Abstract
The potential of bacteriophage as an alternative biocontrol agent has recently been revisited due to the widespread occurrence of antibiotic-resistant bacteria. We isolated a virulent bacteriophage, SPC35, that can infect both Salmonella enterica serovar Typhimurium and Escherichia coli. Morphological analysis by transmission electron microscopy and analysis of its 118,351-bp genome revealed that SPC35 is a T5 group phage belonging to the family Siphoviridae. BtuB, the outer membrane protein for vitamin B12 uptake, was found to be a host receptor for SPC35. Interestingly, resistant mutants of both E. coli and S. Typhimurium developed faster than our expectation when the cultures were infected with SPC35. Investigation of the btuB gene revealed that it was disrupted by the IS2 insertion sequence element in most of the resistant E. coli isolates. In contrast, we could not detect any btuB gene mutations in the resistant S. Typhimurium isolates; these isolates easily regained sensitivity to SPC35 in its absence, suggesting phase-variable phage resistance/sensitivity. These results indicate that a cocktail of phages that target different receptors on the pathogen should be more effective for successful biocontrol.
Bacteriophages (phages) are viruses that specifically infect bacteria. They exist everywhere that bacteria thrive at an estimated ratio of at least 10 phage particles per bacterial cell (50). Phages are highly specific for a species; therefore, they are able to target a pathogen without disrupting the natural microflora (25, 39). As multidrug-resistant bacteria have become more common, phages have attracted attention as a potential alternative to antibiotics. Their advantages include their lower cost compared to that for the development of new antibiotics (39), ability to replicate in the presence of host bacteria, lack of side effects, and high host specificity. The clinical use of phages, known as phage therapy (39), has been reported for human and animal diseases caused by Escherichia coli (5, 13), Pseudomonas aeruginosa (3, 51), Salmonella spp. (8, 52), and Staphylococcus aureus (40).
Recently, the use of phages to prevent food-borne infectious diseases has also been proposed (21, 25, 27). The impact of food-borne illness on public health is considerable and has great economic significance. These diseases still occur, despite dramatic improvements in hygiene and sanitation in food processing. Phages have been used to control food-borne diseases caused by Salmonella spp. (20, 41), Listeria monocytogenes (12, 23), Campylobacter jejuni (20, 28, 53), and E. coli O157:H7 (44, 48). The status of generally recognized as safe (GRAS) was recently given to the L. monocytogenes-specific phage Listex P100, which was approved as a food preservative by the U.S. Food and Drug Administration (9). This has motivated many researchers to investigate the development of phages for biocontrol.
Several limitations must be overcome before phages can be successfully used as therapeutic and/or biocontrol agents. The first limitation is the narrow host range of phages. The high host specificity of most phages is a double-edged sword; high specificity is necessary for the specific targeting of unwanted bacteria, but the narrow host range renders phage biocontrol ineffective for non-host strains (22). The use of broad-host-range phages isolated by multiple-host enrichment methods (7, 11, 29) or a phage cocktail composed of multiple phage types (33, 44, 55) has shown promise in overcoming this problem. Another limitation is the rapid emergence of phage-resistant mutants (6, 11, 18, 32). A phage cocktail can control or delay the emergence of resistant mutants (27, 33); however, some of the underlying mechanisms for the appearance of phage-resistant mutants are not fully elucidated.
In this context, we isolated and characterized the novel lytic bacteriophage SPC35. This bacteriophage has a broad host range that includes the genus Salmonella and the genus Escherichia, which are important food-borne pathogens of humans and animals (10, 43). Bacterial challenge assays revealed distinctive patterns of growth inhibition and emergence of phage-resistant mutants in these two genera.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains used in this study are listed in Table 1. All Salmonella enterica serovar Typhimurium and E. coli mutants were derived from S. Typhimurium SL1344 and E. coli MG1655, respectively. E. coli isolates were kindly provided by Dong-Hyun Kang at Seoul National University, Seoul, South Korea. Bacteria were cultured at 37°C in Luria-Bertani (LB) broth and plates supplemented with 25 μg/ml of chloramphenicol where appropriate.
TABLE 1.
Bacterial strains and SPC35 host range
| Strain (genotype) | Plaque formationa | Source or referenceb |
|---|---|---|
| Salmonella Typhimurium | ||
| UK1 | CC | Laboratory collection |
| LT2 | CC | Laboratory collection |
| SL1344 | CC | Laboratory collection |
| ΔfhuA (SL1344 ΔfhuA) | CC | 25 |
| ΔfepA (SL1344 ΔfepA) | CC | 25 |
| ΔbtuB (SL1344 ΔbtuB) | − | 25 |
| ΔbtuB(pMS100) (ΔbtuB harboring pMS100) | CC | This study |
| 14028s | CC | Laboratory collection |
| ATCC 19586 | CC | ATCC |
| ATCC 43174 | CC | ATCC |
| DT 104 | CC | Laboratory collection |
| Salmonella Enteritidis ATCC 13078 | C | ATCC |
| Salmonella enterica isolates | ||
| 3068 | CC | Laboratory collection |
| 3605 | TT | Laboratory collection |
| 3792 | CC | Laboratory collection |
| 4509 | CC | Laboratory collection |
| Escherichia coli | ||
| ATCC 25922 | (+) | ATCC |
| B/1 | (+) | * |
| B/4 | − | * |
| BE4a | − | * |
| C600 | − | * |
| DH5α | CC | Laboratory collection |
| DH10B | CC | Laboratory collection |
| FS575 | T | * |
| GM1 | CC | * |
| GM119 | CC | * |
| HB101 | (+) | * |
| JM109 | CC | * |
| K-12 substrain MC4100 | CC | Laboratory collection |
| K-12 substrain MG1655 | CC | Laboratory collection |
| K-12 substrain W3110 | T | * |
| K-12 2B | − | * |
| K802 | (+) | * |
| LE392 | − | * |
| RP1 | C | * |
| SY327 | CC | * |
| TGI | CC | * |
| Escherichia coli isolates | ||
| C-660 | CC | * |
| E-2g | (+) | * |
| E-2j | − | * |
| E-7b | − | * |
| E-34 | − | * |
| WADDL 2701 | CC | * |
| WADDL 2735 | − | * |
| WADDL 2983 | − | * |
| WADDL 2902 | − | * |
| WADDL 3463 | C | * |
| WADDL 3476 | (+) | * |
| WADDL 3502 | − | * |
| WADDL 3811 | − | * |
| WADDL 3977 | − | * |
| WADDL 4036 | − | * |
| WADDL 4064 | (+) | * |
| WADDL 4083 | CC | * |
| WADDL 4190 | − | * |
| WADDL 4241 | − | * |
CC, a relatively clearer plaque; C, clear plaque; T, turbid plaque; TT, relatively more turbid plaque; (+), growth inhibition zone; −, no plaque.
ATCC, American Type Culture Collection; *, collection of Food Hygiene laboratory at Seoul National University, Seoul, South Korea.
Plasmid construction.
The plasmid used in the complementation assay was constructed by cloning the complete btuB gene and its promoter region from Salmonella SL1344 into the low-copy-number plasmid pACYC184. The btuB gene and its promoter region were amplified by PCR using primers btuB-CF-2 (5′-TTG TAG GGC ATG CTC AGT GGA TGT-3′) and btuB-CR-2 (5′-ATA CAA GCT TGG TGG GAC GTG GTT-3′). The PCR product was digested with restriction enzymes SphI and HindIII and then inserted into the corresponding restriction sites of plasmid pACYC184. The DNA insertion was verified by sequencing, and the resultant plasmid was named pMS100.
Isolation of bacteriophages.
Ten samples of chicken feces and two samples of chicken internal organs were obtained from the Mo-ran traditional market, Seoul, South Korea. The 25-g samples were homogenized in 225 ml sterile Butterfield's phosphate-buffered dilution water (0.25 M KH2PO4 adjusted to pH 7.2 with NaOH) for 90 s with a blender (BacMixer 400; Interscience Laboratory Inc., St. Nom, France). The 10-ml samples were then mixed with 50 ml LB broth and incubated at 37°C for 24 h with agitation (220 rpm). After centrifugation of the culture (9,000 × g, 10 min, 4°C) and filtration of the clear supernatant (0.22-μm-pore-size filter; Millipore, Ireland), the filtrate (10 ml) was mixed with 50 ml LB broth and 500 μl overnight culture of S. Typhimurium SL1344. This bacterium-phage mixture was incubated at 37°C for 8 h (at 220 rpm), followed by centrifugation and filtration as described above. Tenfold serial dilutions of these filtrates were used in the spotting assay with S. Typhimurium SL1344 or E. coli MG1655 to confirm the presence of bacteriophages. To isolate and purify the bacteriophages, the overlay assay was carried out as previously described (2). Each plaque showing a unique morphology was picked with a sterile tip, followed by elution with sodium chloride-magnesium sulfate (SM) buffer (50 mM Tris-HCl [pH 7.5], 100 mM NaCl, 10 mM MgSO4). This process was repeated at least three times.
Spotting assay and overlay assay.
The host bacteria were cultured overnight, and then 100 μl of them was inoculated into 5 ml of soft LB agar (0.4% agar), which had been heated to 42°C in a water bath. After gentle vortexing of this mixture, it was poured into prepared LB agar plates (1.5% agar with 25 μg/ml chloramphenicol, if necessary) and allowed to solidify at room temperature for 30 min to produce bacterial lawns. Then, 10 μl of phage stock dilutions (10-fold serial dilutions in SM buffer) was spotted onto the top agar layer, and the plates were dried at room temperature for 30 min. These plates were incubated overnight at 37°C and inspected on the next day for single plaques or bacterial growth inhibition zones.
The overlay assay was carried out with modifications. Briefly, phage stock dilutions (100 μl) were mixed with the same volume of overnight bacterial culture in 5 ml soft LB agar (0.4% agar), and the mixture was then poured into plates. After the medium was allowed to solidify for 30 min at room temperature, the plates were incubated at 37°C and plaques were examined on the next day.
Propagation of SPC35.
Bacteriophage propagation and concentration were carried out according to the methods of Sambrook and Russell (47), with some modifications. Briefly, the lysate of a single plaque of phage SPC35 was added to an E. coli MG1655 culture (optical density at 600 nm [OD600] = 0.5 to 0.6), which was then incubated at 37°C for 4 h. The cleared culture with cell debris (host bacteria lysate) was treated with chloroform (1% of final volume), incubated at 37°C for 5 min, and then centrifuged (15,000 × g, 10 min, 4°C). Bacteriophage particles in the filtered supernatant (0.22-μm-pore-size filter) were precipitated with 10% (wt/vol) polyethylene glycol (PEG) 6000 in 1 M NaCl at 4°C for 10 h. After centrifugation (10,000 × g, 15 min, 4°C), precipitated phages were resuspended in SM buffer and separated by CsCl density gradient ultracentrifugation (78,500 × g, 2 h, 4°C). The phage band fraction was separated by puncturing the centrifuge tube bottom with a heated needle and dialyzed against 1 liter of dialysis buffer (50 mM Tris-HCl [pH 8.0], 10 mM NaCl, 10 mM MgSO4) in a Spectra/Por dialysis membrane (molecular weight cutoff, 12,000 to 14,000; Spectrum Laboratories, Inc.). The dialysis buffer was changed after 1 h, and the phage fraction was dialyzed for an additional 1 h before it was transferred into a sterilized glass ampoule. The titers of these concentrated phage stocks were determined by the overlay assay, and the phage stocks were stored at 4°C until further use.
Morphological analysis by TEM.
Drops of SPC35 stock dilution (4 μl, approximately 1 × 1010 PFU/ml) were placed on carbon-coated copper grids; after 1 min, the excess phage suspension was removed with filter paper. Equal volumes of 2% aqueous uranyl acetate (pH 4.0) were added for 20 s to negatively stain the phage particles, and excess solution was removed as described above. Phages were examined by transmission electron microscopy (TEM; LEO 912AB transmission electron microscope; Carl Zeiss) at an 80-kV accelerating voltage, and images were scanned with a Proscan 1,024- by 1,024-pixel charge-coupled device camera at the National Academy of Agricultural Science (Suwon, South Korea). Phages were classified according to the International Committee on Taxonomy of Viruses (ICTV) classification (16).
Sequencing of SPC35 genomic DNA and genome analysis.
Extraction of phage DNA was carried out using proteinase K and SDS as previously described (47). Whole-genome sequencing of SPC35 was performed with a Genome Sequencer FLX titanium sequencer (Roche, Mannheim, Germany) and assembled with GS de novo assembler software (Roche) at Macrogen Inc., South Korea. Open reading frames (ORFs) of the SPC35 genome were determined using GeneMark.hmm software (37) and the BLAST algorithm (4). The tRNAscan-SE program (36) was used to search for tRNA-coding sequences.
Phage adsorption assay.
Exponentially growing cells in early log phase (1 ml, OD600 = 1.0 to 1.5) were harvested by centrifugation (13,000 × g, 1 min, 4°C), washed with phosphate-buffered saline (pH 7.2), and resuspended in 10 ml LB broth. This bacterial suspension was aliquoted into 10 microtubes (1 ml suspension per 1.75-ml tube) and preincubated at 37°C for 5 min. Then, SPC35 was added (multiplicity of infection [MOI] = 0.01) to each of the 10 tubes with 1-min intervals, and the tubes were incubated until 10 min incubation of the first tube. Cells were immediately removed by centrifugation (13,000 × g, 1 min, 4°C) and filtration (0.22-μm-pore-size filter), and the number of unbound free phage particles in each tube was determined by overlay assay. The baseline number was determined by adding the same concentration of SPC35 to fresh LB broth that was not inoculated with host bacteria and incubating the broth at 37°C for 10 min.
Bacterial challenge assay.
For the bacterial challenge assay, 50 ml fresh LB broth was inoculated with an overnight culture of S. Typhimurium SL1344 or E. coli MG1655 (1% inoculum), followed by incubation at 37°C at 220 rpm until the OD600 was 0.5. SPC35 stock dilutions (100 μl) were then added (MOI = 0.1, 1, or 10) to these cultures, which were incubated for another 24 h. Bacterial growth was monitored by measuring the OD600 at various time points. As a negative control, one bacterial culture was inoculated with 100 μl SM buffer instead of SPC35.
Isolation and subculturing of SPC35-resistant mutants.
To isolate SPC35-resistant mutants, the high-titer overlay assay was performed as previously described (14, 30). Briefly, overnight cultures of the host bacteria were serially diluted (10-fold), and 100 μl of each dilution was added to 5 ml soft LB agar (0.4% agar). Then, 100 μl of SPC35 suspension (about 2 × 1010 PFU/ml) was added (MOI ≥ 10), and these mixtures were poured onto LB agar plates and incubated for 24 h at 37°C for the standard overlay assay.
Five colonies from each resultant plate of S. Typhimurium SL1344 and E. coli MG1655 were picked with sterilized loops. Each colony was streaked onto a fresh LB agar plate and simultaneously inoculated into 3 ml LB broth, followed by overnight incubation at 37°C. The spotting assay was used to verify the phage resistance. These procedures were repeated at least three times to isolate a single colony and evaluate maintenance of phage resistance.
Amplification and DNA sequencing of btuB.
The btuB genes of E. coli MG1655 and S. Typhimurium were amplified by PCR using the following primers: for E. coli MG1655, primers MG btuB-CF (5′-GCA TGC TCA TCA GAT GTC CAG ATC T-3′) and MG btuB-CR (5′-AAG CTT ACC AGC ACG GTG GGA C-3′), and for S. Typhimurium, primers btuB-CF (5′-GCA TGC TCA GTG GAT GTC CAG C-3′) and btuB-CR (5′-AAG CTT ACC AGC ACG GTG GGA C-3′). PCR was carried out using a GeneAmp PCR system 9700 apparatus (Applied Biosystems, Foster City, CA) with the following steps: a predenaturation at 95°C for 7 min, followed by amplification for 30 cycles at 95°C for 45 s, 60°C for 45 s, and 72°C for 140 s and a final extension at 72°C for 7 min. The PCR products were analyzed by 1% agarose gel electrophoresis with 0.5× TAE buffer (20 mM Tris-acetate, 0.5 mM EDTA [pH 8.0]).
The btuB genes from SPC35-resistant E. coli mutants were sequenced by primer walking using the oligonucleotides SPC35-1F (5′-GGG TGA GTG GTT CTG CC-3′), SPC35-1R (5′-CCG TAG GAA GCA ATG AAG CG-3′), SPC35-2F (5′-CCC TGA ATC TCC AGA CAA CC-3′), and SPC35-2R (5′-GCT CTC TGC TAT TCC GTT ACTC-3′) and primers MG btuB-CF and MG btuB-CR, described above. DNA sequencing was performed with an ABI Prism 3730XL DNA analyzer (Applied Biosystems) at Macrogen Inc., South Korea.
Nucleotide sequence accession number.
The GenBank accession number of SPC35 is HQ406778.
RESULTS
Isolation of SPC35 and host range determination.
Nine bacteriophages that formed clear plaques on S. Typhimurium SL1344 were isolated. The ability of these phages to infect both S. Typhimurium SL1344 and E. coli MG1655 was confirmed by spotting phage lysate onto bacterial lawns. One phage, SPC35, formed clear plaques against both bacteria and was selected for further experiments. E. coli MG1655 was chosen to propagate SPC35 because plaques produced in this strain were larger and clearer than those produced in S. Typhimurium SL1344.
An SPC35 stock (about 2.8 × 1011 PFU/ml) was diluted 10-fold in SM buffer and analyzed by the spotting assay to determine the host range. All seven S. Typhimurium strains, one Salmonella Enteritidis isolate, and four Salmonella enterica isolates tested were sensitive to SPC35. In addition, SPC35 produced single plaques or bacterial growth inhibition zones with 23 of 40 E. coli strains (Table 1).
SPC35 morphology.
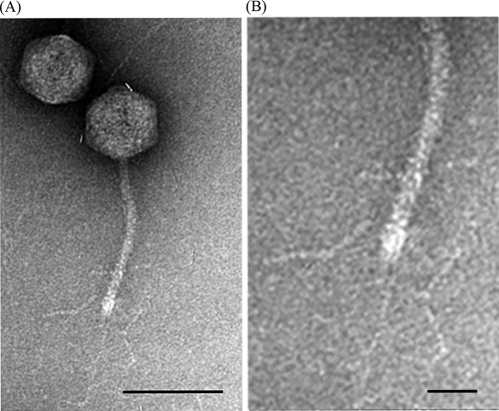
Morphological analysis by TEM revealed that phage SPC35 has a head with an icosahedral symmetry and a long tail without a contractile sheath, indicating that it belongs to the family Siphoviridae. The isometric head of SPC35 had a mean diameter of 70 nm, and the long noncontractile tail was 154 nm by 10 nm (Fig. 1), which closely resembled the morphology of bacteriophage T5 (head diameter, 60 nm; tail, 150 nm by 8 nm).
FIG. 1.
Transmission electron micrographs of phage SPC35. Bars, 100 nm (A) and 25 nm (B).
Sequencing and organization of the SPC35 genome.
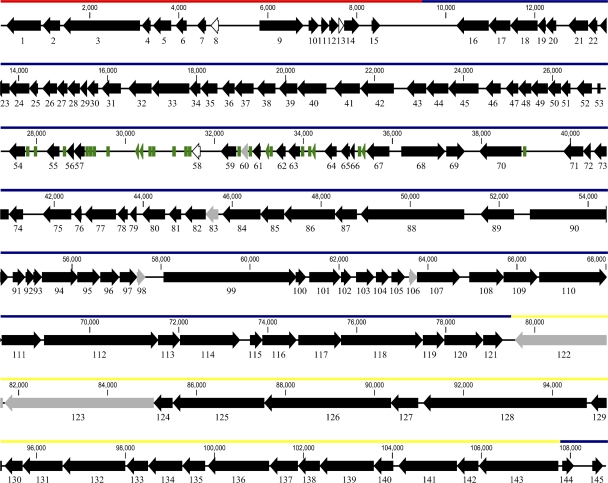
Only one contig of 108,778 bp (average G+C content, 39.24%) was constructed with 99.97% of bases with a quality score of ≥40. However, the 4,548 nucleotides at the left end of the contig and the 5,025 nucleotides at the right end of the contig were covered by an average read depth of 125.05, while the nucleotides in the other region were covered by an average read depth of 69.18, suggesting that SPC35 contains terminally redundant DNA at both ends of its genome. Accordingly, the SPC35 genome was composed of 118,351 bp with a terminally repeated 9,573 bp and exhibited an average G+C content of 39.39%. One hundred thirty-seven of the 145 predicted putative ORFs were closely related to those of T5 or T5-like phages; thus, phage SPC35 was classified to be a T5 group phage (Fig. 2; see Fig. S1 and Table S1 in the supplemental material). Similar to the genomes of these phages, the SPC35 genome was divided into pre-early, early, and late regions by gene function (Fig. 2).
FIG. 2.
SPC35 genome map excluding the terminally redundant DNA sequence at the right end. The predicted ORFs with their directions of transcription and tRNA-coding sequences are indicated by colored arrows: black, homologous to T5 group phages (T5, BF23, H8, and EPS7); gray, homologous to other microorganisms; white, no homology; green, tRNA-coding sequences. The red, blue, and yellow lines above the arrows indicate the pre-early, early, and late regions, respectively.
The pre-early region of the SPC35 genome, which appears to be transferred into the host bacteria in the first-step transfer, contains 15 ORFs that encode various host enzyme inhibitors, including inhibitors for host DNA, RNA, and protein synthesis and gene products that degrade host DNA. ORF10, ORF11, ORF12, ORF14, and ORF15 showed high similarities to ORFs in the pre-early region of T5 and EPS7, which encode these inhibitory proteins (26, 54). In addition, the gene products of ORF1, ORF3, ORF4, and ORF5 were highly homologous to deoxynucleoside 5′-monophosphatase (DMP; 97%), A1 protein (98%), probable A1 protein precursor (98%), and A2 protein (97%) of T5, respectively. These proteins are required for the second-step transfer of the T5 phage genome (54).
The two-step genome transfer mechanism of T5 is facilitated by terminally redundant DNA sequences spanning the pre-early region, a common feature of T5-like phages (26). Similar to T5 or T5-like phages, the terminally repeated 9,573 nucleotides of the SPC35 genome spanned the pre-early region (Fig. 2; see Fig. S1 in the supplemental material) and appeared to facilitate the two-step transfer of the SPC35 genome.
The early region of the SPC35 genome encodes a variety of proteins thought to participate in DNA replication, transcription, recombination, metabolism, signal transduction, and cell lysis; most of these genes are highly homologous to T5 genes. For instance, the putative gene products of ORF110, ORF111, and ORF112 are replicative DNA helicase, DNA replication primase, and DNA polymerase, respectively, with 99% identity to the corresponding T5 genes. ORF39, ORF40, and ORF73 encode phage lysis-related proteins, including lysozyme (endolysin, 80% similarity to the T5 gene), putative holin (97% similarity to the T5 gene), and spore-cortex-lytic enzyme precursor (97% similarity to the T5 gene), respectively. In addition, the repertory of 24 identified tRNA-coding sequences in the early region of the SPC35 genome was identical to that of the T5 genome, except that the tRNA-coding sequence for Val was absent and an additional tRNA-coding sequence for Lys was present in the SPC35 genome. We could not find any genes involved in lysogen formation.
The late region of the SPC35 genome was composed of 24 ORFs. The majority of these ORFs encode structural and morphogenesis proteins, such as head and tail proteins, a portal protein, and a receptor-binding protein. The putative prohead protease, portal protein, terminase (large subunit), and putative SciB protein, encoded by ORF137, ORF139, ORF141, and ORF142, respectively, were nearly identical to the corresponding T5 proteins, whereas tail proteins Pb3 and Pb4, major tail protein, and major head protein precursor, encoded by ORF126, ORF125, ORF132, and ORF136, respectively, showed greater homology to EPS7 proteins. In addition, ORF143, encoding a receptor-binding protein, and ORF144, encoding a receptor-blocking protein, were highly homologous to those of BF23 (84% and 99% identities, respectively), suggesting a common host receptor for BF23 and SPC35.
BtuB acts as a receptor for SPC35.
Because the morphological characteristics and genome of SPC35 were similar to those of T5 group phages, we investigated several known receptors of T5-like phages for SPC35 infection. S. Typhimurium SL1344 deletion mutants, lacking fhuA, btuB, or fepA (26), were tested for susceptibility to SPC35 by the spotting assay. The only mutant that SPC35 could not infect was the btuB deletion mutant, indicating a crucial role for BtuB in the infection process of SPC35. Complementation of the btuB mutation by pMS100 harboring the wild-type (WT) btuB gene restored the susceptibility of SL1344 to SPC35.
The requirement of BtuB in the initial step of SPC35 infection was further assessed by the phage adsorption assay of WT, mutant, and complemented strains. As shown in Fig. 3, phage adsorption by S. Typhimurium SL1344 was completely abolished by the btuB mutation. Complementation of the btuB mutation with pMS100 restored infection to the levels seen with WT btuB, resulting in the gradual reduction of free phage particles. These results strongly suggest that BtuB is the host receptor for phage SPC35.
FIG. 3.
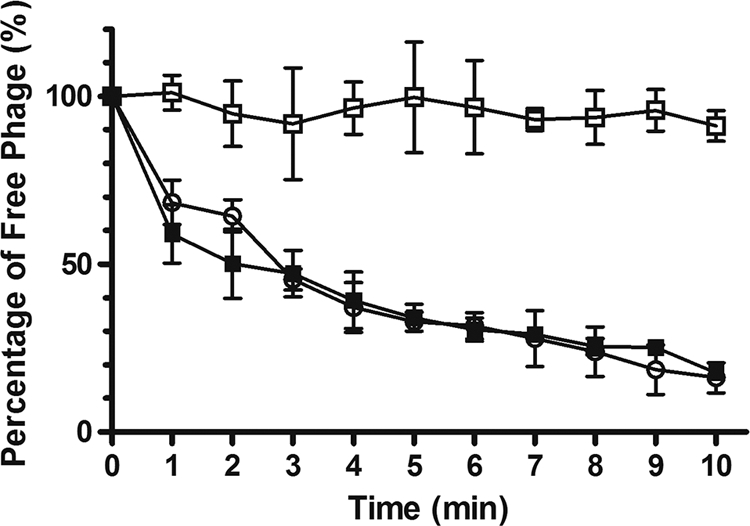
SPC35 adsorption of S. Typhimurium SL1344 (open circles), ΔbtuB (open squares), and ΔbtuB(pMS100) (closed squares). Exponentially growing cells were infected with SPC35 (MOI, 0.01) and incubated at 37°C. After centrifugation and filtration, the phage titer in the filtrate was determined by standard overlay assay. The percentage of free phage particles was calculated as follows: (phage titer in the supernatant of SPC35-infected bacterial suspension/phage titer in the supernatant of SPC35-inoculated LB broth) × 100. The results are expressed as means and standard deviations of triplicate assays.
Differential responses of S. Typhimurium and E. coli to SPC35 infection.
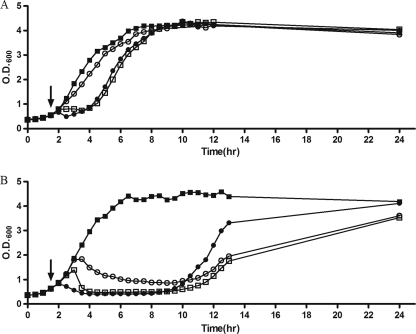
To assess the ability of SPC35 to lyse host bacteria in broth, we evaluated the growth of host bacteria in the presence of SPC35. The effect of SPC35 infection on the growth of S. Typhimurium SL1344 differed markedly from that on the growth of E. coli MG1655 (Fig. 4). SPC35 caused growth retardation in S. Typhimurium SL1344 rather than growth inhibition. Infection of SL1344 at an MOI of 0.1 produced only slight growth retardation, and an MOI of 1 or 10 produced growth retardation of about 2 h (Fig. 4A). However, in E. coli MG1655, infection with SPC35 at an MOI of 0.1 began to suppress host growth significantly about 2 h after infection. Growth inhibition was maintained until approximately 10 h after phage infection, after which bacterial growth eventually resumed (Fig. 4B). Several isolates from this culture obtained after sequential streaking on fresh LB agar plates showed an SPC35 insensitivity in the standard spotting assay (data not shown), indicating the generation of phage-resistant mutants. Addition of SPC35 at an MOI of 1 or 10 also suppressed E. coli MG1655 growth, but phage-resistant mutants appeared more rapidly at an MOI of 10.
FIG. 4.
Bacterial challenge assay with SPC35 to S. Typhimurium SL1344 (A) and E. coli MG1655 (B). SPC35 (100 μl) was added at an MOI of 0.1 (open circles), an MOI of 1 (open squares), or an MOI of 10 (closed circles) to the bacterial culture after 1.5 h incubation (vertical arrows, OD600 = 0.5 to 0.6). A bacterial culture inoculated with SM buffer instead of SPC35 was used as the negative control (closed squares).
The differential responses to SPC35 infection by S. Typhimurium and E. coli were also observed on solid media. In the high-titer (ca. 2 × 1010-PFU/ml) overlay assay, colonies of various sizes within the overall confluent lysis zone were observed on the E. coli MG1655 plate. In contrast, the S. Typhimurium SL1344 plate produced a bacterial lawn without any visible lysis zones or plaques (data not shown). However, when 10-fold dilutions of S. Typhimurium were incubated with the same concentration of SPC35, several putative phage-resistant colonies were formed at an MOI of 106.
SPC35 resistance is heritable in E. coli mutants but not in S. Typhimurium mutants.
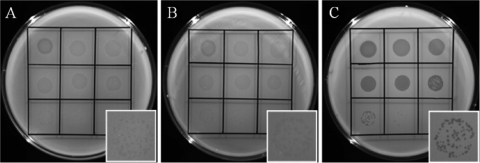
Five of the SPC35-resistant S. Typhimurium colonies and five of the SPC35-resistant E. coli colonies were picked from each of the high-titer overlay plates; these colonies were streaked three times onto fresh LB agar plates in the absence of SPC35 to isolate a single colony. At each streaking step, the newly formed colonies were tested by spotting assay to verify phage resistance. Interestingly, persistence of phage resistance differed between the putative E. coli and S. Typhimurium mutants. All five putative E. coli mutants maintained their resistance throughout the sequential streaking, resulting in no plaques on the spotted lawns. In contrast, all five putative S. Typhimurium mutants showed SPC35 sensitivity after the first streaking. However, SPC35 formed relatively turbid plaques in the spotting assay performed with colonies obtained after the first or second streaking but formed clear plaques when the assay was performed with colonies obtained after the third streaking (Fig. 5).
FIG. 5.
SPC35 was spotted onto plates of SPC35-resistant S. Typhimurium isolates obtained after a single streaking (A), the second streaking (B), and the third streaking (C). Insets at the bottom right of each panel show individual plaques.
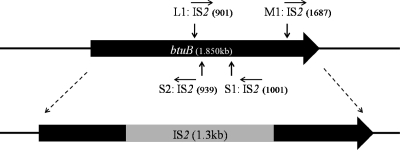
Disruption of the btuB gene by IS2 results in heritable resistance to SPC35 in E. coli.
To determine whether the SPC35-resistant E. coli isolates had lost their ability to synthesize functional BtuB protein because of mutation, we analyzed the btuB genes of these isolates by PCR amplification. Analysis of the PCR products by agarose gel electrophoresis showed fragments of about 3,500 bp from all mutants except SPC35RMG-M2 (Fig. 6). The size of the PCR product in WT E. coli MG1655 was about 2,200 bp; therefore, these results indicated that the btuB gene was mutated by insertion of a 1,300-bp DNA fragment in isolates SPC35RMG-L1, SPC35RMG-M1, SPC35RMG-S1, and SPC35RMG-S2. DNA sequencing of these amplified fragments revealed insertion sequence (IS) IS2 in the btuB genes of all four mutants; however, its orientation and insertion site differed (Table 2 and Fig. 7). IS2 was oriented in the same direction as btuB transcription in SPC35RMG-L1 and SPC35RMG-M1, but the reverse orientation was observed in SPC35RMG-S1 and SPC35RMG-S2. The IS2 insertion in SPC35RMG-L1 was 901 nucleotides from the btuB start codon but 1,687, 1,001, and 939 nucleotides from the start codon in SPC35RMG-M1, SPC35RMG-S1, and SPC35RMG-S2, respectively. In these four mutants, the flanking sequence of IS2 insertion sites had a duplicated sequence of 5 bp, but there is no common noticeable percent G+C content difference in these flanking sequences (Table 2). Complementation of the btuB::IS2 mutant clone, SPC35RMG-L1, with a plasmid containing the intact btuB gene resulted in the formation of clear plaques of SPC35 in the spotting assay (data not shown). This result strongly suggested that the IS2 insertion in the btuB gene provided the SPC35 resistance in E. coli. The amplified btuB fragment of SPC35RMG-M2 was the same size as that of WT E. coli MG1655; however, one nucleotide (G) was deleted among the five G nucleotides from nucleotides 325 to 329 from the start codon.
FIG. 6.
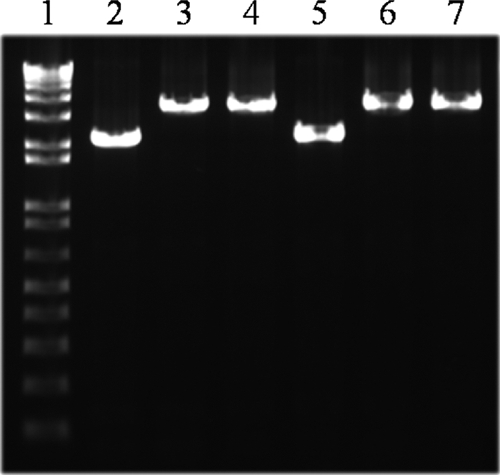
PCR analysis of the btuB gene in SPC35-resistant E. coli mutants. Lane 1, DNA ladder; lane 2, E. coli MG1655; lane 3, SPC35RMG-L1; lane 4, SPC35RMG-M1; lane 5, SPC35RMG-M2; lane 6, SPC35RMG-S1; lane 7, SPC35RMG-S2.
TABLE 2.
DNA sequences at insertion site of IS2
| Strain or IS2 | Flanking sequencea (% G+C content) | Left end-IS2-right end | Flanking sequence (% G+C content) | IS2 orientationb |
|---|---|---|---|---|
| E. coli MG1655 | ATTACAACTACGATCCCCAT | TATGGTCGTTATGAT | ||
| SPC35RMG-L1 | ATTACAACTACGATCCCCAT (40) | TAGACTGGCCCCCTG . . . ATAGGGGCAAATCCA | CCCATTATGGTCGTTATGAT (40) | + |
| SPC35RMG-M1 | ATCAAACCGTTAAAATGGGC (40) | TAGACTGGCCCCCTG . . . ATAGGGGCAAATCCA | TGGGCGGTGTGAGCTTGTGG (65) | + |
| SPC35RMG-S1 | GTTATGTTGAGGATGGATAT (35) | TGGATTTGCCCCTAT . . . CAGGGGGCCAGTCTA | GATATGATCAACGTAATACC (35) | − |
| SPC35RMG-S2 | TCGGCGACGCTCGATGAGAT (60) | TGGATTTGCCCCTAT . . . CAGGGGGCCAGTCTA | GAGATGAAGCAATACACCGT (45) | − |
| IS2Ac | ACGAATAAACGCATAATTAC (30) | TAGACTGGCCCCCTG . . . ATAGGGGCAAATCCA | ATTACCTATCAGGCAGTTTG (40) | + |
| IS2D | ACATCATGGGACAAGAACCC (50) | TGGATTTGCCCCTAT . . . CAGGGGGCCAGTCTA | AACCCCAGAACTTACTTATG (40) | − |
| IS2Ed | CACTGGATGACGAACTGGCC (60) | Truncated IS2 . . . ATAGGGGCAAATCCA | CAGGTGAACTACGCTCCTCT (55) | + |
| IS2F | AACAAATCCGCATTCGTGGC (50) | TGGATTTGCCCCTAT . . . CAGGGGGCCAGTCTA | GTGGCGAAGCATCCTCCCGT (65) | − |
| IS2H | AGTAAATTCCAATTGTTTAT (20) | TGGATTTGCCCCTAT . . . CAGGGGGCCAGTCTA | AACTTGCTCTTTTCTTCTGG (40) | − |
| IS2I | TATTGTTCATTATCCGTTAT (25) | TAGACTGGCCCCCTG . . . ATAGGGGCAAATCCA | GTTATCATCATTGGTTGTGC (40) | + |
| IS2K | AGGGATTTGATGGTCCCTTG (50) | TAGACTGGCCCCCTG . . . ATAGGGGCAAATCCA | CCTTGTGCTGATATGAATAC (40) | + |
The 5-bp flanking sequence duplications are underlined. Note that IS2E and IS2H do not have these duplication sequences.
+, IS2 inserted in the same orientation as the direction of btuB transcription; −, IS2 inserted in the opposite orientation as the direction of btuB transcription.
Sequences for IS2 elements have been retrieved from GenBank accession no. U00096.
IS2E is a truncated element that does not contain an insC (orfA) gene.
FIG. 7.
Schematic representation of IS2 insertion sites in the btuB gene of SPC35-resistant E. coli mutants. Vertical arrows indicate insertion of IS2, and the corresponding insertion sites are shown in parentheses. Horizontal arrows indicate the orientation of IS2.
In contrast, PCR analysis and sequencing of the btuB gene of SPC35-resistant S. Typhimurium isolates using primers btuB-CF and btuB-CR revealed no mutation in the btuB gene.
DISCUSSION
Using S. Typhimurium SL1344 as the host bacterium, we isolated a novel bacteriophage, SPC35, which can infect Salmonella species and E. coli. Morphological analysis by TEM revealed that SPC35 has an icosahedral head and noncontractile tail, indicating that it belongs to the family Siphoviridae (Fig. 1). Sequencing and genome analysis revealed that the SPC35 genome was highly homologous to the genomes of T5 group phages T5, BF23, H8, and EPS7 (26, 45).
Attachment to the host cell is the initial step of phage infection. Various components of the host bacteria, such as flagella, fimbriae, lipopolysaccharide (LPS), and outer membrane proteins, are used as bacteriophage receptors (50). Phage T5 uses the ferrichrome outer membrane transporter FhuA as a receptor (45). Phages BF23 and EPS7 utilize BtuB, the outer membrane protein involved in vitamin B12 uptake (26), and H8, which binds to the host ferric enterobactin receptor FepA (45). In the present study, mutant and genome analysis revealed that BtuB is the host receptor for SPC35. Although the SPC35 genome was highly homologous to the T5 genome (see Fig. S1 in the supplemental material), the putative receptor-binding protein of SPC35, which interacts with its corresponding host receptor, was more similar to that of BF23 (amino acid homology for BF23, 84% identity and 91% positive; amino acid homology for T5, 29% identity and 43% positive).
Several strains of bacteria, including S. enterica isolate 3605, E. coli FS575, and E. coli W3110, as well as colonies obtained after streaking of the SPC35-resistant S. Typhimurium strain once or twice in the absence of SPC35, produced turbid plaques, even though the SPC35 genome does not appear to contain genes involved in lysogen formation (see Table S1 in the supplemental material). These results may be the result of inefficient lysis of the host bacteria or some unknown phage resistance-related factor(s).
The bacterial challenge assay with S. Typhimurium and E. coli showed that host bacteria could not be completely eliminated at any MOI tested (Fig. 4). This result can be explained by the possibility that a proportion of the initial bacterial population was resistant to the phage or by the rapid appearance of resistant mutants. Carey-Smith et al. (11) and Fischer et al. (17) proposed that the growth retardation in the presence of bacteriophage occurs when only a subpopulation of the host bacteria is susceptible to phage infection, such as the 15% of E. coli O157:H7 MuL isolates that were resistant to phage PP01 (17). In contrast, Kocharunchitt et al. (32) suggested that incomplete lysis of phage-treated bacteria was due to physiologic and/or genetic modification of host cells during phage infection. In the present study, we found that the physiologic and/or genetic modification of host cells during phage infection differed between genera of host bacteria. In most of the SPC35-resistant E. coli isolates, an IS2 insertion disrupted the btuB gene, whereas the SPC35-resistant S. Typhimurium isolates had intact btuB genes and only transient phage resistance.
Insertion sequences are transposable elements (typically 700 to 2,000 bp) present in many bacterial chromosomes and plasmids (15). IS2, which naturally occurs in E. coli K-12, is a 1,327-bp insertion sequence (19, 46) of the IS3 family, which consists of IS3, IS150, IS911, and other elements (38). IS3 family members contain only two open reading frames, orfA and orfB, from which the fusion protein transposase, OrfAB, is generated by a −1 translational frameshift; its transposition is mediated by this transposase via circular and linear intermediates (24, 34). Mutations by IS2 insertion have been reported in both regulatory regions and coding regions of target genes; for example, the promoter regions of the galOPETK and lac operons and the coding sequences of galK, relA, and hemB have been mutated by IS2 (35). In the present study, all of the IS2 elements in SPC35-reistant E. coli mutants were inserted in the btuB-coding region (Fig. 7). In these mutants, IS2 was flanked by 5-bp sequence duplications, which normally form during IS2 integration into its target gene (19); IS2 showed no apparent preference for percent G+C content in target sequences (Table 2).
Only a few studies have reported phage-resistant mutants with IS insertions in their receptor genes. The tonB gene of E. coli, which encodes a protein that assists in phage T1 infection as well as the colicin B reaction, was spontaneously inactivated by IS1, IS2, and IS10 in the presence of colicin B (31). Vibrio cholerae phage K139 uses the O1 antigen of LPS as the host receptor for infection. K139-resistant V. cholerae mutants were reported to have IS1004 inserted into the O-antigen biosynthesis gene cluster (manB, involved in the biosynthesis of perosamine, and wbeW, which encodes a glycosyltransferase) (42).
Salmonella Typhimurium LT2 contains only one type of IS element, IS200, whereas E. coli K-12 contains 8 to 12 different IS elements, including IS2. Moreover, IS200 transposes less frequently than E. coli K-12 IS elements (15). Thus, nonheritable phase-variable physiologic changes stimulated by phage infection may be a more common source of resistance than IS200 insertion in S. Typhimurium.
IS elements are widespread; therefore, mutants spontaneously resistant to phage due to IS element insertion into genes required for phage infection are a problem. However, the low transposition frequency of IS elements suggests that this problem could be overcome with phage cocktails consisting of various virulent phages that use different host receptors. Simultaneous insertion of IS elements into every gene encoding a host receptor targeted by the phage cocktail is unlikely. Thus, identification of the phage receptor is useful for the production of better phage cocktails.
The transience of SPC35 resistance in S. Typhimurium strains was not due to pseudolysogeny, as described by Ackermann and DuBow (1), because we did not observe spontaneous production of phages from the SPC35-resistant S. Typhimurium isolates. PCR analysis also revealed that these isolates did not contain the SPC35 genome (data not shown). Differential resistance is not specific to SPC35; E. coli and Salmonella also showed differential resistance to several other phages isolated in our lab (data not shown). Transient phage resistance similar to that observed in S. Typhimurium SL1344 was also reported in E. coli O157:H7 and other Salmonella spp. (32, 49), but the underlying mechanisms for this phenomenon are not known. The mechanism is under investigation in our laboratory.
Supplementary Material
Acknowledgments
This work was supported by a National Research Foundation of Korea (NRF) grant funded by MEST (no. 20090078983), and M. Kim was supported by a grant (no. S2-2009-000-00126-2) from the Korea Student Aid Foundation (KOSAF), Republic of Korea.
Footnotes
Published ahead of print on 21 January 2011.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Ackermann, H. W., and M. S. DuBow. 1987. Viruses of prokaryotes, vol. 1. General properties of bacteriophages. CRC Press, Inc., Boca Raton, FL.
- 2.Adams, M. H. 1959. Methods of study of bacterial viruses, p. 443-457. In A. D. Hershey (ed.), Bacteriophages. Interscience Publishers Inc., New York, NY.
- 3.Ahmad, S. I. 2002. Treatment of post-burns bacterial infections by bacteriophages, specifically ubiquitous Pseudomonas spp. notoriously resistant to antibiotics. Med. Hypotheses 58:327-331. [DOI] [PubMed] [Google Scholar]
- 4.Altschul, S. F., et al. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barrow, P., M. Lovell, and A. Berchieri, Jr. 1998. Use of lytic bacteriophage for control of experimental Escherichia coli septicemia and meningitis in chickens and calves. Clin. Diagn. Lab. Immunol. 5:294-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bastias, R., G. Higuera, W. Sierralta, and R. T. Espejo. 2010. A new group of cosmopolitan bacteriophages induce a carrier state in the pandemic strain of Vibrio parahaemolyticus. Environ. Microbiol. 12:990-1000. [DOI] [PubMed] [Google Scholar]
- 7.Bielke, L., S. Higgins, A. Donoghue, D. Donoghue, and B. M. Hargis. 2007. Salmonella host range of bacteriophages that infect multiple genera. Poult. Sci. 86:2536-2540. [DOI] [PubMed] [Google Scholar]
- 8.Borie, C., et al. 2009. Aerosol spray treatment with bacteriophages and competitive exclusion reduces Salmonella Enteritidis infection in chickens. Avian Dis. 53:250-254. [DOI] [PubMed] [Google Scholar]
- 9.Bren, L. 2007. Bacteria-eating virus approved as food additive. FDA Consum. 41:20-22. [PubMed] [Google Scholar]
- 10.Buzby, J. C., and T. Roberts. 2009. The economics of enteric infections: human foodborne disease costs. Gastroenterology 136:1851-1862. [DOI] [PubMed] [Google Scholar]
- 11.Carey-Smith, G. V., C. Billington, A. J. Cornelius, J. A. Hudson, and J. A. Heinemann. 2006. Isolation and characterization of bacteriophages infecting Salmonella spp. FEMS Microbiol. Lett. 258:182-186. [DOI] [PubMed] [Google Scholar]
- 12.Carlton, R. M., W. H. Noordman, B. Biswas, E. D. de Meester, and M. J. Loessner. 2005. Bacteriophage P100 for control of Listeria monocytogenes in foods: genome sequence, bioinformatic analyses, oral toxicity study, and application. Regul. Toxicol. Pharmacol. 43:301-312. [DOI] [PubMed] [Google Scholar]
- 13.Chibani-Chennoufi, S., et al. 2004. In vitro and in vivo bacteriolytic activities of Escherichia coli phages: implications for phage therapy. Antimicrob. Agents Chemother. 48:2558-2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chibeu, A., et al. 2009. The adsorption of Pseudomonas aeruginosa bacteriophage phiKMV is dependent on expression regulation of type IV pili genes. FEMS Microbiol. Lett. 296:210-218. [DOI] [PubMed] [Google Scholar]
- 15.Deonier, R. C. 1996. Native insertion sequence elements: locations, distributions, and sequence relationships, p. 2000-2011. In Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, DC.
- 16.Fauquet, C., M. Mayo, J. Maniloff, U. Desselberger, and L. Ball. 2005. Virus taxonomy: eighth report of the International Committee on Taxonomy of Viruses, p. 359-367. Elsevier, San Diego, CA.
- 17.Fischer, C. R., M. Yoichi, H. Unno, and Y. Tanji. 2004. The coexistence of Escherichia coli serotype O157:H7 and its specific bacteriophage in continuous culture. FEMS Microbiol. Lett. 241:171-177. [DOI] [PubMed] [Google Scholar]
- 18.Garcia, P., C. Madera, B. Martinez, A. Rodriguez, and J. Evaristo Suarez. 2009. Prevalence of bacteriophages infecting Staphylococcus aureus in dairy samples and their potential as biocontrol agents. J. Dairy Sci. 92:3019-3026. [DOI] [PubMed] [Google Scholar]
- 19.Ghosal, D., H. Sommer, and H. Saedler. 1979. Nucleotide sequence of the transposable DNA-element IS2. Nucleic Acids Res. 6:1111-1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goode, D., V. M. Allen, and P. A. Barrow. 2003. Reduction of experimental Salmonella and Campylobacter contamination of chicken skin by application of lytic bacteriophages. Appl. Environ. Microbiol. 69:5032-5036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Greer, G. G. 2005. Bacteriophage control of foodborne bacteria. J. Food Prot. 68:1102-1111. [DOI] [PubMed] [Google Scholar]
- 22.Greer, G. G., and B. D. Dilts. 1990. Inability of a bacteriophage pool to control beef spoilage. Int. J. Food Microbiol. 10:331-342. [DOI] [PubMed] [Google Scholar]
- 23.Guenther, S., D. Huwyler, S. Richard, and M. J. Loessner. 2009. Virulent bacteriophage for efficient biocontrol of Listeria monocytogenes in ready-to-eat foods. Appl. Environ. Microbiol. 75:93-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haas, M., and B. Rak. 2002. Escherichia coli insertion sequence IS150: transposition via circular and linear intermediates. J. Bacteriol. 184:5833-5841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hagens, S., and M. J. Loessner. 2007. Application of bacteriophages for detection and control of foodborne pathogens. Appl. Microbiol. Biotechnol. 76:513-519. [DOI] [PubMed] [Google Scholar]
- 26.Hong, J., et al. 2008. Identification of host receptor and receptor-binding module of a newly sequenced T5-like phage EPS7. FEMS Microbiol. Lett. 289:202-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hudson, J. A., C. Billington, G. Carey-Smith, and G. Greening. 2005. Bacteriophages as biocontrol agents in food. J. Food Prot. 68:426-437. [DOI] [PubMed] [Google Scholar]
- 28.Hwang, S., et al. 2009. Isolation and characterization of bacteriophages specific for Campylobacter jejuni. Microbiol. Immunol. 53:559-566. [DOI] [PubMed] [Google Scholar]
- 29.Jensen, E. C., et al. 1998. Prevalence of broad-host-range lytic bacteriophages of Sphaerotilus natans, Escherichia coli, and Pseudomonas aeruginosa. Appl. Environ. Microbiol. 64:575-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johnson, K., and S. Lory. 1987. Characterization of Pseudomonas aeruginosa mutants with altered piliation. J. Bacteriol. 169:5663-5667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kitamura, K., Y. Torii, C. Matsuoka, and K. Yamamoto. 1995. DNA sequence changes in mutations in the tonB gene on the chromosome of Escherichia coli K12: insertion elements dominate the spontaneous spectra. Jpn. J. Genet. 70:35-46. [DOI] [PubMed] [Google Scholar]
- 32.Kocharunchitt, C., T. Ross, and D. L. McNeil. 2009. Use of bacteriophages as biocontrol agents to control Salmonella associated with seed sprouts. Int. J. Food Microbiol. 128:453-459. [DOI] [PubMed] [Google Scholar]
- 33.Kunisaki, H., and Y. Tanji. 2010. Intercrossing of phage genomes in a phage cocktail and stable coexistence with Escherichia coli O157:H7 in anaerobic continuous culture. Appl. Microbiol. Biotechnol. 85:1533-1540. [DOI] [PubMed] [Google Scholar]
- 34.Lewis, L. A., N. Gadura, M. Greene, R. Saby, and N. D. Grindley. 2001. The basis of asymmetry in IS2 transposition. Mol. Microbiol. 42:887-901. [DOI] [PubMed] [Google Scholar]
- 35.Lewis, L. A., D. Lewis, V. Persaud, S. Gopaul, and B. Turner. 1994. Transposition of IS2 into the hemB gene of Escherichia coli K-12. J. Bacteriol. 176:2114-2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lowe, T. M., and S. R. Eddy. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25:955-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lukashin, A. V., and M. Borodovsky. 1998. GeneMark.hmm: new solutions for gene finding. Nucleic Acids Res. 26:1107-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mahillon, J., and M. Chandler. 1998. Insertion sequences. Microbiol. Mol. Biol. Rev. 62:725-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Matsuzaki, S., et al. 2005. Bacteriophage therapy: a revitalized therapy against bacterial infectious diseases. J. Infect. Chemother. 11:211-219. [DOI] [PubMed] [Google Scholar]
- 40.Matsuzaki, S., et al. 2003. Experimental protection of mice against lethal Staphylococcus aureus infection by novel bacteriophage phi MR11. J. Infect. Dis. 187:613-624. [DOI] [PubMed] [Google Scholar]
- 41.Modi, R., Y. Hirvi, A. Hill, and M. W. Griffiths. 2001. Effect of phage on survival of Salmonella Enteritidis during manufacture and storage of cheddar cheese made from raw and pasteurized milk. J. Food Prot. 64:927-933. [DOI] [PubMed] [Google Scholar]
- 42.Nesper, J., D. Kapfhammer, K. E. Klose, H. Merkert, and J. Reidl. 2000. Characterization of Vibrio cholerae O1 antigen as the bacteriophage K139 receptor and identification of IS1004 insertions aborting O1 antigen biosynthesis. J. Bacteriol. 182:5097-5104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Newell, D. G., et al. 2010. Food-borne diseases—the challenges of 20 years ago still persist while new ones continue to emerge. Int. J. Food Microbiol. 139:S3-S15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.O'Flynn, G., R. P. Ross, G. F. Fitzgerald, and A. Coffey. 2004. Evaluation of a cocktail of three bacteriophages for biocontrol of Escherichia coli O157:H7. Appl. Environ. Microbiol. 70:3417-3424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rabsch, W., et al. 2007. FepA- and TonB-dependent bacteriophage H8: receptor binding and genomic sequence. J. Bacteriol. 189:5658-5674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ronecker, H. J., and B. Rak. 1987. Genetic organization of insertion element IS2 based on a revised nucleotide sequence. Gene 59:291-296. [DOI] [PubMed] [Google Scholar]
- 47.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 48.Sharma, M., J. R. Patel, W. S. Conway, S. Ferguson, and A. Sulakvelidze. 2009. Effectiveness of bacteriophages in reducing Escherichia coli O157:H7 on fresh-cut cantaloupes and lettuce. J. Food Prot. 72:1481-1485. [DOI] [PubMed] [Google Scholar]
- 49.Sheng, H., H. J. Knecht, I. T. Kudva, and C. J. Hovde. 2006. Application of bacteriophages to control intestinal Escherichia coli O157:H7 levels in ruminants. Appl. Environ. Microbiol. 72:5359-5366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Skurnik, M., and E. Strauch. 2006. Phage therapy: facts and fiction. Int. J. Med. Microbiol. 296:5-14. [DOI] [PubMed] [Google Scholar]
- 51.Soothill, J. S. 1994. Bacteriophage prevents destruction of skin grafts by Pseudomonas aeruginosa. Burns 20:209-211. [DOI] [PubMed] [Google Scholar]
- 52.Toro, H., et al. 2005. Use of bacteriophages in combination with competitive exclusion to reduce Salmonella from infected chickens. Avian Dis. 49:118-124. [DOI] [PubMed] [Google Scholar]
- 53.Wagenaar, J. A., M. A. Van Bergen, M. A. Mueller, T. M. Wassenaar, and R. M. Carlton. 2005. Phage therapy reduces Campylobacter jejuni colonization in broilers. Vet. Microbiol. 109:275-283. [DOI] [PubMed] [Google Scholar]
- 54.Wang, J., et al. 2005. Complete genome sequence of bacteriophage T5. Virology 332:45-65. [DOI] [PubMed] [Google Scholar]
- 55.Zhang, J., et al. 2010. Development of an anti-Salmonella phage cocktail with increased host range. Foodborne Pathog. Dis. 7:1415-1419. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.