En 1314 (Nod−) | En 6500 (Nod++) | Sakukei 4 (Nod++) | |
| Statistics | ||||||||||
| No. of sequences | 182 | 159 | 161 | 90 | 128 | 124 | 163 | 160 | 138 | 112 |
| Library coverage (%)a | 94.5 | 95.6 | 97.5 | 92.2 | 91.4 | 94.4 | 96.9 | 98.1 | 92.0 | 95.5 |
| No. of OTUsb | 27 | 17 | 14 | 14 | 18 | 15 | 17 | 15 | 21 | 17 |
| Diversity indexes | ||||||||||
| Chao1 | 34.5 | 24 | 15.5 | 24.5 | 36.3 | 25.5 | 19.5 | 16 | 39.3 | 19.5 |
| ACE | 40.8 | 28.2 | 17.0 | 35.2 | 68.8 | 28.0 | 21.7 | 17.2 | 51.4 | 21.0 |
| Shannon index | 2.6 | 2.0 | 1.8 | 2.0 | 1.6 | 1.8 | 1.6 | 2.0 | 2.2 | 1.9 |
| Simpson index (1/D) | 8.7 | 5.1 | 4.3 | 6.5 | 2.7 | 3.9 | 2.5 | 4.3 | 5.9 | 3.7 |
Cx = 1 − (n/N), where nx is the number of singletons that are encountered only once in a library and N is the total number of clones.
OTUs were defined at 97% sequence identity.
Phylogenetic diversities of leaf-associated bacteria with different nodulation phenotypes under standard nitrogen fertilization conditions.
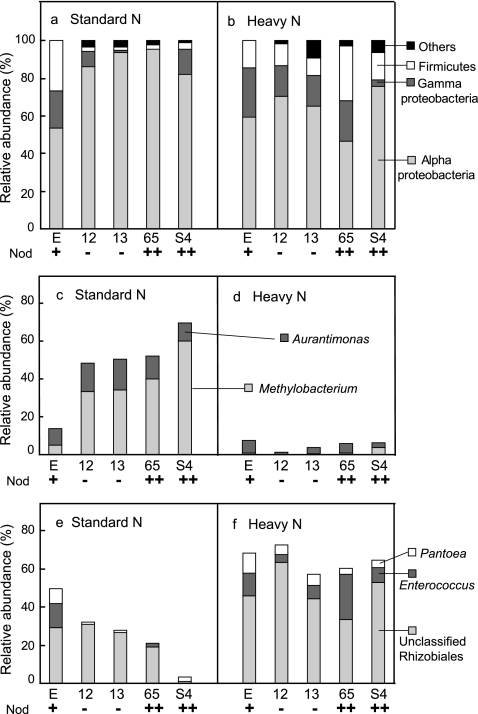
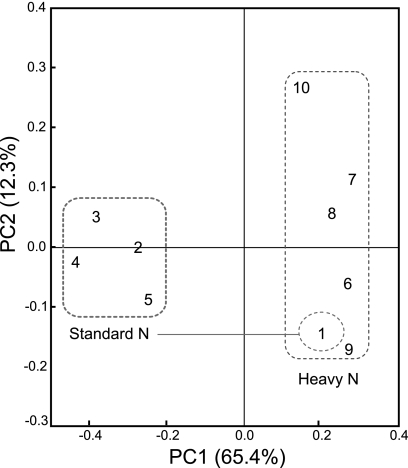
Assessment of the phylogenetic composition by using RDP Classifier revealed that the relative abundance of Alphaproteobacteria in Nod+ soybeans (53.8%) was noticeably lower than those in Nod− and Nod++ soybeans (82.0% to 95.6%) under SN fertilization, whereas those of the Gammaproteobacteria and Firmicutes showed the opposite patterns (19.8% and 26.4%, respectively, in Nod+, and 0% to 13.3% and 1.9% to 3.9%, respectively, in the mutant soybeans) (Fig. 2 a). Further analyses at lower taxonomic levels showed that a population shift of Methylobacterium sp. was mainly responsible for the low abundance of Alphaproteobacteria in Nod+ soybeans (4.9% for Methylobacterium sp.) compared with the mutants (Nod− and Nod++) (33.3% to 60.2%) (Fig. 2c). In contrast, two genera, Pantoea and Enterococcus, were mainly responsible for the greater abundance of Gammaproteobacteria and Firmicutes in Nod+ soybeans (7.7% and 13.2%, respectively) than in the mutants (Nod− and Nod++) (Fig. 2e). The results of PCoA clearly showed a tight cluster, including both Nod− and Nod++ soybeans under SN fertilization, whereas the Nod+ soybeans were distinctly separated from the mutants (Fig. 3).
FIG. 2.
Phylogenetic compositions of 16S rRNA gene clone libraries of soybean leaf-associated bacteria with different nodulation phenotypes under standard and heavy nitrogen fertilization conditions. (a and b) Phylogenetic compositions at the phylum level. (c and d) Relative abundances of Aurantimonas sp. and Methylobacterium sp. (e and f) Relative abundances of Pantoea sp., Enterococcus sp., and unclassified Rhizobiales bacteria. E, Enrei; 12, En 1282; 13, En 1314; 65, En 6500; S4, Sakukei 4. Nod, nodulation phenotype: wild type (+), nonnodulated (−) and hypernodulated (++).
FIG. 3.
Principal-coordinates analysis (PCoA) of 16S rRNA gene clone libraries of soybean leaf-associated bacteria with different nodulation genotypes. The plot was constructed by using UniFrac distances weighted by the relative abundances. Each number represents the phylogenetic composition of soybean leaf-associated bacteria with different soybean nodulation genotypes under standard and heavy nitrogen conditions. Numbers 1 to 5 and 6 to 10 were sampled under standard and heavy nitrogen conditions, respectively. Soybean genotypes: 1 and 6, wild type (Enrei); 2 and 7, nonnodulated (En 1282); 3 and 8, nonnodulated (En 1314); 4 and 9, hypernodulated (En 6500); 5 and 10, hypernodulated (Sakukei 4). Dashed line indicates bacterial communities with similar phylogenetic compositions (see the text). Notice the position of the wild-type Enrei under standard nitrogen conditions; it is located inside the cluster for all genotypes under heavy nitrogen conditions.
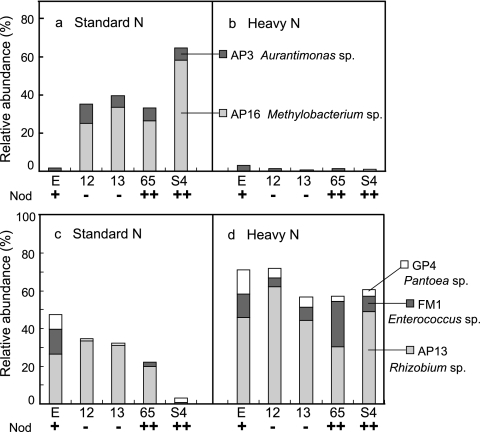
The cluster analysis identified two OTUs (AP3 and AP16) that had noticeably lower abundance in Nod+ soybeans than in both the Nod− and the Nod++ mutants under SN conditions (Fig. 4 a). The sequences of representative clones for these two OTUs showed high levels of identity to those of Aurantimonas ureilytica (98%) (DQ883810) and Methylobacterium extorquens (99%) (GU992354), respectively (Fig. 5). In contrast, the relative abundances of the two OTUs GP4 and FM1 were higher in Nod+ soybean (7.7% and 13.2%, respectively) than in the mutants (Fig. 4c). The representative sequences of these two OTUs showed high levels of identity to those of Pantoea ananatis (100%) (GQ383910) and Enterococcus mundtii (99%) (Y18340), respectively (Fig. 5).
FIG. 4.
Relative abundances of the operational taxonomic units (OTUs) that occurred in the 16S rRNA gene clone libraries of soybean leaf-associated bacteria and responded to nodulation phenotype and nitrogen fertilization conditions. The abundance of each OTU (defined by ≥97% identity) corresponds to the data in Fig. 5. E, Enrei; 12, En 1282; 13, En 1314; 65, En 6500; S4, Sakukei 4.
FIG. 5.
Phylogenetic distributions of OTUs that occurred in the 16S rRNA gene clone libraries of soybean leaf-associated bacteria and responded to nodulation phenotype and nitrogen fertilization conditions. The dendrogram indicates the phylogenetic relationships among the representative sequences of OTUs (defined by ≥97% identity). The table indicates the relative abundances of clones belonging to each OTU in each library and the results of a BLAST search using the representative sequences. E, Enrei; 12, En 1282; 13, En 1314; 65, En 6500; S4, Sakukei 4, Acc, accession. Gray shading indicates OTUs responsible for modulation and N fertilization (see text).
Comparisons of phylogenetic diversities of leaf-associated bacteria between standard and heavy nitrogen fertilization conditions.
Assessment of phylogenetic compositions using RDP Classifier showed that the effect of the nodulation phenotype on leaf-associated bacterial communities in soybeans was obscured under HN fertilization. However, the relative abundance of Alphaproteobacteria was dramatically lower in all mutants (Nod− and Nod++) (46.4% to 75.9%) under HN fertilization than under SN fertilization (82% to 95.6%) (Fig. 2b). Conversely, the proportions of Gammaproteobacteria and/or Firmicutes were higher in all mutants (Nod− and Nod++) under HN fertilization (3.6% to 25.8% for Gammaproteobacteria and 9.4% to 29% for Firmicutes) (Fig. 2b). Further analyses at lower taxonomic levels revealed that the relative abundances of two genera (Aurantimonas and Methylobacterium) in the Alphaproteobacteria were lower in all mutants (0% to 3.6%) under HN fertilization (Fig. 2d). In contrast, the abundances of Pantoea in the Gammaproteobacteria and Enterococcus in the Firmicutes were higher in all mutants (Nod− and Nod++) under HN fertilization (Fig. 2f). Unexpectedly, the analyses also revealed an increase in the abundance of the unclassified Rhizobiales bacterial population on all nodulation phenotypes under HN fertilization (range, 33.3% to 63.2%) compared with the SN fertilization (1.6% to 31.4%) (Fig. 2f). Surprisingly, the phylogenetic compositions of Nod+ soybeans under both SN and HN fertilization were highly similar to each other (Fig. 2 and 4), except for a slight increase in the unclassified Rhizobiales bacterial population under HN fertilization (Fig. 2e and f). The results of PCoA clearly showed that dramatic shifts in the community structures of leaf-associated bacteria in all mutants occurred in response to HN fertilization, as explained by the PC1 axis (65.4%) on the PCoA plot (Fig. 3). Interestingly, these results also revealed that the community structure of leaf-associated bacteria on Nod+ soybeans under SN conditions was very similar to those on all nodulation phenotypes under HN conditions.
The cluster analysis revealed that the same four OTUs (AP3, AP16, GP4, and FM1) that were responsive to the nodulation phenotype under SN fertilization were markedly responsive to the effects of HN fertilization in all mutants (Fig. 4 and 5). The relative abundances of two OTUs, AP3 and AP16, were dramatically lower in all mutants (Fig. 4b) under HN fertilization, whereas conversely, those of the OTUs GP4 and FM1 were higher (Fig. 4d and 5). Unexpectedly, the OTU AP13 was identified as responsible for the increase in the relative abundance of the unclassified Rhizobiales bacteria in all nodulation phenotypes under HN fertilization (Fig. 4d and 5). The representative sequence of OTU AP13 was classified as Rhizobium sp.; its closest relative among known species was Rhizobium oryzae (EU056822), with 98% identity.
DISCUSSION
Dramatic impacts of both the nodulation phenotype and the nitrogen fertilization level were observed on leaf-associated bacterial communities in soybeans (Fig. 3). Although a similar experimental design was employed in a previous study of stem-associated bacteria in soybeans (21), the results here provide more interesting insights into how underground events, such as nodulation and fertilization, affect the diversity of the microbial communities associated with aboveground tissues in plants. Interestingly, the populations of the two genera Methylobacterium and Aurantimonas on the leaves of all mutants responded more dramatically to the nodulation phenotype under SN fertilization and to HN fertilization than was the case in the stems (21) (see Fig. S4 in the supplemental material). Cluster analysis with a combined data set from leaves and stems revealed that the same two taxonomic groups (Methylobacterium sp. and Aurantimonas sp.) were responsive to the nodulation phenotype and nitrogen fertilization (OTUs MET1 for Methylobacterium sp. and AUR1 and AUR2 for Aurantimonas sp. in Fig. S5 in the supplemental material). These results clearly indicate that these two subpopulations of Methylobacterium sp. and Aurantimonas sp. were stable community members in the aboveground tissues (leaves and stems) of the mutant soybeans examined (see Fig. S4 in the supplemental material).
Surprisingly, the phylogenetic compositions of leaf-associated bacteria in Nod+ soybeans were virtually unaffected by the nitrogen fertilization level (Fig. 2 to 5), except for a slight increase in an unclassified Rhizobiales bacterial population (Fig. 2f and 4d). The results of PCoA further revealed that the diversities of leaf-associated bacteria in the wild-type Enrei under SN and HN fertilization were highly similar, in terms of not only simple phylogenetic composition, but also community structure, including both species abundance and phylogenetic diversity (Fig. 3). More importantly, the PCoA results in Fig. 3 imply that the leaves in the wild-type-nodulated soybean under SN fertilization were in a physiological state of nitrogen saturation, regardless of the exogenous nitrogen levels in soils. These results may suggest that tighter regulation of leaf-associated bacteria was present in the wild-type soybean than in the nodulation mutants examined and also indicate that soybean-associated bacterial communities were more regulated in leaves than in stems. These findings support the hypothesis that the systems that autoregulate nodulation are responsible for the shifts seen in the community of symbiotic bacteria here and in previous studies (21), because, under the field conditions examined, autoregulation was activated only in the wild-type Enrei. Furthermore, leaf tissue is currently believed to be the site of production of SDF, which suppresses rhizobial infection of roots (26, 34, 47). Therefore, the effects of SDF could be reflected more directly in the community structures of leaf-associated bacteria than in those of stem-associated bacteria. Although the exact mechanisms involved are still unknown, the previous and present studies clearly indicate that nodule regulation systems and nitrogen-sensing systems in plants affect the diversity of bacterial communities, not only in the roots (18), in which nodule formation actually occurs, but also systemically in the leaves and stems in a similar manner.
The analyses of phylogenetic composition and the results of PCoA also revealed that soybeans with two very different nodulation phenotypes, Nod− and Nod++, hosted microbial communities with highly similar structures in their leaves under SN fertilization (Fig. 2 and 3), as previously shown for stems (21). A possible explanation for this could be the absence of mycorrhization in Nod+ soybeans under the experimental conditions examined, presumably due to the rotation of paddy and upland fields. Therefore, the autoregulation systems were not activated in Nod− soybeans, and the autoregulation systems of Nod++ soybeans were genetically impaired. Thus, both Nod− and Nod++ soybeans could be considered equivalent hosts with no activated autoregulation.
Cluster analysis of 16S rRNA gene sequences identified, to the species level, OTUs responsible for the population shifts of leaf-associated bacteria (Fig. 4). The OTU AP3 had a high level of identity to A. ureilytica (98%); Aurantimonas sp. was identified as a population highly sensitive to the nodulation phenotype and nitrogen level (see Fig. S4A in the supplemental material), as has been observed in the stems of soybeans (21). The genus Aurantimonas was only recently established (8), and its biology in relation to plant symbiosis is largely unknown. However, Aurantimonas sp. has been isolated from the surface and inside of rice leaves (28), as well as from the stems of Nod++ soybeans (M. Anda, S. Ikeda, S. Eda, T. Okubo, S. Sato, S. Tabata, H. Mitsui, and K. Minamisawa, unpublished data). A detailed phylogenetic analysis has suggested that a phylogenetically related subpopulation of the genus Aurantimonas exists as plant-associated bacteria (Anda et al., unpublished data), and further study of these unknown plant-associated bacteria should be conducted.
The OTU AP16 had a high level of identity to M. extorquens (99%), which has also been identified as a bacterial group responding to the nodulation phenotype and nitrogen level in the stems of soybeans (21). However, in the leaves, Methylobacterium sp. in all mutants was more sensitive to the nodulation phenotype and nitrogen level than it was in the stems (21) (see Fig. S4B in the supplemental material). Members of the genus Methylobacterium are well known as plant-associated bacteria (7) and include both epiphytes and endophytes; scavenging of monocarbon wastes, such as methanol, in plant tissues is a putative role of this bacterial group (7). Therefore, population shifts of Methylobacterium sp. could indicate changes in plant physiological status. In fact, both nodulation and nitrate sensing in plants activate the autoregulation system, which causes a dramatic shift in their internal hormone balance (5, 34).
The OTUs GP4, FM1, and AP13 were the main bacterial groups in Nod+ soybeans under SN fertilization, and their abundances in Nod+ soybeans were relatively unchanged under HN fertilization compared to SN fertilization, except for a slight increase in OTU AP13. However, the relative abundances of these three OTUs in all mutants were dramatically higher under HN fertilization (Fig. 4c and d). The representative sequence of OTU GP4 was identical to that of P. ananatis, which is a plant-associated bacterium isolated from diverse plants, including soybeans (14). The representative sequence of OTU FM1 had a high level of identity to that of E. mundtii (99%), which has also been isolated from soybeans as a biopreserver producing bacteriocin (48), suggesting that the phytosphere could be a good source of beneficial bacteria for the fermentation industry. Unexpectedly, the detailed phylogenetic and cluster analyses identified OTU AP13 as an increased population of Alphaproteobacteria under HN fertilization in all nodulation phenotypes. The representative sequences of OTU AP13 had a high level of identity to those of R. oryzae (98%), which was recently isolated from the roots of a wild rice as an endophytic bacterium and which can form nodules on soybeans (37).
It is well known that exogenous nitrate suppresses infection with rhizobia and inhibits nodule formation (4, 6, 34). Recently, Okamoto et al. (34) suggested that a nitrate-induced CLE gene in Lotus japonica is involved in autoregulation systems through the HAR1 gene, which is a homolog of GmNARK in soybeans. These findings suggest that HN fertilization gives rise to nitrate-induced nodulation inhibition through the soybean autoregulation system in Nod− mutants. Thus, a GmNARK-mediated signaling pathway may regulate a subpopulation of Alphaproteobacteria, which may be closely related to rhizobia, beyond the regulation of nodule formation by Bradyrhizobium japonicum in soybeans. This may have resulted in the low ratio of Alphaproteobacteria in the wild-type soybeans under both SN and HN fertilizations and also in the Nod− soybeans under HN fertilization. In fact, the two Rhizobiales genera (Aurantimonas and Methylobacterium) found in the present study are closely related to Bradyrhizobium spp., endosymbionts of soybeans (16); these two genera were also responsible for the community shifts in stems with different nodulation phenotypes and under different soil nitrogen levels (see Table S1 in the supplemental material) (21). It is also interesting that, under HN fertilization, the community structures of Nod++ soybeans were highly similar to those of wild-type and Nod− soybeans (Fig. 3). As Nod++ soybeans do not harbor a functional GmNARK gene (31), the GmNARK-mediated nitrate sensing systems are not functional (34). Nevertheless, the effects of HN fertilization on Nod++ soybeans occurred systemically in the leaves, and the pattern of the community shifts in Nod++ soybeans was very similar to that in Nod− soybeans. Therefore, as has previously been shown with stem-associated bacteria (21), the GmNARK gene is not essential for the population shifts of leaf-associated bacteria responding to HN fertilization. These present and previous results strongly imply the presence of GmNARK-independent nitrogen-sensing systems that could regulate the structures of bacterial communities on the aboveground tissues of soybeans (20). In addition, surprisingly, the results of PCoA with combined data sets from both stem and leaf samples clearly indicated that the nitrogen fertilization level could be a dominant force in shaping the community structures of plant-associated bacteria for aboveground plant tissues, over and above the nodulation phenotype and the tissue specificity of bacterial colonization (see Fig. S6 in the supplemental material).
Besides the impacts of the nodulation phenotype and nitrogen level, the comparisons of stem- and leaf-associated bacterial communities revealed that the stems and leaves have distinct bacterial communities even at the phylum or class level (see Fig. S7 in the supplemental material). As expected from the harsher environmental conditions of leaves than of stems, the numbers of OTUs and the diversity indexes of leaf-associated bacteria (Table 2) were considerably lower than those of stem-associated bacteria (21). The bacterial community in stems consisted mainly of Proteobacteria (Alpha-, Beta-, and Gammaproteobacteria) (21). Meanwhile, the majority of the community in the leaves consisted of Alpha- and Gammaproteobacteria and Firmicutes, with almost no Betaproteobacteria. Furthermore, in the Alpha- and Gammaproteobacteria, the Bradyrhizobium, Devosia, Sphingomonadaceae, and Acinetobacter bacterial groups were exclusively found in the stems and not in the leaves (see Fig. S7 in the supplemental material). Interestingly, in the stems of wild-type soybeans under SN fertilization, the most dominant OTU was a group of Agrobacterium sp. (23% relative abundance) (21), whereas an unclassified Rhizobiales bacterial group represented by OTU AUR4 in Fig. S5 in the supplemental material, was highly abundant in the leaves (27%) compared to stems. Interestingly, the phylogenetic analysis of the representative sequences of OTUs revealed that this unclassified bacterial group is closely related to Aurantimonas sp. (Fig. 5; see Fig. S5 in the supplemental material). These results clearly indicate the presence of distinct tissue specificity—even between stems and leaves—for the colonization of symbiotic bacteria in the phytosphere at levels from phylum to genus. The data in the present study also highlighted the noticeable differences in results obtained from the culture-dependent and -independent analyses for the diversity of soybean leaf-associated bacteria. While only a trace amount of leaf bacterial populations was classified as Pseudomonas sp. (1.6%), May et al. (30) reported that approximately 20% of culturable epiphytic bacteria on healthy soybean leaves were Pseudomonas syringae pv. glycinea. These differences may reflect a bias in the culturability of leaf-associated bacterial communities (35, 39). Alternatively, the bacterial cell enrichment method employed in the present study may have a bias regarding the efficiencies of cell extraction from plant tissues for different taxonomic groups.
In conclusion, this study showed that the community structure of leaf-associated bacteria was affected by the nodulation phenotype and nitrogen fertilization level more dramatically than that of stem-associated bacteria. The nodulation phenotype and nitrogen level caused similar directional changes in the leaf-associated bacterial communities, with a shift of subpopulation in Alphaproteobacteria. The cluster analyses revealed that the relative abundances of Aurantimonas sp. and Methylobacterium sp. were especially sensitive to the nodulation phenotype and nitrogen level; they were lower in the wild-type nodulation phenotype and in all the nodulation mutants under HN fertilization than under SN fertilization. These results imply that there is an important link between the nitrogen signaling pathways and symbiotic regulation systems in plants for interaction with plant-symbiotic microbes other than rhizobia (20).
Aurantimonas and Methylobacterium sp. bacteria responding to the nodulation phenotype and soil N level have been isolated (Anda et al. unpublished data), and tissue localization and quantitative analyses of these bacteria in the phytosphere are ongoing. These efforts should facilitate our understanding of how plants regulate and shape the structures of microbial communities.
Supplementary Material
Acknowledgments
We thank M. Kokubun and T. Minami (Tohoku University) for providing the soybean seeds and for analysis of nodulation phenotypes, respectively.
This work was supported in part by Special Coordination Funds for Promoting Science and Technology; in part by PROBRAIN; in part by a grant from the Ministry of Agriculture, Forestry, and Fisheries of Japan (Genomics for Agricultural Innovation, PMI-0002); and in part by Grant-in-Aid for Scientific Research (C) 22580074.
Footnotes
Published ahead of print on 14 January 2011.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Akao, S., and H. Kouchi. 1992. A supernodulating mutant isolated from soybean cultivar Enrei. Soil Sci. Plant Nutr. 38:183-187. [Google Scholar]
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Ashelford, K. E., N. A. Chuzhanova, J. C. Fry, A. J. Jones, and A. J. Weightman. 2006. New screening software shows that most recent large 16S rRNA gene clone libraries contain chimeras. Appl. Environ. Microbiol. 72:5734-5741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barbulova, A., A. Rogato, E. D'Apuzzo, S. Omrane, and M. Chiurazzi. 2007. Differential effects of combined N sources on early steps of the Nod factor-dependent transduction pathway in Lotus japonicus. Mol. Plant Microbe Interact. 20:994-1003. [DOI] [PubMed] [Google Scholar]
- 5.Caba, J. M., M. L. Centeno, B. Fernandez, P. M. Gresshoff, and F. Ligero. 2000. Inoculation and nitrate alter phytohormone levels in soybean roots: differences between a supernodulating mutant and the wild type. Planta 211:98-104. [DOI] [PubMed] [Google Scholar]
- 6.Carroll, B. J., D. L. McNeil, and P. M. Gresshoff. 1985. Isolation and properties of soybean [Glycine max (L.) Merr.] mutants that nodulate in the presence of high nitrate concentrations. Proc. Natl. Acad. Sci. U. S. A. 82:4162-4166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Corpe, W. A. 1985. A method for detecting methylotrophic bacteria on solid surfaces. J. Microbiol. Methods 3:215-221. [Google Scholar]
- 8.Denner, E. B. M., et al. 2003. Aurantimonas coralicida gen. nov., sp. nov., the causative agent of white plague type II on Caribbean scleractinian corals. Int. J. Syst. Evol. Microbiol. 53:1115-1122. [DOI] [PubMed] [Google Scholar]
- 9.Ewing, B., and P. Green. 1998. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 8:186-194. [PubMed] [Google Scholar]
- 10.Ewing, B., L. Hillier, M. C. Wendl, and P. Green. 1998. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 8:175-185. [DOI] [PubMed] [Google Scholar]
- 11.Fehr, R. W., C. E. Caviness, D. T. Burmood, and J. S. Pennington. 1971. Stage of development descriptions for soybeans, Glycine Max (L.) Merrill. Crop Sci. 11:929-931. [Google Scholar]
- 12.Felsenstein, J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783-791. [DOI] [PubMed] [Google Scholar]
- 13.Francisco, P. B., Jr., and S. Akao. 1993. Autoregulation and nitrate inhibition of nodule formation in soybean cv. Enrei and its nodulation mutants. J. Exp. Bot. 44:547-553. [Google Scholar]
- 14.Gitaitis, R., et al. 2002. Recovery of Pantoea ananatis, causal agent of center rot of onion, from weeds and crops in Georgia, U. S. A. Crop Prot. 21:983-989. [Google Scholar]
- 15.Good, I. J. 1953. The population frequencies of species and the estimation of population parameters. Biometrika 40:237-264. [Google Scholar]
- 16.Gupta, R. S., and A. Mok. 2007. Phylogenomics and signature proteins for the alpha Proteobacteria and its main groups. BMC Microbiol. 7:106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hamady, M., C. Lozupone, and R. Knight. 2010. Fast UniFrac: facilitating high-throughput phylogenetic analyses of microbial communities including analysis of pyrosequencing and PhyloChip data. ISME J. 4:17-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hardoim, P. R., L. S. Van Overbeek, and J. D. Van Elsas. 2008. Properties of bacterial endophytes and their proposed role in plant growth. Trends Microbiol. 16:463-471. [DOI] [PubMed] [Google Scholar]
- 19.Ikeda, S., et al. 2009. Development of a bacterial cell enrichment method and its application to the community analysis in soybean stems. Microb. Ecol. 58:703-714. [DOI] [PubMed] [Google Scholar]
- 20.Ikeda, S., et al. 2010. Community- and genome-based views of plant-associated bacteria: plant-bacterial interactions in soybean and rice. Plant Cell Physiol. 51:1398-1410. [DOI] [PubMed] [Google Scholar]
- 21.Ikeda, S., et al. 2010. Community shifts of soybean stem-associated bacteria responding to different nodulation phenotypes and N levels. ISME J. 4:315-326. [DOI] [PubMed] [Google Scholar]
- 22.Ikeda, S., et al. 2008. Microbial community analysis of field-grown soybeans with different nodulation phenotypes. Appl. Environ. Microbiol. 74:5704-5709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kemp, P. F., and J. Y. Aller. 2004. Bacterial diversity in aquatic and other environments: what 16S rDNA libraries can tell us. FEMS Microbiol. Ecol. 47:161-177. [DOI] [PubMed] [Google Scholar]
- 24.Kouchi, H., et al. 2010. How many peas in a pod? Legume genes responsible for mutualistic symbioses underground. Plant Cell Physiol. 51:1381-1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-175. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley & Sons Ltd., Chichester, United Kingdom.
- 26.Lin, Y. H., B. J. Ferguson, A. Kereszt, and P. M. Gresshoff. 2010. Suppression of hypernodulation in soybean by a leaf-extracted, NARK- and Nod factor-dependent, low molecular mass fraction. New Phytol. 185:1074-1086. [DOI] [PubMed] [Google Scholar]
- 27.Lozupone, C., and R. Knight. 2005. UniFrac: a new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 71:8228-8235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mano, H., F. Tanaka, C. Nakamura, H. Kaga, and H. Morisaki. 2007. Culturable endophytic bacterial flora of the maturing leaves and roots of rice plants (Oryza sativa) cultivated in a paddy field. Microbes Environ. 22:175-185. [Google Scholar]
- 29.Matsunami, T., A. Kaihatsu, T. Maekawa, M. Takahashi, and M. Kokubun. 2004. Characterization of vegetative growth of a supernodulating soybean genotype. Sakukei 4. Plant Prod. Sci. 7:165-171. [Google Scholar]
- 30.May, R., B. Völksch, and G. Kampmann. 1997. Antagonistic activities of epiphytic bacteria from soybean leaves against Pseudomonas syringae pv. glycinea in vitro and in planta. Microb. Ecol. 34:118-124. [DOI] [PubMed] [Google Scholar]
- 31.Nishimura, R., et al. 2002. HAR1 mediates systemic regulation of symbiotic organ development. Nature 420:426-429. [DOI] [PubMed] [Google Scholar]
- 32.Offre, P., et al. 2007. Identification of bacterial groups preferentially associated with mycorrhizal roots of Medicago truncatula. Appl. Environ. Microbiol. 73:913-921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oka-Kira, E., and M. Kawaguchi. 2006. Long-distance signaling to control root nodule number. Curr. Opin. Plant Biol. 9:496-502. [DOI] [PubMed] [Google Scholar]
- 34.Okamoto, S., et al. 2009. Nod factor/nitrate-induced cle genes that drive HAR1-mediated systemic regulation of nodulation. Plant Cell Physiol. 50:67-77. [DOI] [PubMed] [Google Scholar]
- 35.Okubo, T., et al. 2009. Nodulation-dependent communities of culturable bacteria endophytes from stems of field-grown soybeans. Microbes Environ. 24:253-258. [DOI] [PubMed] [Google Scholar]
- 36.Page, R. D. M. 1996. TreeView: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12:357-358. [DOI] [PubMed] [Google Scholar]
- 37.Peng, G., Q. Yuan, H. Li, Z. Zhang, and Z. Tan. 2008. Rhizobium oryzae sp. nov., isolated from the wild rice Oryza alta. Int. J. Syst. Evol. Microbiol. 58:2158-2163. [DOI] [PubMed] [Google Scholar]
- 38.Radutoiu, S., et al. 2003. Plant recognition of symbiotic bacteria requires two LysM receptor-like kinases. Nature 425:585-592. [DOI] [PubMed] [Google Scholar]
- 39.Saito, A., S. Ikeda, H. Ezura, and K. Minamisawa. 2007. Microbial community analysis of the phytosphere using culture-independent methodologies. Microbes Environ. 22:93-105. [Google Scholar]
- 40.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 41.Scheublin, T. R., I. R. Sanders, K. Keel, and J. R. van der Meer. 2010. Characterization of microbial communities colonising the hyphal surfaces of arbuscular mycorrhizoal fungi. ISME J. 4:752-763. [DOI] [PubMed] [Google Scholar]
- 42.Schloss, P. D., and J. Handelsman. 2005. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl. Environ. Microbiol. 71:1501-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schloss, P. D., and J. Handelsman. 2006. Introducing SONS, a tool for operational taxonomic unit-based comparisons of microbial community memberships and structures. Appl. Environ. Microbiol. 72:6773-6779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Searle, I. R., et al. 2003. Long-distance signaling in nodulation directed by a CLAVATA1-like receptor kinase. Science 299:109-112. [DOI] [PubMed] [Google Scholar]
- 45.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang, Q., G. M. Garrity, J. M. Tiedje, and J. R. Cole. 2007. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73:5261-5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yamaya, H., and Y. Arima. 2010. Evidence that a shoot-derived substance is involved in regulation of the super-nodulation trait in soybean. Soil Sci. Plant Nutr. 56:115-122. [Google Scholar]
- 48.Zendo, T., et al. 2005. Identification and production of a bacteriocin from Enterococcus mundtii QU 2 isolated from soybean. J. Appl. Microbiol. 99:1181-1190. [DOI] [PubMed] [Google Scholar]
- 49.Zhou, J., et al. 2002. Spatial and resource factors influencing high microbial diversity in soil. Appl. Environ. Microbiol. 68:326-334. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.