Abstract
Ammonia oxidation, the first step in nitrification, is performed by certain Beta- and Gammaproteobacteria and Crenarchaea to generate metabolic energy. Ammonia monooxygenase (amoA) genes from both Bacteria and Crenarchaea have been found in a variety of marine ecosystems, but the relative importance of Bacteria versus Crenarchaea in ammonia oxidation is unresolved, and seasonal comparisons are rare. In this study, we compared the abundance of betaproteobacterial and crenarchaeal amoA genes in the coastal Arctic Ocean during summer and winter over 2 years. Summer and winter betaproteobacterial amoA clone libraries were significantly different, although the gene sequences were similar to those found in temperate and polar environments. Betaproteobacterial and crenarchaeal amoA genes were 30- to 115-fold more abundant during the winter than during the summer in both years of the study. Archaeal amoA genes were more abundant than betaproteobacterial amoA genes in the first year, but betaproteobacterial amoA was more abundant than archaeal amoA the following year. The ratio of archaeal amoA gene copies to marine group I crenarchaeal 16S rRNA genes averaged 2.9 over both seasons and years, suggesting that ammonia oxidation was common in Crenarchaea at this location. Potential nitrification rates, as well as the total amoA gene abundance, were highest in the winter when competition with phytoplankton was minimal and ammonium concentrations were the highest. These results suggest that ammonium concentrations were important in determining the rates of ammonia oxidation and the abundance of ammonia-oxidizing Betaproteobacteria and Crenarchaea.
Ammonia oxidation is the first, rate-limiting step in nitrification, the microbially mediated process in which ammonium is oxidized to nitrite and then to nitrate (57). Previously, only certain groups within the Beta- and Gammaproteobacteria were believed to perform ammonia oxidation, but then the ammonia monooxygenase subunit A (amoA) gene was detected in Crenarchaea (49, 55), and ammonia oxidation by an archaeon was experimentally demonstrated (33). Since then, archaeal amoA has been found in a variety of environments, such as soils (38), marine sediments (2), and the oceans (3, 18), including the Arctic Ocean (19). A third archaeal phylum, the Thaumarchaeota, has been proposed to include the ammonia-oxidizing archaea currently classified among the Crenarchaea (6). Archaeal amoA is much more abundant than bacterial amoA in many marine environments (3, 11, 40), including coastal Antarctic waters and the central Arctic Ocean (27). The archaeal amoA gene was also found to be more abundant than the bacterial amoA gene in some estuarine sediments (2), but in San Francisco Bay sediments, betaproteobacterial amoA outnumbered archaeal amoA at higher-salinity locations (42), which raises the question of whether betaproteobacterial amoA genes are more abundant than archaeal amoA genes in other environments. Archaeal ammonia oxidizers may be favored over betaproteobacterial ammonia oxidizers in low-nutrient environments (16).
The fraction of marine Crenarchaea that are ammonia oxidizers has been explored by examining the ratio of archaeal amoA genes to 16S rRNA genes (see, for example, references 11, 40, and 60). This ratio is thought to be 1:1 for ammonia-oxidizing Crenarchaea based on the sequenced genomes of Cenarchaeum symbiosum and Nitrosopumilus maritimus (22, 56). Higher ratios of archaeal amoA to 16S rRNA genes have been found in environmental samples, including 2:1 in Circumpolar Deep Water west of the Antarctic Peninsula (27) and 8:1 in the eastern Canadian Arctic (19). More information is needed to establish whether the capacity for ammonia oxidation exists among all mesophilic marine Crenarchaea.
Measurements of both amoA abundance and nitrification rates from the same samples are rare. Ammonium oxidation rates were correlated with amoA and marine group I crenarchaeal 16S rRNA gene copies throughout the water column of the Gulf of California (3). In contrast, rates were not correlated with bacterial, archaeal, or total amoA gene abundance in the Central California Current (48), nor were archaeal amoA abundance and potential nitrification rates correlated in sediments of the Plum Island Sound Estuary in Massachusetts (5). Nitrification rates range from 3.1 nmol liter−1 day−1 in the oligotrophic Atlantic (8) to 45 nmol liter−1 day−1 in Monterey Bay and Southern California Bight (58, 59) and can exceed 100 nmol liter−1 day−1 at the base of the euphotic zone of the North Pacific Ocean (13). The abundance of amoA genes was not examined in these studies. No estimates of nitrification are available from polar waters.
Nitrification rates and the abundance of ammonia oxidizers may differ seasonally in polar regions where summer and winter conditions vary greatly. These rates and organisms depend on ammonium generated by heterotrophic processes, such as excretion and microbial degradation of organic nitrogen (57), which are coupled to primary production, suggesting that nitrification and ammonia oxidizer abundance should be highest in the summer. However, ammonia oxidation is thought to be inhibited by light (20) and by competition with phytoplankton and heterotrophic bacteria for ammonium (57, 59), both of which are minimized during winter. These data lead to the alternative hypothesis that nitrification and ammonia oxidizer abundance are highest in the winter. Higher ammonium oxidation in the winter would help to explain the seasonal variation of marine Crenarchaea in the Arctic Ocean and Antarctic coastal waters (7, 43) if most marine Crenarchaea oxidize ammonia. In Franklin Bay in the Canadian Arctic, archaea were 16% of total prokaryotes in surface waters during winter but decreased to less than 10% in spring and summer (1). More data on ammonium oxidation and amoA abundance in summer and winter should provide insights into possible controls of nitrification rates and nitrifying communities in the oceans.
In the present study we examined winter and summer communities of ammonia-oxidizing prokaryotes in the coastal Arctic Ocean near Barrow, Alaska. Surface waters were sampled to determine the abundance of betaproteobacterial and archaeal amoA genes and marine group I Crenarchaea 16S rRNA genes in the summer and winter over 2 years. Clone libraries were prepared to examine partial amoA gene sequences and to compare coastal arctic ammonia-oxidizing communities to those found elsewhere. Potential nitrification rates and other physical data were measured to understand how these communities vary with such seasonal extremes and to provide more information about an understudied ocean. We found that potential nitrification rates and the abundance of betaproteobacterial and crenarchaeal amoA genes varied seasonally, reaching their highest levels in winter.
MATERIALS AND METHODS
Sample collection.
Seawater samples were obtained from the coastal Arctic Ocean near Barrow, Alaska, over 2 years. Table S1 in the supplemental material gives individual sample locations, dates, and times. Sample sites were in nearshore waters of the Chukchi and Beaufort Seas on either side of Barrow Point. Summer samples were obtained from surface waters (∼2-m depth) via a manual pump from small boats in July 2007 and July 2008. Winter samples were extracted by drilling a hole in the sea ice and manually pumping water from directly below the ice (∼2 m) in January 2008 and January 2009. The seawater was maintained at nearly in situ temperatures until filtration. Between 1.3 and 1.8 liters of seawater were vacuum filtered through 0.22-μm-pore-size Durapore membrane filters (Millipore) within a few hours after collection at a land-based facility, with three to six replicates per sampling location. Filters were then preserved in a CTAB (cetyltrimethylammonium bromide) buffer and stored at −80°C until DNA extractions were performed.
Environmental parameters.
Nutrient concentrations, chlorophyll a concentration, prokaryotic production, and microscopic direct counts of prokaryotic abundance were examined by standard methods as described previously (10). Briefly, nutrient concentrations were determined by using the sodium nitroprusside method for ammonium or an automated, segmented flow colorimetric analysis for nitrite + nitrate, phosphate, or silicate. The chlorophyll a concentration was measured by extraction of cells captured by using GF/F filters (Whatman) in 90% acetone and subsequent measurement of fluorescence. Prokaryote abundance was measured microscopically from 0.2-μm-pore-size polycarbonate filters stained with DAPI (4′,6′-diamidino-2-phenylindole) by using semiautomated image analysis (9). Prokaryotic production was determined by a 1.5- to 2.5-h incubation with [3H]leucine (20 nM added concentration) at in situ temperatures in the dark (29). Leucine incorporation rates were converted to prokaryotic production by assuming 1.5 kg of C per mol of incorporated leucine.
DNA extraction and PCR screening.
DNA was extracted from the filters by using a modified CTAB protocol involving two chloroform extractions and a high-salt isopropanol precipitation (12). A Quant-iT PicoGreen dsDNA assay kit (Invitrogen) was used to measure DNA concentration via fluorescence. Samples were initially screened for presence of betaproteobacterial, gammaproteobacterial, and archaeal amoA genes by PCR, followed by examination of PCR products on 1.5% agarose gels. Primers used are listed in Table 1 .
TABLE 1.
PCR primers used for screening and cloning of environmental DNA and for determining gene abundance using qPCR
| Gene | Group | Primer |
Purpose | Reference | |
|---|---|---|---|---|---|
| Name | Sequence (5′-3′) | ||||
| amoA | Betaproteobacteria | AmoA-1F* | GGGGHTTYTACTGGTGGT | Screening | 53 |
| AmoA-2R | CCCCTCKGSAAAGCCTTCTTC | qPCR | 47 | ||
| amoA | Gammaproteobacteria | amoA-3F | GGTGAGTGGGYTAACMG | Screening | 46 |
| amoB-4R | GCTAGCCACTTTCTGG | ||||
| amoA | Crenarchaea | AmoA-19F | ATGGTCTGGCTWAGACG | Screening | 38 |
| AmoA-643R | TCCCACTTWGACCARGCGGCCATCCA | ||||
| amoA | Crenarchaea group A | Arch-amoAFA | ACACCAGTTTGGYTACCWTCDGC | qPCR | 3 |
| Arch-amoAR | GCGGCCATCCATCTGTATGT | 18 | |||
| amoA | Crenarchaea group B | Arch-amoAFB | CATCCRATGTGGATTCCATCDTG | qPCR | 3 |
| Arch-amoAR | GCGGCCATCCATCTGTATGT | 18 | |||
| 16S rRNA | Total Bacteria | w49dir | CGGTCCAGACTCCTACGGG | qPCR | 37 |
| w34rev | TTACCGCGGCTGCTGGCAC | ||||
| 16S rRNA | Crenarchaea marine group I | GI_751F | GTCTACCAGAACAYGTTC | qPCR | 40 |
| GI_956R | HGGCGTTGACTCCAATTG | ||||
| 16S rRNA | Crenarchaea PSL12 group | pSL12_750F | GGTCCRCCAGAACGCGC | qPCR | 40 |
| pSL12_876R | GTACTCCCCAGGCGGCAA | ||||
Betaproteobacterial amoA PCR products from one summer 2007 Chukchi Sea sample (Chuk-1 in Table S1 in the supplemental material) and three pooled winter 2008 Chukchi Sea samples (Chuk-3, Chuk-4, and Chuk-5) were cloned by using a TOPO TA cloning kit (Invitrogen). The pooled winter 2008 winter samples were collected on different days from the same location. Archaeal amoA products from pooled winter 2008 Chukchi Sea samples were also cloned; however, summer archaeal amoA genes were not. The clones were sequenced via the Sanger method by the High Throughput Genomics Unit of the University of Washington, Seattle, WA. Initial betaproteobacterial amoA sequences were 492 bp long, and the library contained 92 clones from summer 2007 and 91 clones from winter 2008. The archaeal amoA sequences were 624 bp long, and the library contained 45 sequences from winter 2008 samples. Sequences of poor quality or short reads and primer sequences were excluded. The sequences have been submitted to the GenBank database under accession numbers GU453692 to GU453916.
Phylogenetic analysis.
Sequences were aligned by using BioEdit (21; www.mbio.ncsu.edu/BioEdit) or MEGA version 4 (54; www.megasoftware.net) using CLUSTAL W. Clustering and operational taxonomic unit (OTU) analysis was performed using DOTUR (50). OTUs were defined by sequences having <5% divergence in nucleotides. Coverage ratios for each library were calculated as one minus the quotient of the number of OTUs containing only one sequence divided by the total number of sequences. Summer and winter betaproteobacterial libraries were compared by using ∫-Libshuff (51). Phylogenetic trees were prepared using MEGA4. Nucleotide sequences, including all three codon positions, were analyzed with 500 bootstrap replicates and were based on the Jukes-Cantor model of DNA substitution. Sequences from Barrow and other environments were first trimmed to 447 bp in the case of betaproteobacterial amoA and 591 bp in the case of archaeal amoA to exclude primer sites and so that the analysis was performed on sequences of equal lengths. The amino acid-based tree for betaproteobacterial amoA was based on distances adjusted by the Poisson correction.
Quantification of amoA and 16S rRNA genes by qPCR.
Quantitative PCR (qPCR) reactions were performed on a Rotor-Gene 6000 real-time rotary qPCR analyzer (Corbett Research) using SYBR Green PCR master mix (Applied Biosystems). Primers used for each quantified gene are given in Table 1. The primer concentrations were 0.16 μM in each 12.5-μl reaction, and the following cycling times were used: 95°C for 10 min, followed by 45 individual cycles of 95°C for 15 s, 50°C for 15 s, and 72°C for 15 s. Optimal annealing temperatures are typically lower with the Rotor-Gene system than with other qPCR systems (data not shown). Plasmids made by cloning environmental samples were used as standards. For each gene and primer set, the qPCRs were run with three replicates of both standards and DNA from each sampling location. DNA concentrations for standards and samples were quantified via Pico Green analysis using the Rotor-Gene 6000. Standard curve coefficients of variation and efficiencies were as follows: betaproteobacterial amoA (r2 = 0.994, efficiency = 83%), archaeal amoA group A (r2 = 0.997, efficiency = 77%), Crenarchaea marine group I 16S rRNA (r2 = 0.999, efficiency = 71%), and pSL12 group 16S rRNA (r2 = 0.999, efficiency = 86%). Melting curve and sequence analyses of qPCR products were used to test for nonspecific amplification. Gene abundance was normalized per ng of extracted DNA, per ml of filtered seawater, and per total 16S rRNA gene abundance.
Potential nitrification rates.
Potential nitrification rates were estimated by tracing 15N-labeled NH4+ into the nitrite plus the nitrate pool in four samples from summer 2008 (Beau-5, Beau-6, Chuk-6, and Chuk-7) and four samples from winter 2009 (Chuk-8, Beau-7, Beau-8, and Beau-9). Briefly, 15N-labeled NH4+ was added to reach a final concentration of 1 μM, and the samples were incubated in triplicate 250-ml acid-washed polycarbonate bottles in the dark at in situ temperatures for 24 h. The high concentration of the 15N-labeled NH4+ added was used to minimize isotope dilution. Initial samples and samples taken after 24 h were filtered through 0.2-μm-pore-size polycarbonate filters, and the filtrate was stored at −80°C until later analysis. The accumulation of 15N in NO2− and NO3− by oxidation of 15N-NH4+ was determined by using the denitrifier method (52), which converts NO2− and NO3− (NOx−) to nitrous oxide (N2O). The δ15N value of N2O was analyzed by using previously described methods (14, 45). The accuracy and precision of this method was less than ±0.5‰ (±0.00018 atom percent 15N [at% 15N]) for samples containing >2.5 nmol of nitrate as determined by multiple analysis of a sodium nitrate solution for which the δ15N value of the solid NaNO3 had been previously determined using an on-line carbon-nitrogen analyzer coupled with an isotope ratio mass spectrometer (Finnigan ConFlo II/Delta-Plus). Apparent 15N-ammonia oxidation rates (15Rox) were calculated using the following equation:
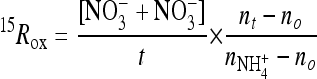 |
where nt is the at% 15N in the NOx− pool measured at time t and [NO3− + NO2−] is the measured concentration of the NOx− pool. The 15N activity of the initial NH4+ pool (no) was assumed to be 0.3663% at% 15N.
RESULTS
Summer and winter environmental conditions in the coastal Arctic Ocean near Barrow, Alaska, are very different (Table 2). The ocean is ice covered during much of the winter, and polar darkness persists for 2 months. Water temperatures varied between −1.7°C in winter to as high as 5.1°C in summer. Nutrient concentrations also varied with the seasons and were highest in the winter. Nutrient levels declined each summer once sunlight and photosynthesis return. Chlorophyll a concentrations, while never reaching zero during the winter, were about 10 times higher during the summer. Both prokaryotic abundance and production were also highest during the summer months. Prokaryote abundance ranged between 0.22 × 106 and 0.55 × 106 cells per ml in the winter, increasing to 0.93 × 106 to 1.91 × 106 cells per ml in the summer. Prokaryotic production followed a similar pattern and was 19 to 103 nmol of C liter−1 h−1 in the winter, increasing to 156 to 280 nmol of C liter−1 h−1 in the summer. Although there were significant differences between seasons for each environmental parameter (P < 0.05, Student t test), the Chukchi Sea and Beaufort Sea values were not significantly different (P > 0.05, Student t test).
TABLE 2.
Environmental parameters in the coastal Chukchi and Beaufort Seas near Barrow, Alaska
| Parameter | Summer 2007 |
Winter 2008 |
Summer 2008 |
Winter 2009 |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Chukchi |
Beaufort |
Chukchia |
Chukchi |
Beaufort |
Chukchi |
Beaufort |
|||||||
| Mean | SD | Mean | SD | Mean | SD | Mean | SD | Mean | SD | Mean | Mean | SD | |
| No. of locations | 2 | 2 | 3 | 2 | 4 | 1 | 3 | ||||||
| Prokaryotes (106 cells/ml) | 1.75 | 0.25 | 1.91 | 0.25 | 0.55 | 0.18 | 0.93 | 0.00 | 1.21 | 0.11 | 0.22 | 0.52 | 0.05 |
| Water temp (°C) | 5.1 | 0.6 | 3.2 | 1.0 | -0.5 | 2.2 | 4.2 | 2.9 | 2.5 | 1.5 | -1.7 | -1.7 | 0.0 |
| Salinity | 32.0 | 0.0 | 22.0 | 2.8 | 35.3 | 0.6 | 32.0 | 0.0 | 30.3 | 0.5 | 30.0 | 34.7 | 0.6 |
| Chlorophyll a concn (μg/liter) | 0.55 | 0.06 | 0.41 | 0.11 | 0.06 | 0.01 | 1.62 | 0.38 | 1.03 | 0.12 | 0.14 | 0.04 | 0.00 |
| Ammonium concn (μM) | 0.39 | 0.02 | 0.25 | 0.01 | 1.88 | 0.23 | 0.74 | 0.35 | 0.66 | 0.48 | 4.72 | 3.60 | 0.14 |
| Nitrate/nitrite concn (μM) | 0.90 | 0.26 | 0.25 | 0.02 | 11.35 | 0.19 | 0.63 | 0.29 | 1.47 | 1.21 | 6.86 | 6.98 | 0.58 |
| Phosphate concn (μM) | 0.53 | 0.00 | 0.22 | 0.06 | 1.95 | 0.08 | 0.72 | 0.14 | 0.72 | 0.07 | 1.30 | 0.99 | 0.09 |
| Silica concn (μM) | 9.92 | 1.06 | 4.65 | 0.21 | 23.0 | 0.85 | 9.61 | 0.27 | 9.01 | 0.40 | 18.2 | 15.9 | 0.40 |
| Prokaryotic production (nmol of C liter−1 h−1) | 237 | 39 | 280 | 35 | 20 | 4 | 156 | 97 | 180 | 22 | 103 | 19 | 1 |
The Beaufort Sea was not sampled in winter 2008.
Phylogeny of bacterial and archaeal amoA.
Both betaproteobacterial and archaeal amoA genes were detected in the coastal Arctic Ocean during both seasons; however, partial amoA gene sequences found in clone libraries were not very diverse. Of 183 betaproteobacterial amoA sequences from both seasons, only six OTUs were found (Table 3), defined by <5% divergence in nucleotides. The Chao1 statistic was 7 for the entire set of summer and winter betaproteobacterial amoA. Individual betaproteobacterial amoA libraries contained only three and five OTUs in the summer and winter, respectively. Only four OTUs were present in a library containing 45 archaeal amoA clones from winter, but the Shannon and Simpson diversity indices were similar for both betaproteobacterial and archaeal amoA sequences from winter. The amoA gene from Gammaproteobacteria was not detected.
TABLE 3.
Composition and diversity statistics for amoA clone librariesa
| Parameter |
Betaproteobacteria |
Archaea (winter) | ||
|---|---|---|---|---|
| Summer | Winter | Totalb | ||
| No. of clones | 92 | 91 | 183 | 45 |
| No. of OTUs | 3 | 5 | 6 | 4 |
| % Coverage | 100.0 | 97.8 | 98.9 | 100.0 |
| Chao1 score | 3 | 6 | 7 | 4 |
| Chao1 score 95% CIc | 3-3 | 5.1-18.5 | 6.1-19.7 | 4-4 |
| Shannon score | 0.36 | 1.16 | 1.10 | 1.16 |
| Shannon score 95% CI | 0.18-0.53 | 1.04-1.28 | 0.98-1.21 | 0.99-1.33 |
| Simpson | 0.84 | 0.33 | 0.40 | 0.33 |
Groups are defined at 5% nucleotide acid divergence.
That is, the total of both summer and winter.
CI, confidence interval.
Most betaproteobacterial amoA sequences from the present study separated into three large clades, one OTU (summer group 1) containing mostly summer sequences and two OTUs (winter groups 1 and 2) containing mostly winter sequences (Fig. 1). The summer and winter betaproteobacterial amoA sequences were different from each other (P < 0.01, ∫-Libshuff test). The large summer OTU (summer group 1) contained 84 summer sequences and 21 winter sequences. Another more distant summer clade (summer group 2) contained four sequences. There were four predominantly winter OTUs, with the largest (winter group 1) containing 40 winter sequences and no summer sequences. The second largest winter OTU (winter group 2) contained 28 winter sequences and 4 summer sequences. Sequences in each of these OTUs were similar to sequences in other marine environments, including water column and sediments in both polar and temperate seas (27, 44). Winter group 1 formed a unique clade with other polar sequences sharing >97% identity in nucleotides. However, sequences from other more temperate environments become a part of this clade with these sequences if the cutoff is widened to a level of >95% identity (results not shown). Most of the Barrow betaproteobacterial amoA sequences were more closely related to cultured Nitrosomonas species, such as Nitrosomonas cryotolerans, also isolated from Alaskan waters (26), than to species of Nitrosospira.
FIG. 1.
Neighbor-joining phylogenetic tree of betaproteobacterial amoA. The phylogeny is based on nucleotide sequences. Bootstrap values lower than 50% are omitted. GenBank accession numbers are shown for sequences from other studies.
Betaproteobacterial amoA sequences at the amino acid level are less diverse than the nucleotide sequences (see Fig. S1 in the supplemental material). While the two large winter nucleotide OTUs differed by 30 or 50 nucleotides from the large summer group 1, most of these substitutions are synonymous. Winter group 2 was nearly identical to the large summer group 1 on an amino acid basis. Winter group 1 differed from the summer group 1 by only three to five amino acids.
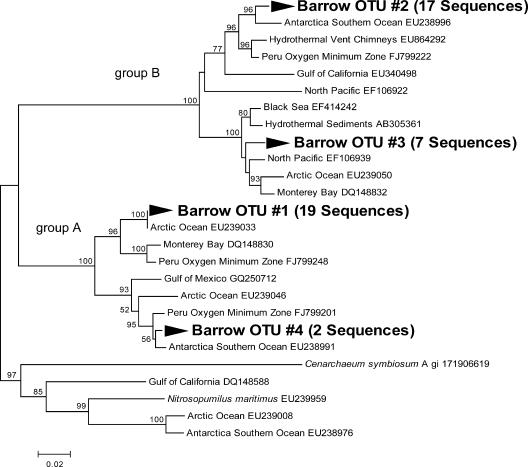
Archaeal amoA sequences form two distinct groups (Fig. 2) similar to the shallow water group A and deep water group B clades reported previously (3). These sequences were also similar to other reported sequences from a variety of nonpolar environments, including the Black Sea, North Pacific, and Gulf of California (3, 35, 40). Although amoA sequences from both group A and group B were found in clone libraries, only group A amoA was quantifiable via qPCR, so the abundance data discussed below are only for group A.
FIG. 2.
Neighbor-joining phylogenetic tree of archaeal amoA. The phylogeny is based on nucleotide sequences. Bootstrap values lower than 50% are omitted. GenBank accession numbers are shown for sequences from other studies.
Abundance of amoA and 16S rRNA genes.
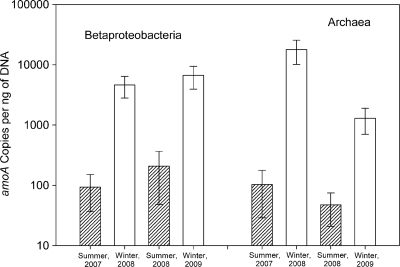
Both betaproteobacterial and archaeal group A amoA genes (Fig. 3) were more abundant during the winter than the summer (P < 0.01, Student t test); archaeal amoA genes from group B, typically seen in deeper waters, were not detected. The differences between summer and winter abundance varied between betaproteobacteria and archaea and between the 2 years. Betaproteobacterial amoA gene copies per ng of DNA increased by 30-fold to nearly 50-fold from summer 2007 to winter 2008 and between summer 2008 and winter 2009 (P < 0.01, Student t test). Group A archaeal amoA increased by >170-fold from summer 2007 to winter 2008, but the increase was much smaller (nearly 30-fold) from summer 2008 to winter 2009 (P < 0.01, Student t test).
FIG. 3.
Copies of amoA genes per ng of DNA for betaproteobacteria and archaea. Error bars represent the standard deviations of all triplicate qPCR measurements from each sample location during the season. There were three replicate measurements for each sample location, and the numbers of sample locations were four (summer 2007 and winter 2009), three (winter 2008), and six (summer 2008).
Bacterial amoA genes were significantly more abundant than archaeal amoA genes in two of the 4 months we examined; there was no significant difference in summer 2007 (Fig. 3). There were significantly more betaproteobacterial amoA genes than archaeal amoA genes in summer 2008 (4-fold) and in winter 2009 (5-fold) (P < 0.01, Student t test). In contrast, archaeal amoA genes were significantly more abundant by nearly 4-fold than betaproteobacterial amoA genes in winter 2008 (P < 0.01, Student t test). Similar results were obtained when amoA abundance was normalized per ml of filtered seawater (see Fig. S2 in the supplemental material) and per total 16S rRNA genes (data not shown).
Betaproteobacterial amoA gene copies were correlated with both ammonium concentrations (r = 0.79, P < 0.01) and nitrate plus nitrite concentrations (r = 0.74, P < 0.01) but were negatively correlated with chlorophyll a concentrations (r = −0.69, P < 0.01) and prokaryotic production (r = −0.82, P < 0.01). Archaeal amoA gene copies were not significantly correlated with ammonium concentrations (r = 0.16, P > 0.05) but were correlated with nitrate plus nitrite concentrations (r = 0.78, P < 0.01) and negatively correlated with prokaryotic production (r = −0.55, P < 0.05).
The 16S rRNA genes from Crenarchaea within marine group I (Table 4) were also more abundant during the winter than in the summer (P < 0.01, Student t test), following a pattern similar to that of archaeal amoA genes. Marine group I 16S rRNA genes per ng of DNA increased by nearly 200-fold from summer 2007 to winter 2008. The following year, 16S rRNA genes from this archaeal group increased only by ∼15-fold from summer 2008 to winter 2009. Marine archaea in the pSL12 group were not common in our samples, being represented by only 44 to 311 copies of rRNA genes per ng of DNA in winter. rRNA genes from the pSL12 group were higher during summer months, but nonspecific DNA amplification occurred in the summer samples. Cloning and sequencing revealed that the PCR products were not from 16S rRNA genes (data not shown). As a result, pSL12 data were excluded from all further analysis.
TABLE 4.
Abundance of Bacteria and Crenarchaea in the coastal Chukchi Sea and Beaufort Sea near Barrow, Alaskaa
| Sample period and location | No. of prokaryotes (103 cells/ml or 103 gene copies/ng) |
|||||
|---|---|---|---|---|---|---|
| Total |
Bacteria |
Marine group I |
||||
| Mean | SD | Mean | SD | Mean | SD | |
| Summer 2007 | ||||||
| Chuk-1 | 1,576 | 432 | 481 | 33 | 0.01 | 0.00 |
| Chuk-2 | 1,923 | 152 | 519 | 35 | 0.01 | 0.01 |
| Beau-1 | 2,088 | 302 | 482 | 66 | 0.02 | 0.01 |
| Beau-2 | 1,737 | 193 | 987 | 16 | 0.06 | 0.01 |
| Winter 2008 | ||||||
| Chuk-3 | 422 | 77 | 687 | 65 | 9.94 | 1.85 |
| Chuk-4 | 467 | 132 | 151 | 14 | 1.71 | 0.50 |
| Chuk-5 | 748 | 159 | 460 | 36 | 4.99 | 0.19 |
| Summer 2008 | ||||||
| Beau-3 | 1,066 | 268 | 538 | 11 | 0.05 | 0.01 |
| Beau-4 | 1,271 | 305 | 720 | 58 | 0.05 | 0.00 |
| Chuk-6 | 929 | 133 | 662 | 22 | 0.04 | 0.01 |
| Chuk-7 | 935 | 233 | 607 | 57 | 0.02 | 0.00 |
| Beau-5 | 1,183 | 417 | 425 | 17 | 0.01 | 0.00 |
| Beau-6 | 1,331 | 210 | 544 | 131 | 0.01 | 0.00 |
| Winter 2009 | ||||||
| Chuk-8 | 220 | 77 | 212 | 3 | 0.15 | 0.01 |
| Beau-7 | 524 | 106 | 409 | 21 | 0.58 | 0.07 |
| Beau-8 | 568 | 156 | 399 | 50 | 0.53 | 0.04 |
| Beau-9 | 475 | 55 | 366 | 31 | 0.56 | 0.07 |
Estimates of total prokaryote abundance are based on microscopically enumerated direct counts. All other estimates are based on qPCR quantification of 16S rRNA genes. Chuk, Chukchi Sea; Beau, Beaufort Sea.
Ammonia oxidizers made up a larger proportion of the prokaryotic population in winter than in summer, but in neither season was the percentage high. Total amoA genes were 0.04% of total 16S rRNA genes in the summer versus 3.8% in winter. Copies of archaeal marine group I 16S rRNA genes were 0.6% of total bacterial 16S rRNA genes during the winter but were less than 0.01% during the summer (Table 4).
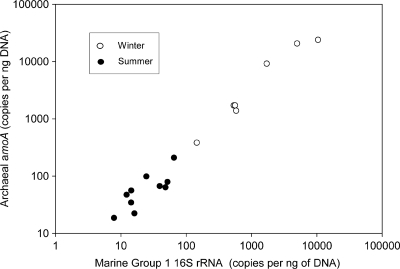
The ratio of archaea amoA genes to Marine group I 16S rRNA genes averaged 2.87 over all samples (Fig. 4). The amoA/16S rRNA ratio for archaea was similar in both seasons, averaging 2.57 ± 1.10 in the summer and 3.31 ± 1.09 in the winter, which is not significantly different (P > 0.05, Student t test). Archaea amoA gene copies correlated with marine group I 16S rRNA copies when data from all locations and seasons were examined (r = 0.96, P < 0.01).
FIG. 4.
Ratio of archaea amoA copies to marine group I copies of the 16S rRNA gene. The 16S rRNA gene data do not include archaea in the pSL12 group.
Potential nitrification rates and amoA abundance.
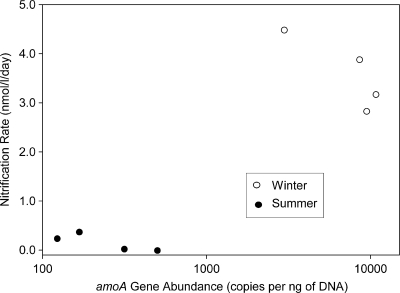
Potential nitrification rates were >20-fold higher in the winter than in the summer (Fig. 5). Rates from four locations averaged 0.15 nmol liter−1 day−1 in summer 2008. Rates the following winter (2009), again from four locations, ranged from 2.82 to 4.48 nmol liter−1 day−1, with a mean of 3.59 nmol liter−1 day−1. The total amoA gene copies from the same locations averaged 300 copies per ng of DNA in summer 2008 and nearly 8,000 in winter 2009. When potential nitrification rates are plotted against total betaproteobacterial and archaeal amoA (Fig. 5), separate winter and summer groups appear. Potential nitrification rates correlate with total amoA copies per ng of DNA (r = 0.74, P < 0 0.05, n = 8). The correlation between rates and amoA copies is similar for betaproteobacterial amoA (r = 0.75, P < 0.05) and archaeal amoA (r = 0.70, P > 0.05) when analyzed separately.
FIG. 5.
Potential nitrification rate versus amoA gene copies. Estimates of gene copies include both betaproteobacterial and archaeal amoA.
DISCUSSION
The coastal Arctic Ocean is ice-covered during the winter, and the sun stays below the horizon for 2 months each year. Photosynthesis by phytoplankton in the water column or algae in sea ice can fuel food webs during the sunlit months of summer, but photosynthesis is drastically reduced during the fall and then ceases altogether during the dark, winter months. Chemoautotrophy, such as ammonia oxidation, may become important during the winter when light-driven primary production and competition from phytoplankton is low. The objectives of the present study were to examine ammonia-oxidizing prokaryotes, in particular their abundance and activity in the summer versus in the winter, and to relate these data to environmental parameters such as ammonium concentrations. Our hypothesis was that ammonia oxidizers would be more abundant and active in the winter when competition from primary producers for ammonium was at a minimum.
Our samples were taken from nearshore surface waters of the Arctic Ocean on both the Chukchi Sea and Beaufort Sea sides of Barrow Point. Summer and winter environments in these waters differed in physical properties, prokaryotic abundance and production, and chlorophyll a and nutrient concentrations. Low winter primary production was reflected in lower prokaryotic abundance and production. Abundance declined only 3-fold on average from summer to winter, which is much less than the corresponding drop in prokaryotic production of almost 7-fold. However, potential nitrification rates increased by 24-fold, implying a greater role for chemoautotrophy during the winter. In Monterey Bay, nitrification rates from all depths varied only 4.5-fold in bimonthly measurements over 2 years (59); ammonium concentrations were always low, with a maximum of only 0.08 μM, which is much lower than the concentrations we observed. The highest potential nitrification rates in the coastal Arctic in the winter (3.6 nmol liter−1 day−1) were lower than rates (20 to 35 nmol liter−1 day−1) in Monterey Bay surface waters (44) and in the Gulf of California (3) but were similar to rates (1 to 10 nmol liter−1 day−1) in the oligotrophic North Atlantic Ocean (8). Low water temperatures explain why nitrification in the coastal Arctic Ocean is similar to rates in the oligotrophic North Atlantic Ocean, in spite of high ammonium concentrations in the Arctic.
Partial bacterial and crenarchaeal amoA gene sequences from our Arctic samples were similar to sequences found in other environments. These Arctic amoA sequences were closely related to sequences found in the central Arctic Ocean (27), but they were also similar to sequences from several low latitude regions (28, 36, 41, 44), including sediments (4, 17, 42). Gammaproteobacterial amoA was not detected in these coastal waters, and yet the total gammaproteobacterial abundance determined by fluorescence in situ hybridization comprised between 8 and 21% of total prokaryotes in western Arctic waters, while the total betaproteobacterial abundance ranged from 1 to 6.5% of total prokaryotes (1, 15, 30). Archaeal amoA genes from both shallow-water group A and deep-water group B appeared in our clone libraries, but the abundance of group B was low, below the qPCR detection limits.
The summer and winter communities of betaproteobacterial ammonia oxidizers were significantly different. One of the winter betaproteobacterial amoA clades (winter group 1) formed an OTU with other polar sequences at >97% nucleotide identity; however, sequences from more temperate regions enter this group if the cutoff is relaxed to >95% similarity. Winter sequences differed from summer sequences by 30 to 50 nucleotides. However, differences in amino acid sequences were much lower, suggesting that while there are distinct summer and winter populations of ammonia-oxidizing Betaproteobacteria, the proteins encoded by amoA genes may be functionally equivalent.
The abundance of both betaproteobacterial and archaeal ammonia oxidizers, as well as potential nitrification rates, were higher in the winter than in the summer, probably because of higher ammonium concentrations, the lack of competition with phytoplankton and other microbes for ammonium, and no inhibition by light in the winter. This hypothesis is supported by results from previous studies. Crenarchaeal abundance as a percentage of total prokaryote abundance correlated with ammonium concentrations in the Chukchi Sea and the Canadian Basin of the Arctic Ocean (31) and in the North Sea (24). The negative relationship between archaea and phytoplankton has also been observed in nearshore Antarctic waters (43). Similar to our observations, potential nitrification rates were correlated with both crenarchaeal 16S rRNA copies and amoA gene copies in the Gulf of California (3). However, potential nitrification rates and the ratio of potential nitrification rates to amoA gene copies were 10- and 50-fold higher, respectively, in the Gulf of California than in the coastal Arctic Ocean during winter. The difference in rates reflects overall lower microbial activity in these polar waters due to lower water temperatures and rates of primary production, but it remains unclear why the ratio of nitrification to amoA gene copies is lower in the Arctic.
Betaproteobacterial ammonia oxidizers were more abundant in the coastal Arctic than has been observed previously in marine waters (3, 27, 40), although betaproteobacterial amoA also outnumbered archaeal amoA in much of the San Francisco Bay (42). In the Central Arctic Ocean, archaeal amoA was more than 900 times more abundant than betaproteobacterial amoA (27), a much larger difference than we observed even in winter 2008 when archaeal amoA were most abundant. Ammonium concentrations may explain these results. Concentrations of ammonium in the coastal waters we examined ranged from 0.3 μM in summer to nearly 5 μM in winter, much higher than observed in other marine studies of amoA gene abundance (3, 35), except for the estuarine studies (2, 17). The ammonium concentrations were 2.5 times higher during the winter when betaproteobacterial amoA was more abundant than the prior year when crenarchaeal amoA was most abundant. Crenarchaeal ammonia oxidizers may be better adapted to oligotrophic environments such as the central Arctic, since the half-saturation constant (Km) for ammonia oxidation by Nitrosopumilus maritimus SCM1 is lower than that found in other microbes (39). Some crenarchaeal ammonia oxidizers may even be partially inhibited by high ammonium concentrations (23).
Estimates of the amoA to 16S rRNA gene ratio suggest that all oceanic Crenarchaea are ammonia oxidizers. If so, it would help explain why we and others observed higher crenarchaeal abundance in the winter of polar waters (1, 7, 30, 43). In Arctic coastal waters of the Beaufort Sea and Chukchi Sea, the ratio of archaeal amoA genes to marine group I 16S rRNA genes averaged 2.87 over all samples. This ratio is similar to that found in other environments (24, 35, 60) but much lower than the ratio of eight observed in Canadian Arctic waters (19). A ratio greater than one suggests that that some marine Crenarchaea possess multiple copies of the amoA gene as do some bacteria (25, 32). Further work is necessary to determine whether the amoA to 16S rRNA gene ratio is affected by problems with qPCR primers (34, 40). The primers used here are the same as those in used in other studies.
The abundance of betaproteobacterial and archaeal ammonia oxidizers and nitrification rates in the coastal Arctic Ocean are driven by the interactions between competition with phytoplankton for ammonium, light inhibition, and fluxes of ammonium from sediments. All of these factors lead to nitrification being seasonally uncoupled from primary production and explain the seasonal differences in abundance and rates we observed in these coastal Arctic waters. Ammonium concentrations alone may be sufficient to explain why bacterial ammonia oxidizers were unexpectedly more abundant than observed in other marine waters during the winter. Most, if not all, marine group I Crenarchaea appear capable of ammonia oxidation in these waters, which would explain why they are most abundant during the winter in these Arctic coastal waters and in other polar regions.
Supplementary Material
Acknowledgments
This study was supported by U.S. National Science Foundation grants OPP0632233 and NSF-OCE0824997 (to J. M. Beman and B.N.P.).
We thank Barbara Campbell and Liying Yu for technical assistance and collaboration and Glenn Sheehan and Lewis Brower for logistical support at the Barrow Arctic Science Consortium.
This is SOEST contribution number 8080.
Footnotes
Published ahead of print on 14 January 2011.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Alonso-Sáez, L., O. Sánchez, J. M. Gasol, V. Balagué, and C. Pedrós-Alio. 2008. Winter-to-summer changes in the composition and single-cell activity of near-surface Arctic prokaryotes. Environ. Microbiol. 10:2444-2454. [DOI] [PubMed] [Google Scholar]
- 2.Beman, J. M., and C. A. Francis. 2006. Diversity of ammonia-oxidizing archaea and bacteria in the sediments of a hypernutrified subtropical estuary: Bahia del Tobari, Mexico. Appl. Environ. Microbiol. 72:7767-7777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beman, J. M., B. N. Popp, and C. A. Francis. 2008. Molecular and biogeochemical evidence for ammonia oxidation by marine Crenarchaeota in the Gulf of California. ISME J. 2:429-441. [DOI] [PubMed] [Google Scholar]
- 4.Bernhard, A. E., T. Donn, A. E. Giblin, and D. A. Stahl. 2005. Loss of diversity of ammonia-oxidizing bacteria correlates with increasing salinity in an estuary system. Environ. Microbiol. 7:1289-1297. [DOI] [PubMed] [Google Scholar]
- 5.Bernhard, A. E., et al. 2010. Abundance of ammonia-oxidizing archaea and bacteria along an estuarine salinity gradient in relation to potential nitrification rates. Appl. Environ. Microbiol. 76:1285-1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brochier-Armanet, C., B. Boussau, S. Gribaldo, and P. Forterre. 2008. Mesophilic crenarchaeota: proposal for a third archaeal phylum, the Thaumarchaeota. Nat. Rev. Microb. 6:245-252. [DOI] [PubMed] [Google Scholar]
- 7.Church, M. J., et al. 2003. Abundance and distribution of planktonic Archaea and Bacteria in the waters west of the Antarctic Peninsula. Limnol. Oceanogr. 48:1893-1902. [Google Scholar]
- 8.Clark, D. R., A. P. Rees, and I. Joint. 2008. Ammonium regeneration and nitrification rates in the oligotrophic Atlantic Ocean: implications for new production estimates. Limnol. Oceanogr. 53:52-62. [Google Scholar]
- 9.Cottrell, M. T., and D. L. Kirchman. 2003. Contribution of major bacterial groups to bacterial biomass production (thymidine and leucine incorporation) in the Delaware Estuary. Limnol. Oceanogr. 48:168-178. [Google Scholar]
- 10.Cottrell, M. T., and D. L. Kirchman. 2009. Photoheterotrophic microbes in the Arctic Ocean in summer and winter. Appl. Environ. Microbiol. 75:4958-4966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De Corte, D., T. Yokokawa, M. M. Varela, H. Agogue, and G. J. Herndl. 2009. Spatial distribution of Bacteria and Archaea and amoA gene copy numbers throughout the water column of the Eastern Mediterranean Sea. ISME J. 3:147-158. [DOI] [PubMed] [Google Scholar]
- 12.Dempster, E. L., K. V. Pryor, D. Francis, J. E. Young, and H. J. Rogers. 1999. Rapid DNA extraction from ferns for PCR-based analyses. Biotechniques 27:66-68. [DOI] [PubMed] [Google Scholar]
- 13.Dore, J. E., and D. M. Karl. 1996. Nitrification in the euphotic zone as a source for nitrite, nitrate, and nitrous oxide at Station ALOHA. Limnol. Oceanogr. 41:1619-1628. [Google Scholar]
- 14.Dore, J. E., B. N. Popp, D. M. Karl, and F. J. Sansone. 1998. A large source of atmospheric nitrous oxide from subtropical North Pacific surface waters. Nature 396:63-66. [Google Scholar]
- 15.Elifantz, H., A. I. Dittel, M. T. Cottrell, and D. L. Kirchman. 2007. Dissolved organic matter assimilation by heterotrophic bacterial groups in the western Arctic Ocean. Aquat. Microb. Ecol. 50:39-49. [Google Scholar]
- 16.Erguder, T. H., N. Boon, L. Wittebolle, M. Marzorati, and W. Verstraete. 2009. Environmental factors shaping the ecological niches of ammonia-oxidizing archaea. FEMS Microbiol. Rev. 33:855-869. [DOI] [PubMed] [Google Scholar]
- 17.Francis, C. A., G. D. O'Mullan, and B. B. Ward. 2003. Diversity of ammonia monooxygenase (amoA) genes across environmental gradients in Chesapeake Bay sediments. Geobiology 1:129-140. [Google Scholar]
- 18.Francis, C. A., K. J. Roberts, J. M. Beman, A. E. Santoro, and B. B. Oakley. 2005. Ubiquity and diversity of ammonia-oxidizing archaea in water columns and sediments of the ocean. Proc. Natl. Acad. Sci. U. S. A. 102:14683-14688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Galand, P. E., et al. 2009. Archaeal diversity and a gene for ammonia oxidation are coupled to oceanic circulation. Environ. Microbiol. 11:971-980. [DOI] [PubMed] [Google Scholar]
- 20.Guerrero, M. A., and R. D. Jones. 1996. Photoinhibition of marine nitrifying bacteria. 1. Wavelength-dependent response. Mar. Ecol. Prog. Ser. 141:183-192. [Google Scholar]
- 21.Hall, T. A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41:95-98. [Google Scholar]
- 22.Hallam, S. J., et al. 2006. Pathways of carbon assimilation and ammonia oxidation suggested by environmental genomic analyses of marine crenarchaeota. PLoS Biol. 4:520-536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hatzenpichler, R., et al. 2008. A moderately thermophilic ammonia-oxidizing crenarchaeote from a hot spring. Proc. Natl. Acad. Sci. U. S. A. 105:2134-2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Herfort, L., et al. 2007. Variations in spatial and temporal distribution of Archaea in the North Sea in relation to environmental variables. FEMS Microbiol. Ecol. 62:242-257. [DOI] [PubMed] [Google Scholar]
- 25.Hommes, N. G., L. A. Sayavedra-Soto, and D. J. Arp. 2001. Transcript analysis of multiple copies of amo (encoding ammonia monooxygenase) and hao (encoding hydroxylamine oxidoreductase) in Nitrosomonas europaea. J. Bacteriol. 183:1096-1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jones, R. D., R. Y. Morita, H. P. Koops, and S. W. Watson. 1988. A new marine ammonium-oxidizing bacterium, Nitrosomonas cryotolerans sp. nov. Can. J. Microbiol. 34:1122-1128. [Google Scholar]
- 27.Kalanetra, K. M., N. Bano, and J. T. Hollibaugh. 2009. Ammonia-oxidizing Archaea in the Arctic Ocean and Antarctic coastal waters. Environ. Microbiol. 11:2434-2445. [DOI] [PubMed] [Google Scholar]
- 28.Kim, O.-S., P. Junier, J. F. Imhoff, and K.-P. Witzel. 2008. Comparative analysis of ammonia monooxygenase (amoA) genes in the water column and sediment-water interface of two lakes and the Baltic Sea. FEMS Microbiol. Ecol. 66:367-378. [DOI] [PubMed] [Google Scholar]
- 29.Kirchman, D. L. 2001. Measuring bacterial biomass production and growth rates from leucine incorporation in natural aquatic environments, p. 227-237. In J. H. Paul (ed.), Marine microbiology, vol. 30. Academic Press, Inc., San Diego, CA. [Google Scholar]
- 30.Kirchman, D. L., M. T. Cottrell, and C. Lovejoy. 2010. The structure of bacterial communities in the western Arctic Ocean as revealed by pyrosequencing of 16S rRNA genes. Environ. Microbiol. 12:1132-1143. [DOI] [PubMed] [Google Scholar]
- 31.Kirchman, D. L., H. Elifantz, A. I. Dittel, R. R. Malmstrom, and M. T. Cottrell. 2007. Standing stocks and activity of archaea and bacteria in the western Arctic Ocean. Limnol. Oceanogr. 52:495-507. [Google Scholar]
- 32.Klotz, M. G., and J. M. Norton. 1998. Multiple copies of ammonia monooxygenase (amo) operons have evolved under biased AT/GC mutational pressure in ammonia-oxidizing autotrophic bacteria. FEMS Microbiol. Lett. 168:303-311. [DOI] [PubMed] [Google Scholar]
- 33.Könneke, M., et al. 2005. Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature 437:543-546. [DOI] [PubMed] [Google Scholar]
- 34.Konstantinidis, K. T., J. Braff, D. M. Karl, and E. F. DeLong. 2009. Comparative metagenomic analysis of a microbial community residing at a depth of 4,000 meters at Station ALOHA in the North Pacific Subtropical Gyre. Appl. Environ. Microbiol. 75:5345-5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lam, P., et al. 2007. Linking crenarchaeal and bacterial nitrification to anammox in the Black Sea. Proc. Natl. Acad. Sci. U. S. A. 104:7104-7109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lam, P., et al. 2009. Revising the nitrogen cycle in the Peruvian oxygen minimum zone. Proc. Natl. Acad. Sci. U. S. A. 106:4752-4757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee, D. H., Y. G. Zo, and S. J. Kim. 1996. Nonradioactive method to study genetic profiles of natural bacterial communities by PCR-single-strand-conformation polymorphism. Appl. Environ. Microbiol. 62:3112-3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Leininger, S., et al. 2006. Archaea predominate among ammonia-oxidizing prokaryotes in soils. Nature 442:806-809. [DOI] [PubMed] [Google Scholar]
- 39.Martens-Habbena, W., P. M. Berube, H. Urakawa, J. R. de la Torre, and D. A. Stahl. 2009. Ammonia oxidation kinetics determine niche separation of nitrifying Archaea and Bacteria. Nature 461:976-979. [DOI] [PubMed] [Google Scholar]
- 40.Mincer, T. J., et al. 2007. Quantitative distribution of presumptive archaeal and bacterial nitrifiers in Monterey Bay and the North Pacific Subtropical Gyre. Environ. Microbiol. 9:1162-1175. [DOI] [PubMed] [Google Scholar]
- 41.Molina, V., et al. 2007. Ammonia-oxidizing beta-Proteobacteria from the oxygen minimum zone off northern Chile. Appl. Environ. Microbiol. 73:3547-3555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mosier, A. C., and C. A. Francis. 2008. Relative abundance and diversity of ammonia-oxidizing archaea and bacteria in the San Francisco Bay estuary. Environ. Microbiol. 10:3002-3016. [DOI] [PubMed] [Google Scholar]
- 43.Murray, A. E., et al. 1998. Seasonal and spatial variability of bacterial and archaeal assemblages in the coastal waters near Anvers Island, Antarctica. Appl. Environ. Microbiol. 64:2585-2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.O'Mullan, G. D., and B. B. Ward. 2005. Relationship of temporal and spatial variabilities of ammonia-oxidizing bacteria to nitrification rates in Monterey Bay, California. Appl. Environ. Microbiol. 71:697-705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Popp, B. N., F. J. Sansone, T. M. Rust, and D. A. Merritt. 1995. Determination of concentration and carbon isotopic composition of dissolved methane in sediments and nearshore waters. Anal. Chem. 67:405-411. [Google Scholar]
- 46.Purkhold, U., et al. 2000. Phylogeny of all recognized species of ammonia oxidizers based on comparative 16S rRNA and amoA sequence analysis: implications for molecular diversity surveys. Appl. Environ. Microbiol. 66:5368-5382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rotthauwe, J. H., K. P. Witzel, and W. Liesack. 1997. The ammonia monooxygenase structural gene amoA as a functional marker: molecular fine-scale analysis of natural ammonia-oxidizing populations. Appl. Environ. Microbiol. 63:4704-4712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Santoro, A. E., K. L. Casciotti, and C. A. Francis. 2010. Activity, abundance, and diversity of nitrifying archaea and bacteria in the central California Current. Environ. Microbiol. doi: 10.1111/j.1462-2920.2010.02205.x. [DOI] [PubMed]
- 49.Schleper, C., G. Jurgens, and M. Jonuscheit. 2005. Genomic studies of uncultivated archaea. Nat. Rev. Microb. 3:479-488. [DOI] [PubMed] [Google Scholar]
- 50.Schloss, P. D., and J. Handelsman. 2005. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl. Environ. Microbiol. 71:1501-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schloss, P. D., B. R. Larget, and J. Handelsman. 2004. Integration of microbial ecology and statistics: a test to compare gene libraries. Appl. Environ. Microbiol. 70:5485-5492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sigman, D. M., et al. 2001. A bacterial method for the nitrogen isotopic analysis of nitrate in seawater and freshwater. Anal. Chem. 73:4145-4153. [DOI] [PubMed] [Google Scholar]
- 53.Stephen, J. R., et al. 1999. Effect of toxic metals on indigenous soil beta-subgroup proteobacterium ammonia oxidizer community structure and protection against toxicity by inoculated metal-resistant bacteria. Appl. Environ. Microbiol. 65:95-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596-1599. [DOI] [PubMed] [Google Scholar]
- 55.Venter, J. C., et al. 2004. Environmental genome shotgun sequencing of the Sargasso Sea. Science 304:66-74. [DOI] [PubMed] [Google Scholar]
- 56.Walker, C. B., et al. 2010. Nitrosopumilus maritimus genome reveals unique mechanisms for nitrification and autotrophy in globally distributed marine crenarchaea. Proc. Natl. Acad. Sci. U. S. A. 107:8818-8823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ward, B. B. 2000. Nitrification and the marine nitrogen cycle, p. 427-453. In D. L. Kirchman (ed.), Microbial ecology of the oceans. Wiley-Liss, New York, NY.
- 58.Ward, B. B. 1987. Nitrogen transformations in the Southern California Bight. Deep Sea Res. Int. 34:785-805. [Google Scholar]
- 59.Ward, B. B. 2005. Temporal variability in nitrification rates and related biogeochemical factors in Monterey Bay, California, U.S.A. Mar. Ecol. Prog. Ser. 292:97-109. [Google Scholar]
- 60.Wuchter, C., et al. 2006. Archaeal nitrification in the ocean. Proc. Natl. Acad. Sci. U. S. A. 103:12317-12322. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.