Abstract
WHO type III nasopharyngeal carcinoma (NPC) is highly prevalent in Indonesia and 100% associated with Epstein-Barr virus (EBV). NPC tumor cells express viral proteins, including BARF1, which is secreted and is considered to have oncogenic and immune-modulating properties. Recently, we found conserved mutations in the BARF1 gene in NPC isolates. This study describes the expression and purification of NPC-derived BARF1 and analyzes humoral immune responses against prototype BARF1 (B95-8) and purified native hexameric BARF1 in sera of Indonesian NPC patients (n = 155) compared to healthy EBV-positive (n = 56) and EBV-negative (n = 16) individuals. BARF1 (B95-8) expressed in Escherichia coli and baculovirus, as well as BARF1-derived peptides, did not react with IgG or IgA antibodies in NPC. Purified native hexameric BARF1 protein isolated from culture medium was used in enzyme-linked immunosorbent assay (ELISA) and revealed relatively weak IgG and IgA responses in human sera, although it had strong antibody responses to other EBV proteins. Higher IgG reactivity was found in NPC patients (P = 0.015) than in regional Indonesian controls or EBV-negative individuals (P < 0.001). IgA responses to native BARF1 were marginal. NPC sera with the highest IgG responses to hexameric BARF1 in ELISA showed detectable reactivity with denatured BARF1 by immunoblotting. In conclusion, BARF1 has low immunogenicity for humoral responses and requires native conformation for antibody binding. The presence of antibodies against native BARF1 in the blood of NPC patients provides evidence that the protein is expressed and secreted as a hexameric protein in NPC patients.
Epstein-Barr virus (EBV) is a human gammaherpesvirus with tropism for B lymphocytes and epithelial cells. EBV infection occurs worldwide, and about 90% of the world population is persistently infected. EBV is etiologically linked to several lymphoid and epithelial malignancies, with the latter including most undifferentiated and poorly differentiated nasopharyngeal carcinomas (NPC) (WHO types II and III, respectively) (16, 29) and about 10% of gastric adenocarcinomas (GC) worldwide (22, 32, 47). NPC has a well-defined geographical distribution and is particularly prevalent in Southeast Asia. Both genetic and dietary influences are thought to be important in NPC etiology (2, 48). NPC shows latency type II EBV transcription in all tumor cells, with expression of the noncoding small RNAs EBER1 and -2 (EBER1/2), BamHI A rightward transcripts (BARTs), and Epstein-Barr nuclear antigen 1 (EBNA1) (10, 46). Latent membrane protein 1 (LMP1) and LMP2 are more heterogeneously expressed (1, 10, 46, 49). Furthermore, transcription of an additional viral gene in BamHI-A rightward frame 1 (BARF1) was described previously (5, 34, 35, 50). BARF1 mRNA is exclusively expressed in EBV-positive carcinomas and is absent from EBV-positive lymphomas (12, 32, 43). However, it can be activated by switching on the viral lytic cycle (11, 27). Direct demonstration of BARF1 protein expression in carcinoma tissue has proven extremely difficult, although one report described its presence in NPC tumor extracts (5). Recently, it was shown that BARF1 lacking the first 20 amino acids is actively secreted (6, 7, 30), and BARF1 protein was detected in sera of NPC patients in amounts of 500 to 5,000 ng/ml, but not in healthy EBV carriers (13).
The functions assigned to BARF1 are diverse. BARF1 has been shown to have transforming activity and to prevent senescence (33, 44, 45) and apoptosis (3, 42). Secreted BARF1 protein (sBARF1) has been reported to have mitogenic activity on human B cells and primary monkey kidney epithelial cells (30). A possible role for sBARF1 as an immune-modulating protein was suggested, since Fc-tagged BARF1 protein was able to act as an antagonist for macrophage colony-stimulating factor (M-CSF) (4, 36). Parts of the BARF1 protein are homologous to the Ig superfamily of receptors, and a small domain has homology with the T cell receptor costimulatory molecule CD80 (4, 36, 38). However, the exact function of the secreted BARF1 protein in EBV-related carcinoma is still under investigation.
Nasopharyngeal carcinomas are characterized by a significant infiltrate of CD4+ and CD8+ T cells (15). Therefore, BARF1 is expected to trigger immune responses. Indeed, T cell responses against BARF1-derived peptides were recently detected in NPC patients, opening options for immune therapy (18). However, lymphocytes obtained from the NPC tumor environment are functionally impaired (17), suggesting local immune modulation, which may be linked to BARF1. Antibody-dependent cellular cytotoxicity against BARF1-transfected Raji cells using sera of NPC patients has been described (37). Unfortunately, the study does not agree with the more recent knowledge of rapid and complete secretion of BARF1. Aberrant EBV serology is commonly used to support NPC diagnosis and provides an affordable approach for population screening (8, 26). Antibodies targeting the BARF1 protein could be used as a new diagnostic tool to identify NPC patients.
In this study, we analyze the humoral immune response to BARF1 protein in a large group of Indonesian NPC patients. Here, we employ a BARF1 sequence found in NPC with 2 conservative mutations, V29A and H130R, which are not considered to affect the three-dimensional (3D) structure significantly (14). This protein was expressed in human 293 cells and purified as a hexameric protein. The use of “native” hexameric BARF1 protein proved to be essential in detecting anti-BARF1 antibody responses in NPC patients.
MATERIALS AND METHODS
Sera from NPC patients and healthy EBV carriers.
Serum panels from histologically confirmed NPC patients (n = 155) were collected at the Department of Ear, Nose, and Throat (ENT), Dr. Sardjito General Hospital, Yogyakarta, Indonesia. NPC sera were taken prior to treatment. NPC staging was done by ENT examination and computed tomography (CT) scan and classified according to the 1996 Union International Cancer Control (UICC) classification. Sera from EBV-positive healthy individuals (n = 56) were obtained from the Indonesian Red Cross blood bank, Yogyakarta, Indonesia, and the archives of the VU University Medical Center Department of Pathology, respectively, and have been extensively characterized (19, 20, 28, 39). EBV-negative sera (n = 16) were collected from healthy individuals in the Netherlands.
Plasmids.
The B95-8 prototype BARF1 sequence was obtained from JY cells by PCR and cloned into the pCRScript vector (Stratagene, Agilent Technologies, Amstelveen, Netherlands). Subsequently, the sBARF1 B95-8 sequence (amino acid [aa] 21 to aa 221) was cloned into the pHTA donor vector, adding a 6-His tag C terminally (BARF1-His vector). In addition, the sBARF1 sequence (aa 21 to aa 221) was cloned into the pGEX 4T-3 vector (Amersham Biosciences, GE Healthcare, Diegem, Belgium), adding a glutathione S-transferase (GST) tag (BARF1-GST vector). A consensus BARF1 DNA sequence from an Indonesian NPC patient was obtained from S. Hutajulu (14). This BARF1 sequence differs from B95-8 at positions 29 (valine to alanine) and 130 (histidine to arginine) and was used for construction of the BARF1 expression vector pcDNA-BARF1. The amplification product was cloned into a pcDNA4/TO vector (Invitrogen, Breda, Netherlands) (referred to below as native sBARF1).
Cell culture.
293HEK cells were cultured in Dulbecco's modified Eagle's medium (DMEM); an EBV-negative NPC-derived cell line, Hone1, was cultured in RPMI medium (Lonza, Breda, Netherlands); and the gastric cancer cell line AGS was cultured in HAM's nutrient mixture F12 (Sigma-Aldrich, Zwijndrecht, Netherlands) at 37°C and 5% CO2 in a humidified atmosphere. All media contained 10% fetal calf serum (FCS), 100 units/ml penicillin (PEN), 100 μg/ml streptomycin (STR), 2 mM l-glutamine, and 50 μM β-mercaptoethanol. After stable transfection with pcDNA-BARF1, 293HEK-BARF1 cells were cultured under selection with 95 μg/ml phleomycin (Zeocin). Spodoptera frugiperda (Sf9) insect cells were cultured in Sf-900 II SFM with l-glutamine medium (Invitrogen) at 27°C.
BARF1 protein expression and purification.
Recombinant baculovirus (BARF1-His vector) was produced as described previously (21). BARF1-GST was generated by inducing pGEX-BARF1-transformed Escherichia coli with 10 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 4 h and purified with glutathione-Sepharose (Pharmacia, GE Healthcare) as described by the manufacturer. NPC-derived human-secreted BARF1 (native-sBARF1) protein was produced by stable transfection of 293HEK cells with the pcDNA-BARF1 vector using Lipofectamine (Invitrogen). For protein production, cells were washed twice with phosphate-buffered saline (PBS), and serum-free medium was added for 24 h, after which the medium was harvested. Medium (300 ml) was diluted 1:1 in binding buffer (20 mM Tris, pH 7.4, 0.5 M NaCl) and incubated for 1 h with 10 ml concanavalin A bead slurry (Sigma-Aldrich). After being washed, BARF1 was eluted with 500 mM methyl-alpha-d-glucopyranoside in binding buffer. Multiple elutions were concentrated to 1 ml with a Centriprep 50 device (Millipore, Amsterdam, Netherlands) and loaded onto a 25-ml Superdex-200 Sepharose size exclusion column (Pharmacia, GE Healthcare). Fractions containing sBARF1 were pooled and dialyzed against PBS overnight at +4°C. The protein concentration was determined by using a BCA Protein Assay kit (Pierce, Thermo Fisher Scientific, Etten Leur, Netherlands).
Synthetic peptides.
Antigenic peptide epitopes of sBARF1 were derived by computer prediction techniques, as described previously (24, 25, 40), using high scores for hydrophilicity, flexibility, and β-turn probability. Peptides named BARF1 N peptide (aa 40 to 79), M peptide (aa 151 to 188), and C peptide (aa 187 to 221) (Fig. 1A) were synthesized with a peptide synthesizer (433A; Applied Biosystems, Nieuwerkerk a/d IJssel, Netherlands) using 9-fluorenylmethoxy carbonyl (Fmoc) amino acids purchased from Bachem. Peptides were purified by reverse-phase high-performance liquid chromatography.
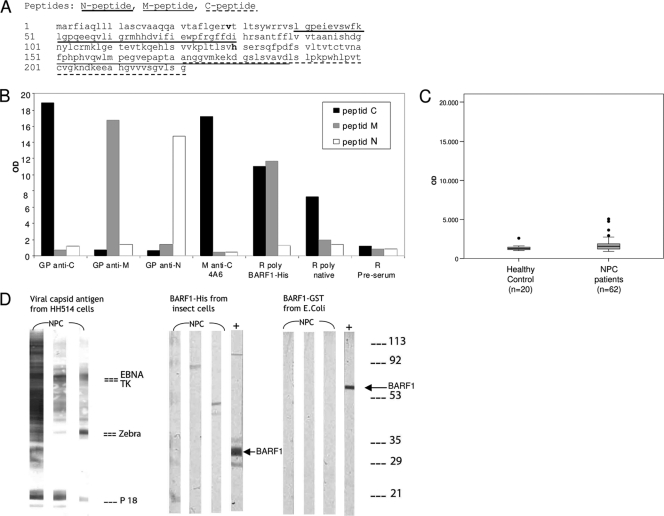
FIG. 1.
BARF1 (peptide) sequence and specific animal antibody reactivity against B95-8-derived recombinant BARF1 protein and antigenic peptides. (A) BARF1 sequence containing 2 common amino acid substitutions, V29A and H130R, found in Indonesian isolates (boldface). Antigenic peptides designed by computer analysis, named N peptide (aa 40 to 79), M peptide (aa 151 to 188), and C peptide (aa 187 to 221), are underlined. (B) Antibody reactivity to peptides measured in ELISA. Guinea pig antibodies against C peptide, M peptide, N peptide, and mouse monoclonal anti-C peptide (4A6) specifically react to their target peptides. The rabbit polyclonal antibody anti-BARF1-His (K150.3) binds to both C and M peptides. Rabbit polyclonal antibody raised against the purified native sBARF1 reacts to the C peptide. (C) Antibody reactivity of healthy individuals and NPC patients to the BARF1 C peptide measured in IgG ELISA, showing slightly elevated levels in NPC patients. (D) IgG immunoblot analysis of 3 representative NPC sera with BARF1-His expressed by the Sf9 insect cell expression system and BARF1-GST expressed by E. coli. HH514-derived viral capsid antigens were used as a positive control for EBV antibody presence in patient sera. Guinea pig anti-C peptide antibody (+) showed a clear band on the blot strips, but no BARF1 reactivity was detected in the patient sera despite the presence of diverse antibody responses to other EBV proteins, as visualized on the HH514 strips.
Monoclonal and polyclonal antibodies.
Monoclonal antibodies (MAb) 4A6 and 6F4 were produced by immunization of mice with synthetic BARF1 C peptide. Polyclonal antibody (PAb) K150.3 was produced by immunization of rabbits with purified recombinant BARF1-His protein and administered in Freund's adjuvant. Guinea pig antibodies specific for defined domains of BARF1 (GPαC, GPαM, and GPαN) were prepared by immunizing animals with synthetic BARF1 peptides coupled with keyhole limpet hemocyanin and administered in Freund's adjuvant. Rabbit polyclonal antibodies to native BARF1 (PAb nativeBARF1) was produced by immunization of a rabbit with purified native sBARF1 from 293HEK-BARF1 cells and administered in Immacel-R adjuvant (Pickcell Laboratories BV, Amsterdam, Netherlands).
SDS-PAGE and Western blot analysis.
Cell samples were lysed in PBS containing 1% Triton X-100 and protease inhibitor cocktail (Roche, Almere, Netherlands) and sonified. Cell debris was removed by centrifugation, and the protein concentration was determined using a BCA Protein Assay kit. For sBARF1, the medium was harvested, and debris was removed by centrifugation. For reduced SDS-PAGE, samples were diluted in 2× loading buffer (Bio-Rad, Veenendaal, Netherlands) with β-mercaptoethanol and heated for 5 min at 95°C. For nonreduced SDS-PAGE, samples were diluted in 2× nonreducing loading buffer (Bio-Rad) and incubated for 10 min at room temperature (RT). The samples were run on a 12.5% SDS-PAGE gel and either transferred to a Hybond ECL nitrocellulose membrane (GE Healthcare) or Coomassie stained with Instant Blue (Expedeon, Westburg, Leusden, Netherlands). After being blotted, the membrane was blocked in PBS containing 3% nonfat dry milk for 1 h at RT. Incubation with 4A6 primary antibody (1:100) in PBS containing 0.05% Tween 20 (PBST) with 5% bovine serum albumin (BSA) was followed by IRDye800CW-conjugated secondary antibodies (1:1,000) (Li-Cor, Westburg, Netherlands) in PBST containing 3% milk. Nitrocellulose blots were visualized using the Odyssey Infrared Imaging System (Li-Cor).
Glycosylation and phosphorylation analysis.
Denaturing buffer (1 μl) was added to 9 μl of sBARF1-containing medium and placed at 95°C for 10 min. After cooling, PNGase-F (NEB, Westburg, Netherlands), O-glycosidase, or neuraminidase (Sigma-Aldrich) and matching buffer were added and incubated for 90 min at 37°C. For blocking BARF1 secretion, cells were exposed for 24 h to either 3 μg/ml brefeldin A or 50 μM monensin (Sigma-Aldrich). Phosphorylation of BARF1 was analyzed by immunoblotting using anti-phosphoserine (1:200; Sigma-Aldrich) and anti-phosphothreonine (1:1,000; Cell Signaling Technology, Bioké, Leiden, Netherlands) antibodies.
EBV immunoblotting.
Viral capsid antigen (VCA) was produced in HH514, and EBV-VCA immunoblot strips were made as described previously (23). For detection of EBV antibodies, blot strips were incubated with human sera (1:100). After 3 washes with PBST, the secondary antibody rabbit anti-human IgG (1:1,000; Dako, Glostrup, Denmark) was added. After three washing steps in PBST followed by PBS, bands were visualized either using 0.06% (wt/vol) 4-chloronaphthol (Sigma-Aldrich) and 0.01% (vol/vol) H2O2 in PBS or with ECL+ detection reagent (Amersham).
Anti-BARF1 ELISA.
Individual wells of Costar-9018 high-binding 96-well enzyme-linked immunosorbent assay (ELISA) plates (Corning, VWR, Amsterdam, Netherlands) were coated with 1 μg/ml BARF1 peptide or 10 μg/ml purified sBARF1 protein in 0.05 M NaHCO3, pH 9.6, overnight at 4°C, followed by blocking with 3% BSA in PBS for 1 h at 37°C. Serum samples were tested in duplicate; diluted 1:100 in PBS containing 0.05% Tween, 0.1% Triton X-100, 1% BSA (PBS-TTB); and incubated for 1 h at 37°C. After 4 washes with PBST, the plate was incubated with rabbit anti-human IgG (1:8,000) or IgA (1:4,000) in PBS-TTB and incubated for 1 h at 37°C. After 4 washes with PBST, color was developed using TMB substrate (bioMérieux, Boxtel, Netherlands), and the optical density at 450 nm (OD450) was measured.
Data analysis.
The Mann-Whitney test was performed with the SPSS statistical program, and P values under 0.05 were considered significant. SPSS was also used to calculate the Pearson correlation coefficient.
RESULTS
Lack of antibody response against nonnative B95-8-derived recombinant BARF1 protein and BARF1 peptides.
Since BARF1 mRNA and BARF1 protein are abundantly expressed in NPC patients (35, 50), antibody reactivity to BARF1 was evaluated as a potential new diagnostic marker. In a first approach to study BARF1-specific antibody reactivity in NPC patients by ELISA, BARF1-derived peptides with predicted antigenic domains were selected (Fig. 1A). Antibodies were generated against the three selected synthetic peptides, as well as against a purified full-length baculovirus-encoded BARF1-His fusion protein expressed in insect cells. The antibodies reacted specifically in ELISA to their respective peptides (Fig. 1B), and all reacted with extracts of BARF1-expressing insect cells by immunocytochemical staining and immunoblot analysis (data not shown). Antibodies raised against full-length BARF1-His fusion protein displayed a stronger reaction to the C peptide. However, none of the human sera displayed reactivity to any of the 3 peptides used, despite having abundant antibody reactivity with a wide range of EBV proteins and EBV peptide sequences (8, 9, 28). Only BARF1 C peptide generated a minor IgG response in 9.5% (4/42) of the NPC patient sera (Fig. 1C).
In order to investigate whether human antibodies might be reactive against other determinants on BARF1, purified full-length BARF1-GST (E. coli) and BARF1-His (insect cells) were used in immunoblot analysis to probe for reactivity in sera of NPC patients (n = 149). EBV reactivity of sera on immunoblot strips with EBV VCA protein extracts revealed antibody responses to multiple EBV proteins, i.e., EBNA1, EAd-TK, EAd-p47/54, ZEBRA, and VCA-p18, as shown in Fig. 1D (9). Whereas BARF1-specific polyclonal control antibodies gave a clear staining pattern on the strips, no specific anti-BARF1 response was observed in any of the human serum samples analyzed using either Sf9 insect cell BARF1-His, E. coli-produced BARF1-GST, or the selected peptide antigens.
BARF1 peptides and BARF1-His or BARF1-GST proteins produced in nonhuman expression systems may not have the proper folding and conformation. Therefore, we subsequently focused on the use of an NPC-derived BARF1 sequence produced in a human cell background.
NPC-derived sBARF1 is secreted as a glycosylated hexameric protein from human cells.
Sequence analysis of the BARF1 gene revealed two prevalent amino acid substitutions (V29A and H130R) in Indonesian NPC patients, which were predicted to be conservative in nature, leading to only minor changes in protein folding (14). A stable NPC-derived BARF1-expressing 293HEK cell line was created. The expressed BARF1 protein was rapidly and completely secreted into the culture medium and was detected as a 27- to 29-kDa band under denaturing conditions by SDS-PAGE and Western blot analysis (Fig. 2A), in agreement with previously reported data (7, 31). Under nonreducing conditions, sBARF1 produced by 293HEK cells revealed a 160- to 180-kDa molecule representing the hexameric form of BARF1 (Fig. 2A). Similar to the 293HEK system, BARF1 expressed transiently in Hone1 (NPC) and AGS (gastric carcinoma) cells was rapidly and completely secreted from the cells (data not shown). Earlier, BARF1 was described as a glycosylated protein (7, 38). To analyze the glycosylation status of secreted BARF1 protein (sBARF1), selective glycosidase analysis was performed. Specific enzymes were used to remove O-linked sugar chains, N-linked sugar chains, and sialic end groups of sBARF1 protein. PNGase F cleavage created a major band shift (Fig. 2B), indicating that sBARF1 contained high-mannose N-linked glycosylation. Neuraminidase was used to cut off sialic end groups from sugars, which resulted in a minor band shift (Fig. 2B). O-Glycosidase can cut off a sugar group only when incubated together with neuraminidase. This combination demonstrates the presence of sialic acid end groups, as well as O-linked glycosylation, in the BARF1 protein (Fig. 2B). We conclude that HEK293-expressed sBARF1 is an N-linked glycosylated protein with additional O-linked glycosylic modifications carrying sialic end groups.
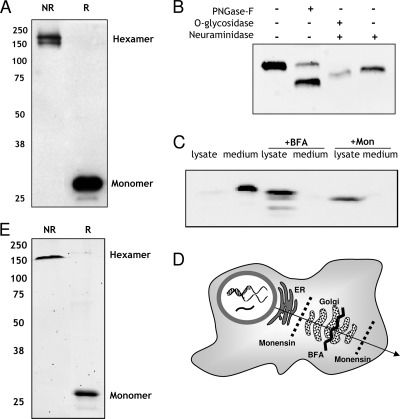
FIG. 2.
BARF1 is secreted as a hexameric glycosylated protein from human HEK293 cells. (A) BARF1 is secreted as a hexamer in the culture supernatant of stably transfected 293HEK-BARF1 cells. Ten microliters of medium was mixed 1:1 with either reduced (R) or nonreduced (NR) loading buffer and separated by SDS-PAGE and Western blotting. A 29-kDa and a 160-kDa band can be seen, representing the monomeric and the hexameric structures of sBARF1, respectively. (B) The glycosylation status of BARF1 in serum-free culture supernatant was determined by direct treatment with several glycosidases. PNGase F cleaves high-mannose N-linked sugar groups. Neuraminidase removes sialic acid groups, and O-glycosidase cleaves O-linked sugar chains. The digested protein was loaded on an SDS-PAGE Western blot and detected using 4A6 anti-BARF1 antibody. (C) Manipulation of BARF1 secretion from HEK293 cells. Under normal growth conditions, hardly any BARF1 can be detected in 293HEK cell lysates; BARF1 is completely secreted in the culture supernatant. Treatment of cells with the transport blockers monensin (Mon) and brefeldin A (BFA) retains sBARF1 in the cell lysate and almost completely blocks secretion into the culture medium. (D) Schematic overview of intercellular transport blockage by brefeldin A and monensin. (E) Native-sBARF1 was purified from culture medium using ConA affinity chromatography followed by size exclusion chromatography and visualized by Coomassie staining as hexameric sBARF1 (NR) and monomeric sBARF1 (R). Purity of ∼98% was routinely reached in repeated experiments.
Phosphorylation of secreted BARF1 protein was examined with anti-phosphothreonine and anti-phosphoserine monoclonal antibodies. Although a band was detected with BARF1-specific antibody (4A6), no band was observed with phosphospecific antibodies (data not shown).
BARF1 protein is rapidly and completely secreted, and therefore, hardly any BARF1 was detected in 293 cell lysate (Fig. 2C). To determine whether BARF1 is indeed secreted via the classical pathway, specific blockers were used. Blockage of BARF1 protein passage at specific stages in the secretory pathway was performed with brefeldin A, which disturbs the Golgi apparatus, and monensin, which halts glycoproteins in the endoplasmic reticulum (ER) (Fig. 2D). Both reagents prevented BARF1 secretion into the medium and allowed protein accumulation inside BARF1-expressing cells (Fig. 2C). Furthermore, brefeldin A and monensin resulted in BARF1 bands at lower molecular weight than BARF1 secreted in medium (Fig. 2C), which can be explained by reduced glycosylation status.
Low antibody responses against native BARF1 exist in NPC patients.
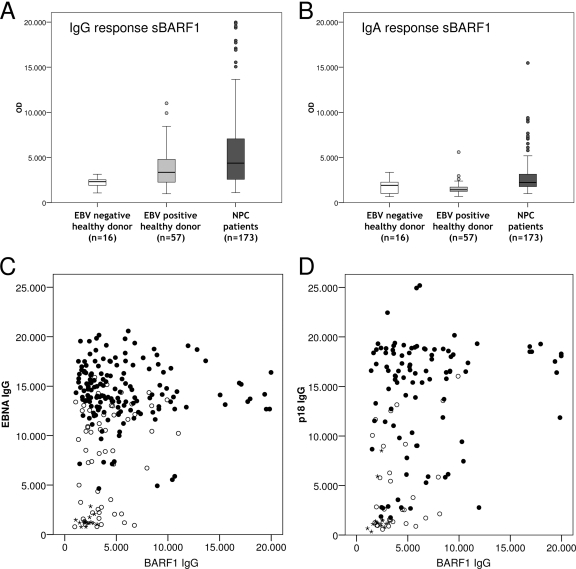
HEK293-produced native sBARF1 protein containing proper glycosylation and hexameric folding was purified with concanavalin A (ConA) and size exclusion chromatography and used to develop an antibody capture ELISA. The purity of the hexameric native sBARF1 was over 98%, as was shown by Coomassie staining (Fig. 2E). Pure native sBARF1 protein was used to develop an antibody capture ELISA. Optimization was performed with anti-BARF1 antibodies and controls (preimmune sera and non-EBV MAbs, as shown in Fig. 1B). Subsequently, sera from Indonesian NPC patients (n = 155) and healthy EBV-positive (n = 55) and EBV-negative (n = 16) Caucasian and Indonesian individuals were tested. A weak but clear IgG response against native sBARF1 was observed in NPC patients, which showed significantly higher reactivity than regional controls (P = 0.015) and EBV-negative individuals (P < 0.001) (Fig. 3A). Healthy Indonesian and Dutch EBV carriers had similar responses. To analyze whether anti-BARF1 responses were related to the level of humoral immunity to other immunodominant EBV proteins, we analyzed anti-VCA and anti-EBNA1 antibody responses in individual NPC patients. The results shown in Fig. 3B and C reveal no correlation between these responses, indicating that anti-BARF1 antibodies are formed independently and at a relatively low level. Although in many NPC patients anti-BARF1 IgA was barely detectable, overall, NPC patients had significantly increased IgA levels against sBARF1 compared to the controls, albeit less pronounced than IgG responses (Fig. 3A and D). Healthy EBV carriers were virtually devoid of anti-BARF1 responses, as shown in Fig. 3A. The proportion of patients with positive IgG-BARF1 test results who were correctly diagnosed with NPC by pathological examination (positive predictive value) was 100%, but due to the limited immunogenicity of BARF1 in vivo, many true NPC patients lack detectable anti-BARF1 responses, thus yielding a negative predictive value of 28%.
FIG. 3.
Low immune responses against native BARF1 in human sera. (A) IgG antibodies against native human sBARF1 detected by antibody capture ELISA. Data are shown as OD450 values for EBV-negative healthy donors (n = 16), EBV-positive healthy donors (n = 56), and NPC patients (n = 155). (B) IgA antibodies against native human sBARF1 were detected by ELISA using EBV-negative healthy donors (n = 15), EBV-positive healthy donors (n = 57), and NPC patients (n = 155). (C) Scatter plot of IgG anti-BARF1 versus IgG anti-EBNA responses. (D) Scatter plot of IgG anti-BARF1 versus IgG anti-VCAp18 responses. Filled circles, NPC patients; open circles, EBV-positive healthy donors; stars, EBV-negative healthy donors.
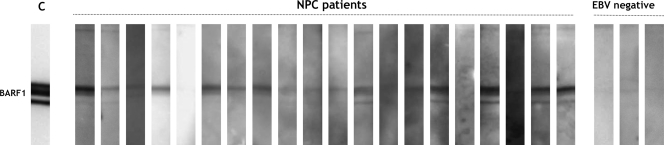
The specificity of the response against native sBARF1 was analyzed by using immunoblot analysis. Immunoblot strips containing purified sBARF1 were incubated with sera from the strongest BARF1 responders in ELISA. A thin BARF1 band was visible for some sera. Only with considerable overexposure was a BARF1 band visualized with most of the sera (Fig. 4), confirming the low-immunogenic nature of BARF1 protein. The staining intensities of the sera on immunoblots did not correlate with OD450 values in antibody capture ELISA, suggesting alternative epitopes may be detected by both techniques (data not shown).
FIG. 4.
Immunoblot confirmation of specific antibody responses to native sBARF1. Native sBARF1 was loaded on a reduced SDS-PAGE gel, blotted to nitrocellulose, and cut into strips. Sera from the highest responders in the NPC patient panel and control sera were analyzed at 1:100 dilution. 4A6 anti-BARF1 antibody was used as a positive control (C). A band at the correct height was detected with most of the selected NPC patient sera only upon considerable overexposure (minutes) when using ECL visualization. The 4A6 signal was revealed in a few seconds.
DISCUSSION
Since transcription of the EBV BARF1 gene was found to be selectively associated with EBV-related malignancies of epithelial origin, substantial research has been done to determine its function and potential role in EBV-driven carcinogenesis. Recently, BARF1 was considered a diagnostic marker, since it was reported to be actively secreted from BARF1-expressing cells in model systems (6, 7). Secreted BARF1 protein was also detected in sera of NPC patients (13). However, the abundance of BARF1 protein in patient sera, as suggested by Houali et al., remains to be confirmed. The expression and secretion of BARF1 in human carcinoma suggest that BARF1 might trigger antibody responses. Using NPC sequence sBARF1 from a human expression system, we showed in this study the presence of anti-BARF1 antibodies in NPC patients and healthy EBV-positive individuals. Despite the use of native NPC-derived sBARF1, the anti-BARF1 antibody response remained low.
Initially, a recombinant B95-8 sequence-derived BARF1 fusion protein expressed in bacterial and insect cell systems did not reveal any reactivity with human sera. Even in a sensitive ELISA, predicted antigenic peptides did not yield detectable anti-BARF1 responses despite excellent results for other EBV markers in our prior work (8, 24, 40, 41). Peptide C is the only BARF1 domain that appeared to generate a minor humoral immune response, as well as for the developed BARF1 monoclonal and polyclonal antibodies. These results favor the C terminus of BARF1 as the most immunogenic BARF1 domain.
Recently, Tarbouriech et al. provided direct evidence that the native form of secreted BARF1 was a glycosylated hexameric protein (38). Since insect cells and bacteria differ from human cells in folding and posttranslational modifications, the need arose for a human expression system of recombinant BARF1 protein, better reflecting the natural form. Sequence analysis of the EBV BARF1 gene of NPC patients revealed two amino acid changes compared to the generally used B95-8 BARF1 sequence (14). These amino acid changes are located at surface-accessible sites on the native BARF1 protein and are suggested to yield only marginal changes in BARF1 protein folding compared to the B95-8 prototype but may affect antibody binding to native BARF1 (14, 38). This NPC-based BARF1 sequence was used for producing a human recombinant BARF1 (HEK293-BARF1). HEK293-BARF1 cells rapidly secrete the sBARF1 protein in the culture medium as a hexameric protein (6, 32). The secreted BARF1 protein is N-linked and O-linked glycosylated, the latter also containing sialic acid modification at the termini. We were not able to confirm phosphorylation of sBARF1 protein, as was suggested previously (6).
Using purified native sBARF1 protein in an antibody capture ELISA, we detected IgG antibody responses against BARF1 in some healthy EBV carriers and most NPC patients. NPC patients responded significantly more strongly to BARF1 than healthy controls (P = 0.015). The presence of a weak response in EBV carriers can be explained by transcription of BARF1 during the lytic cycle in normal EBV infection. The IgA response to sBARF1 was low and difficult to detect.
Our data reveal BARF1 as a marginally immunogenic protein in patients with otherwise strong responses to alternative EBV proteins, such as VCA-p18, EBNA1, and Zebra. The observation that anti-BARF1 antibodies were mainly directed against conformational epitopes on purified native BARF1 protein suggests that BARF1 may circulate in vivo as a free hexameric protein, possibly interacting with M-CSF to modulate immune responses, an interaction that is only minimally interfered with by humoral immune responses.
Acknowledgments
This project was financially supported by the Dutch Cancer Society Project VU2007-3776 and the Nuffic/Netherlands Fellowship Program (NFP-PhD 08/36).
We are grateful to Liesbeth Cairo, Floortje Krechting, and Bo van Kuijk for their technical assistance.
We declare no conflict of interest.
Footnotes
Published ahead of print on 1 December 2010.
REFERENCES
- 1.Brooks, L., Q. Y. Yao, A. B. Rickinson, and L. S. Young. 1992. Epstein-Barr virus latent gene transcription in nasopharyngeal carcinoma cells: coexpression of EBNA1, LMP1, and LMP2 transcripts. J. Virol. 66:2689-2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chang, E. T., and H. O. Adami. 2006. The enigmatic epidemiology of nasopharyngeal carcinoma. Cancer Epidemiol. Biomarkers Prev. 15:1765-1777. [DOI] [PubMed] [Google Scholar]
- 3.Chang, M. S., et al. 2005. Cell-cycle regulators, bcl-2 and NF-kappaB in Epstein-Barr virus-positive gastric carcinomas. Int. J. Oncol. 27:1265-1272. [PubMed] [Google Scholar]
- 4.Cohen, J. I., and K. Lekstrom. 1999. Epstein-Barr virus BARF1 protein is dispensable for B-cell transformation and inhibits alpha interferon secretion from mononuclear cells. J. Virol. 73:7627-7632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Decaussin, G., F. Sbih-Lammali, M. de Turenne-Tessier, A. Bouguermouh, and T. Ooka. 2000. Expression of BARF1 gene encoded by Epstein-Barr virus in nasopharyngeal carcinoma biopsies. Cancer Res. 60:5584-5588. [PubMed] [Google Scholar]
- 6.de Turenne-Tessier, M., P. Jolicoeur, and T. Ooka. 1997. Expression of the protein encoded by Epstein-Barr virus (EBV) BARF1 open reading frame from a recombinant adenovirus system. Virus Res. 52:73-85. [DOI] [PubMed] [Google Scholar]
- 7.de Turenne-Tessier, M., and T. Ooka. 2007. Post-translational modifications of Epstein Barr virus BARF1 oncogene-encoded polypeptide. J. Gen. Virol. 88:2656-2661. [DOI] [PubMed] [Google Scholar]
- 8.Fachiroh, J., et al. 2006. Single-assay combination of Epstein-Barr virus (EBV) E. J. Clin. Microbiol. 44:1459-1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fachiroh, J., et al. 2004. Molecular diversity of Epstein-Barr virus IgG and IgA antibody responses in nasopharyngeal carcinoma: a comparison of Indonesian, Chinese, and European subjects. J. Infect. Dis. 190:53-62. [DOI] [PubMed] [Google Scholar]
- 10.Fåhraeus, R., et al. 1988. Expression of Epstein-Barr virus-encoded proteins in nasopharyngeal carcinoma. Int. J. Cancer 42:329-338. [DOI] [PubMed] [Google Scholar]
- 11.Hatfull, G., A. T. Bankier, B. G. Barrell, and P. J. Farrell. 1988. Sequence analysis of Raji Epstein-Barr virus DNA. Virology 164:334-340. [DOI] [PubMed] [Google Scholar]
- 12.Hayes, D. P., et al. 1999. Expression of Epstein-Barr virus (EBV) transcripts encoding homologues to important human proteins in diverse EBV associated diseases. Mol. Pathol. 52:97-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Houali, K., et al. 2007. A new diagnostic marker for secreted Epstein-Barr virus encoded LMP1 and BARF1 oncoproteins in the serum and saliva of patients with nasopharyngeal carcinoma. Clin. Cancer Res. 13:4993-5000. [DOI] [PubMed] [Google Scholar]
- 14.Hutajulu, S. H., et al. 2010. Conserved mutation of Epstein-Barr virus-encoded BamHI-A Rightward Frame-1 (BARF1) gene in Indonesian nasopharyngeal carcinoma. Infect. Agents Cancer 5:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jayasurya, A., B. H. Bay, W. M. Yap, and N. G. Tan. 2000. Lymphocytic infiltration in undifferentiated nasopharyngeal cancer. Arch. Otolaryngol. Head Neck Surg. 126:1329-1332. [DOI] [PubMed] [Google Scholar]
- 16.Klein, G., et al. 1974. Direct evidence for the presence of Epstein-Barr virus DNA and nuclear antigen in malignant epithelial cells from patients with poorly differentiated carcinoma of the nasopharynx. Proc. Natl. Acad. Sci. U. S. A. 71:4737-4741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li, J., et al. 2007. Functional inactivation of EBV-specific T-lymphocytes in nasopharyngeal carcinoma: implications for tumor immunotherapy. PLoS One 2:e1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martorelli, D., et al. 2008. Spontaneous T cell responses to Epstein-Barr virus-encoded BARF1 protein and derived peptides in patients with nasopharyngeal carcinoma: bases for improved immunotherapy. Int. J. Cancer 123:1100-1107. [DOI] [PubMed] [Google Scholar]
- 19.Meij, P., et al. 2002. Identification and prevalence of CD8(+) T-cell responses directed against Epstein-Barr virus-encoded latent membrane protein 1 and latent membrane protein 2. Int. J. Cancer 99:93-99. [DOI] [PubMed] [Google Scholar]
- 20.Meij, P., et al. 1999. Restricted low-level human antibody responses against Epstein-Barr virus (EBV)-encoded latent membrane protein 1 in a subgroup of patients with EBV-associated diseases. J. Infect. Dis. 179:1108-1115. [DOI] [PubMed] [Google Scholar]
- 21.Meij, P., M. B. Vervoort, C. J. Meijer, E. Bloemena, and J. M. Middeldorp. 2000. Production monitoring and purification of EBV encoded latent membrane protein 1 expressed and secreted by recombinant baculovirus infected insect cells. J. Virol. Methods 90:193-204. [DOI] [PubMed] [Google Scholar]
- 22.Middeldorp, J. M., A. A. Brink, A. J. van den Brule, and C. J. Meijer. 2003. Pathogenic roles for Epstein-Barr virus (EBV) gene products in EBV-associated proliferative disorders. Crit. Rev. Oncol. Hematol. 45:1-36. [DOI] [PubMed] [Google Scholar]
- 23.Middeldorp, J. M., and P. Herbrink. 1988. Epstein-Barr virus specific marker molecules for early diagnosis of infectious mononucleosis. J. Virol. Methods 21:133-146. [DOI] [PubMed] [Google Scholar]
- 24.Middeldorp, J. M., and R. H. Meloen. 1988. Epitope-mapping on the Epstein-Barr virus major capsid protein using systematic synthesis of overlapping oligopeptides. J. Virol. Methods 21:147-159. [DOI] [PubMed] [Google Scholar]
- 25.Modrow, S., et al. 1987. Computer-assisted analysis of envelope protein sequences of seven human immunodeficiency virus isolates: prediction of antigenic epitopes in conserved and variable regions. J. Virol. 61:570-578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ng, M. H., K. H. Chan, S. P. Ng, and Y. S. Zong. 2006. Epstein-Barr virus serology in early detection and screening of nasopharyngeal carcinoma. Ai Zheng 25:250-256. [PubMed] [Google Scholar]
- 27.Ooka, T. 2005. Biological role of the BARF1 gene encoded by Epstein-Barr virus, p. 613. In E. S. Robertson (ed.), Epstein-Barr virus. Caister Academic Press, Wymondham, England.
- 28.Paramita, D. K., et al. 2007. Native early antigen of Epstein-Barr virus, a promising antigen for diagnosis of nasopharyngeal carcinoma. J. Med. Virol. 79:1710-1721. [DOI] [PubMed] [Google Scholar]
- 29.Raab-Traub, N. 2002. Epstein-Barr virus in the pathogenesis of NPC. Semin. Cancer Biol. 12:431-441. [DOI] [PubMed] [Google Scholar]
- 30.Sall, A., et al. 2004. Mitogenic activity of Epstein-Barr virus-encoded BARF1 protein. Oncogene 23:4938-4944. [DOI] [PubMed] [Google Scholar]
- 31.Seto, E., T. Ooka, J. Middeldorp, and K. Takada. 2008. Reconstitution of nasopharyngeal carcinoma-type EBV infection induces tumorigenicity. Cancer Res. 68:1030-1036. [DOI] [PubMed] [Google Scholar]
- 32.Seto, E., et al. 2005. Epstein-Barr virus (EBV)-encoded BARF1 gene is expressed in nasopharyngeal carcinoma and EBV-associated gastric carcinoma tissues in the absence of lytic gene expression. J. Med. Virol. 76:82-88. [DOI] [PubMed] [Google Scholar]
- 33.Sheng, W., G. Decaussin, A. Ligout, K. Takada, and T. Ooka. 2003. Malignant transformation of Epstein-Barr virus-negative Akata cells by introduction of the BARF1 gene carried by Epstein-Barr virus. J. Virol. 77:3859-3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stevens, S. J., et al. 2005. Diagnostic value of measuring Epstein-Barr virus (EBV) DNA load and carcinoma-specific viral mRNA in relation to anti-EBV immunoglobulin A (IgA) and IgG antibody levels in blood of nasopharyngeal carcinoma patients from Indonesia. J. Clin. Microbiol. 43:3066-3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stevens, S. J., et al. 2006. Noninvasive diagnosis of nasopharyngeal carcinoma: nasopharyngeal brushings reveal high Epstein-Barr virus DNA load and carcinoma-specific viral BARF1 mRNA. Int. J. Cancer 119:608-614. [DOI] [PubMed] [Google Scholar]
- 36.Strockbine, L. D., et al. 1998. The Epstein-Barr virus BARF1 gene encodes a novel, soluble colony-stimulating factor-1 receptor. J. Virol. 72:4015-4021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tanner, J. E., et al. 1997. Antibody and antibody-dependent cellular cytotoxicity responses against the BamHI A rightward open-reading frame-1 protein of Epstein-Barr virus (EBV) in EBV-associated disorders. J. Infect. Dis. 175:38-46. [DOI] [PubMed] [Google Scholar]
- 38.Tarbouriech, N., F. Ruggiero, M. de Turenne-Tessier, T. Ooka, and W. P. Burmeister. 2006. Structure of the Epstein-Barr virus oncogene BARF1. J. Mol. Biol. 359:667-678. [DOI] [PubMed] [Google Scholar]
- 39.van Grunsven, W. M., A. Nabbe, and J. M. Middeldorp. 1993. Identification and molecular characterization of two diagnostically relevant marker proteins of the Epstein-Barr virus capsid antigen complex. J. Med. Virol. 40:161-169. [DOI] [PubMed] [Google Scholar]
- 40.van Grunsven, W. M., W. J. Spaan, and J. M. Middeldorp. 1994. Localization and diagnostic application of immunodominant domains of the BFRF3-encoded Epstein-Barr virus capsid protein. J. Infect. Dis. 170:13-19. [DOI] [PubMed] [Google Scholar]
- 41.Verschuuren, E. A., et al. 2002. Treatment of posttransplant lymphoproliferative disease with rituximab: the remission, the relapse, and the complication. Transplantation 73:100-104. [DOI] [PubMed] [Google Scholar]
- 42.Wang, Q., et al. 2006. Anti-apoptotic role of BARF1 in gastric cancer cells. Cancer Lett. 238:90-103. [DOI] [PubMed] [Google Scholar]
- 43.Wang, Y., B. Luo, P. Zhao, and B. H. Huang. 2004. Expression of Epstein-Barr virus genes in EBV-associated gastric carcinoma. Ai Zheng 23:782-787. [PubMed] [Google Scholar]
- 44.Wei, M. X., M. de Turenne-Tessier, G. Decaussin, G. Benet, and T. Ooka. 1997. Establishment of a monkey kidney epithelial cell line with the BARF1 open reading frame from Epstein-Barr virus. Oncogene 14:3073-3081. [DOI] [PubMed] [Google Scholar]
- 45.Wei, M. X., J. C. Moulin, G. Decaussin, F. Berger, and T. Ooka. 1994. Expression and tumorigenicity of the Epstein-Barr virus BARF1 gene in human Louckes B-lymphocyte cell line. Cancer Res. 54:1843-1848. [PubMed] [Google Scholar]
- 46.Young, L. S., et al. 1988. Epstein-Barr virus gene expression in nasopharyngeal carcinoma. J. Gen. Virol. 69:1051-1065. [DOI] [PubMed] [Google Scholar]
- 47.Young, L. S., and A. B. Rickinson. 2004. Epstein-Barr virus: 40 years on. Nat. Rev. Cancer 4:757-768. [DOI] [PubMed] [Google Scholar]
- 48.Yu, M. C., J. H. Ho, S. H. Lai, and B. E. Henderson. 1986. Cantonese-style salted fish as a cause of nasopharyngeal carcinoma: report of a case-control study in Hong Kong. Cancer Res. 46:956-961. [PubMed] [Google Scholar]
- 49.Zheng, X., L. Hu, F. Chen, and B. Christensson. 1994. Expression of Ki67 antigen, epidermal growth factor receptor and Epstein-Barr virus-encoded latent membrane protein (LMP1) in nasopharyngeal carcinoma. Eur. J. Cancer B Oral Oncol. 30B:290-295. [DOI] [PubMed] [Google Scholar]
- 50.zur Hausen. A., et al. 2000. Unique transcription pattern of Epstein-Barr virus (EBV) in EBV-carrying gastric adenocarcinomas: expression of the transforming BARF1 gene. Cancer Res. 60:2745-2748. [PubMed] [Google Scholar]