Abstract
Aichi virus (AiV) genomes were detected in 12 (100%) influent and 11 (92%) effluent wastewater and 36 (60%) river water samples. Among 260 strains identified, 255 were genotype A and 5 were genotype B. This is the first report describing the molecular characterization of AiVs in aquatic environments in Japan.
Aichi viruses (AiVs) are members of the genus Kobuvirus in the family Picornaviridae and possess single-stranded, positive-sense RNA genomes (24). Based on 3C and 3D (3CD) junction region nucleotide sequences, AiVs are currently classified into three genetically distinct genotypes (A to C) (2, 25). They have been identified in Asian (14, 15, 23, 26, 27), European (2, 7, 12, 13, 16, 17), South American (13), and African (19, 20) countries, suggesting their worldwide distribution. Seroprevalence studies performed in several countries demonstrated a high percentage (80 to 99%) of AiV antibodies in adults (3, 13, 18, 21, 24), indicating widespread human exposure. In contrast, AiV infection in patients with sporadic or epidemic gastroenteritis is low (2, 7, 12-17, 19, 20, 23, 27). These findings, however, do not reveal a role of AiVs in human enteric diseases, and thus their actual prevalence, pathogenesis, and molecular epidemiology remain unclear.
Fecal-oral AiV transmission through contaminated food or water is indicated by AiV detection in fecal samples of infected individuals (2, 7, 12-17, 19, 20, 23, 27) and raw and treated sewage (22), sewage-polluted river water (1), and shellfish samples (4, 12, 22). To further elucidate their prevalence and infection route, comprehensive documentation of their environmental occurrence and fate is essential. We investigated the prevalence and genetic diversity of AiVs in wastewater and river water in Japan for 1 year. The presence of AiV genomes in the water samples was determined by nested reverse transcription (RT)-PCR, and the strains were further characterized according to the 3CD junction region sequences.
Between March 2005 and February 2006, influent and effluent samples were collected monthly from a wastewater treatment plant, which utilized a conventional activated sludge process. These samples (100 ml, influent, and 1,000 ml, effluent) were concentrated by the adsorption-elution method using an electronegative filter (catalog no. HAWP-090-00; Millipore, Tokyo, Japan) with an acid rinse procedure (8). Between April 2003 and March 2004, 60 river water samples were collected monthly from five sites (sites 1 to 5) from upstream to downstream along the Tamagawa River in Japan. Further detailed information on the Tamagawa River basin and the locations of sampling sites has been published previously (10). These samples (500 ml) were concentrated using the cation-coated filter method (5, 6). The concentrated wastewater and river water samples were further concentrated using a Centriprep YM-50 device (Millipore) to obtain a 700-μl final volume. Viral RNA was extracted from the concentrated samples using the QIAamp viral RNA minikit (Qiagen, Hilden, Germany), followed by RT using the High Capacity cDNA reverse transcription kit (Applied Biosystems, Foster City, CA).
Nested PCR was performed using the 6261/6779 and C94b/246k primer sets and Hot Start Ex Taq DNA polymerase (TaKaRa Bio Co., Shiga, Japan) to amplify the 266-bp 3CD junction region (25). Second-round PCR products were cloned into the pCR4-TOPO vector (Invitrogen, Carlsbad, CA). Subsequently, eight clones per sample were selected, and both strands were sequenced using the BigDye Terminator v3.1 cycle sequencing kit and the 3130xl Genetic Analyzer (Applied Biosystems). Nucleotide sequences were aligned using ClustalW version 1.83, and distances were calculated using Kimura's two-parameter method (9).
Among 12 influent and 12 effluent wastewater samples tested, AiVs were detected in all influent and 11 (92%) effluent samples by nested RT-PCR, demonstrating high AiV prevalence in the wastewater samples. Nested RT-PCR detected AiVs in 36 of 60 (60%) river water samples from one to four of the five sampling sites in each month, except for July 2003, when no AiVs were detected in any samples. AiVs were not detected in samples from the upstream area (site 1) but were detected in those from mid- to downstream areas (sites 2 to 5) in 8 of 12 (67%) months. Interestingly, the detection rate of AiVs in river water samples was higher than that of noroviruses or sapoviruses (10, 11).
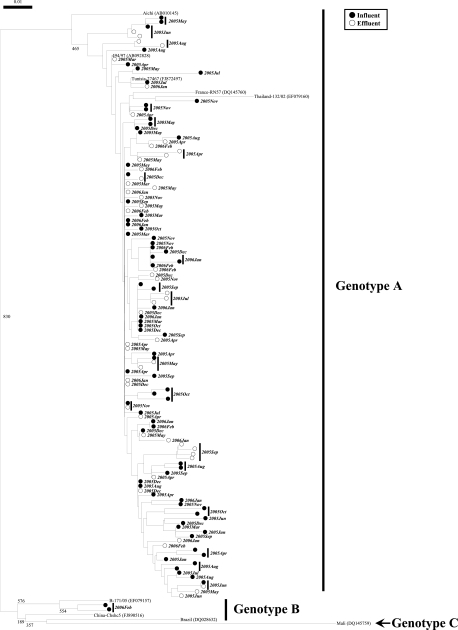
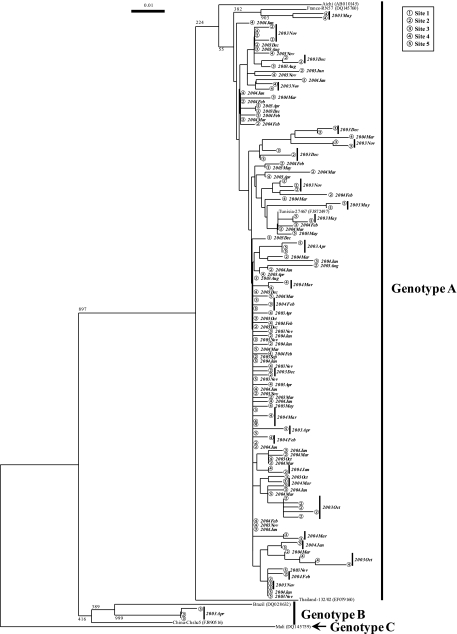
By phylogenetic analysis, we identified 125 and 135 distinct AiV strains from wastewater and river water samples, respectively (Fig. 1 and 2). Genotype A strains were abundant in both wastewater and river water samples, whereas genotype B strains were identified in only one influent sample collected in February 2006 and one river water sample collected in April 2003 at site 3 (Fig. 1 and 2).
FIG. 1.
Phylogenetic tree of AiV strains identified in wastewater samples. The tree was constructed on the basis of sequences of a 224-nucleotide stretch within the 3CD junction region. The numbers on each branch indicate bootstrap values for the genotype, and bootstrap values of 950 or greater were considered statistically significant for grouping. The scale represents nucleotide substitutions per site. Filled and open circles indicate AiV strains detected in influent and effluent wastewater samples, respectively. The year and month are shown in bold italics beside the symbols.
FIG. 2.
Phylogenetic tree of AiV strains identified in water samples from the Tamagawa River. The tree was constructed on the basis of sequences of a 224-nucleotide stretch within the 3CD junction region. The numbers on each branch indicate bootstrap values for the genotype, and bootstrap values of 950 or greater were considered statistically significant for grouping. The scale represents nucleotide substitutions per site. Circled numbers indicate AiV strains detected at each sampling site. The year and month are shown in bold italics beside the symbols.
To date, only a few studies have detected AiVs in water samples. Sdiri-Loulizi et al. (22) identified AiVs in 15 of 250 sewage samples (6%) in Tunisia, much lower than the detection rate reported in the present study. This may be because they performed single-round RT-PCR using the 6261/6779 “outer” primer set. We performed nested PCR utilizing an inner primer set, C94b/246k (25), to increase the sensitivity. Alcalá et al. (1) examined only 11 sewage-polluted river water samples for AiVs, far fewer than were studied here. Furthermore, these studies performed direct nucleotide sequencing without molecular cloning, and both studies identified only a single genotype: genotype A by Sdiri-Loulizi et al. (22) and genotype B by Alcalá et al. (1). In contrast, we cloned the second-round PCR products and sequenced eight clones per sample. Thus, we successfully identified both genotypes A and B even when they coexisted in a single sample (influent wastewater sample in February 2006).
This is the first report describing the molecular detection and characterization of AiVs in aquatic environments in Japan. A total of 260 distinct AiV strains were identified from wastewater and river water samples, with a considerable genetic diversity (Fig. 1 and 2), which was much larger than the number of AiV strains identified in any other previous environmental studies (1, 22). Among the strains identified, 255 were classified into genotype A and 5 were genotype B. Our novel findings based on environmental investigations demonstrate that more genetically diverse AiV strains are prevalent among human populations than previously appreciated. The present study highlights the importance of further studies on AiVs toward a better understanding of their distribution and etiological role, if any, in human enteric diseases.
Nucleotide sequence accession numbers.
The nucleotide sequences determined in this study have been deposited in GenBank under accession numbers AB569644 to AB569903.
Acknowledgments
This work was supported by a Grant-in-Aid for Young Scientists (A) under project number 20686035 from the Ministry of Education, Culture, Sports, Science and Technology, Japan.
We thank Hozue Kuroda at The University of Tokyo for her technical assistance.
Footnotes
Published ahead of print on 21 January 2011.
REFERENCES
- 1.Alcalá, A., et al. 2010. Molecular detection and characterization of Aichi viruses in sewage-polluted waters of Venezuela. Appl. Environ. Microbiol. 76:4113-4115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ambert-Balay, K., et al. 2008. Prevalence and genetic diversity of Aichi virus strains in stool samples from community and hospitalized patients. J. Clin. Microbiol. 46:1252-1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goyer, M., L. S. Aho, J. B. Bour, K. Ambert-Balay, and P. Pothier. 2008. Seroprevalence distribution of Aichi virus among a French population in 2006-2007. Arch. Virol. 153:1171-1174. [DOI] [PubMed] [Google Scholar]
- 4.Hansman, G. S., et al. 2008. Detection of human enteric viruses in Japanese clams. J. Food Prot. 71:1689-1695. [DOI] [PubMed] [Google Scholar]
- 5.Haramoto, E., H. Katayama, K. Oguma, and S. Ohgaki. 2005. Application of cation-coated filter method to detection of noroviruses, enteroviruses, adenoviruses, and Torque Teno viruses in the Tamagawa River in Japan. Appl. Environ. Microbiol. 71:2403-2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haramoto, E., H. Katayama, and S. Ohgaki. 2004. Detection of noroviruses in tap water in Japan by means of a new method for concentrating enteric viruses in large volumes of freshwater. Appl. Environ. Microbiol. 70:2154-2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kaikkonen, S., S. Räsänen, M. Rämet, and T. Vesikari. 2010. Aichi virus infection in children with acute gastroenteritis in Finland. Epidemiol. Infect. 138:1166-1171. [DOI] [PubMed] [Google Scholar]
- 8.Katayama, H., A. Shimasaki, and S. Ohgaki. 2002. Development of a virus concentration method and its application to detection of enterovirus and Norwalk virus from coastal seawater. Appl. Environ. Microbiol. 68:1033-1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kimura, M. 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16:111-120. [DOI] [PubMed] [Google Scholar]
- 10.Kitajima, M., et al. 2010. Detection and genetic analysis of human sapoviruses in river water in Japan. Appl. Environ. Microbiol. 76:2461-2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kitajima, M., et al. 2010. Seasonal distribution and genetic diversity of genogroups I, II, and IV noroviruses in the Tamagawa River, Japan. Environ. Sci. Technol. 44:7116-7122. [DOI] [PubMed] [Google Scholar]
- 12.Le Guyader, F. S., et al. 2008. Aichi virus, norovirus, astrovirus, enterovirus, and rotavirus involved in clinical cases from a French oyster-related gastroenteritis outbreak. J. Clin. Microbiol. 46:4011-4017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oh, D. Y., et al. 2006. Molecular characterization of the first Aichi viruses isolated in Europe and in South America. Arch. Virol. 151:1199-1206. [DOI] [PubMed] [Google Scholar]
- 14.Pham, N. T., et al. 2007. Isolation and molecular characterization of Aichi viruses from fecal specimens collected in Japan, Bangladesh, Thailand, and Vietnam. J. Clin. Microbiol. 45:2287-2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pham, N. T., et al. 2008. Sequence analysis of the capsid gene of Aichi viruses detected from Japan, Bangladesh, Thailand, and Vietnam. J. Med. Virol. 80:1222-1227. [DOI] [PubMed] [Google Scholar]
- 16.Räsänen, S., et al. 2010. Mixed viral infections causing acute gastroenteritis in children in a waterborne outbreak. Epidemiol. Infect. 138:1227-1234. [DOI] [PubMed] [Google Scholar]
- 17.Reuter, G., A. Boldizsár, I. Kiss, and P. Pankovics. 2008. Candidate new species of Kobuvirus in porcine hosts. Emerg. Infect. Dis. 14:1968-1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ribes, J. M., R. Montava, C. J. Téllez-Castillo, M. Fernández-Jiménez, and J. Buesa. 2010. Seroprevalence of Aichi virus in a Spanish population from 2007 to 2008. Clin. Vaccine Immunol. 17:545-549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sdiri-Loulizi, K., et al. 2008. Acute infantile gastroenteritis associated with human enteric viruses in Tunisia. J. Clin. Microbiol. 46:1349-1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sdiri-Loulizi, K., et al. 2009. Detection and genomic characterization of Aichi viruses in stool samples from children in Monastir, Tunisia. J. Clin. Microbiol. 47:2275-2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sdiri-Loulizi, K., et al. 2010. Aichi virus IgG seroprevalence in Tunisia parallels genomic detection and clinical presentation in children with gastroenteritis. Clin. Vaccine Immunol. 17:1111-1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sdiri-Loulizi, K., et al. 2010. First molecular detection of Aichi virus in sewage and shellfish samples in the Monastir region of Tunisia. Arch. Virol. 155:1509-1513. [DOI] [PubMed] [Google Scholar]
- 23.Yamashita, T., et al. 1995. Isolation of cytopathic small round virus (Aichi virus) from Pakistani children and Japanese travelers from Southeast Asia. Microbiol. Immunol. 39:433-435. [DOI] [PubMed] [Google Scholar]
- 24.Yamashita, T., K. Sakae, Y. Ishihara, S. Isomura, and E. Utagawa. 1993. Prevalence of newly isolated, cytopathic small round virus (Aichi strain) in Japan. J. Clin. Microbiol. 31:2938-2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yamashita, T., et al. 2000. Application of a reverse transcription-PCR for identification and differentiation of Aichi virus, a new member of the Picornavirus family associated with gastroenteritis in humans. J. Clin. Microbiol. 38:2955-2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yamashita, T., et al. 1991. Isolation of cytopathic small round viruses with BS-C-1 cells from patients with gastroenteritis. J. Infect. Dis. 164:954-957. [DOI] [PubMed] [Google Scholar]
- 27.Yang, S., et al. 2009. Aichi virus strains in children with gastroenteritis, China. Emerg. Infect. Dis. 15:1703-1705. [DOI] [PMC free article] [PubMed] [Google Scholar]