Abstract
Equine arteritis virus (EAV) replicase consists of two polyproteins (pp1a and pp1ab) that are encoded by open reading frames (ORFs) 1a and 1b of the viral genome. These two replicase polyproteins are posttranslationally processed by three ORF 1a-encoded proteinases to yield at least 13 nonstructural proteins (nsp1 to nsp12, including nsp7α and 7β). These nsps are expressed in EAV-infected cells, but the equine immune response they induce has not been studied. Therefore, the primary purpose of this study was to evaluate the humoral immune response of horses to each of the nsps following EAV infection. Individual nsp coding regions were cloned and expressed in both mammalian and bacterial expression systems. Each recombinant protein was used in an immunoprecipitation assay with equine serum samples from horses (n = 3) that were experimentally infected with three different EAV strains (VB, KY77, and KY84), from stallions (n = 4) that were persistently infected with EAV, and from horses (n = 4) that were vaccinated with the modified live-virus (MLV) vaccine strain. Subsequently, protein-antibody complexes were subjected to Western immunoblotting analysis with individual nsp-specific rabbit antisera, mouse anti-His antibody, or anti-FLAG tag antibody. Nsp2, nsp4, nsp5, and nsp12 were immunoprecipitated by most of the sera from experimentally or persistently infected horses, while sera from vaccinated horses did not react with nsp5 and reacted weakly with nsp4. However, serum samples from vaccinated horses were able to immunoprecipitate nsp2 and nsp12 proteins consistently. Information from this study will assist ongoing efforts to develop improved methods for the serologic diagnosis of EAV infection in horses.
Equine arteritis virus (EAV) is the causative agent of equine viral arteritis (EVA), a respiratory and reproductive disease of horses (51). EAV is a small (approximately 40 to 60 nm in diameter) enveloped virus with a positive-sense, single-stranded RNA genome of ∼12.7 kb and belongs to the family Arteriviridae (genus Arterivirus, order Nidovirales), which also includes porcine reproductive and respiratory syndrome virus (PRRSV), simian hemorrhagic fever virus, and lactate dehydrogenase-elevating virus of mice (14, 46). The EAV genome includes nine known functional open reading frames (ORFs 1a, 1b, and 2 to 7) (45, 46). ORFs 1a and 1b are located in the 5′-proximal three-quarters of the genome and are translated to produce replicase polyproteins pp1a and pp1ab (1,727 and 3,175 amino acids, respectively), with the latter a C-terminally extended version of the former (36). ORF 1b translation depends on a −1 ribosomal frameshift just before termination of ORF 1a translation (21). The two replicase precursor polyproteins are cleaved by three different ORF 1a-encoded proteinases (see below) yielding at least 13 end products named nonstructural proteins 1 to 12 (nsp1 to nsp12), including the recently described nsp7α and nsp7β (54). The three EAV proteinase domains are located in nsp1, nsp2, and nsp4 (46, 63). The remaining seven ORFs (2a, 2b, and 3 to 7) are located in the 3′ quarter of the genome and encode the structural proteins (GP2, E, GP3, GP4, GP5, M, and N, respectively) of the virus.
EAV infection in horses induces long-lasting immunity that protects against reinfection with all strains of the virus (6, 13, 23, 34, 35). Resistance to reinfection is assumed to be mediated by neutralizing antibodies directed against glycoprotein 5 (GP5), the major envelope protein of the virus (4, 6-9, 11, 15, 22, 24, 59). The serum neutralization test, which principally detects antibodies to GP5, remains the most sensitive assay to detect EAV-specific antibodies in horse serum. The antibody responses of horses to individual EAV proteins differ markedly depending on the interval after infection, the infecting virus strain, the individual horse, and the specific serological assay used (6). Until now, the characterization of the humoral antibody response of a horse to EAV has been mainly based on the structural proteins of the virus (16-19, 25-27, 29-31, 33, 38, 50, 58). The serologic response of horses to the individual structural proteins of EAV has been extensively characterized by Western immunoblotting, enzyme-linked immunosorbent assay (ELISA), competitive ELISA (cELISA), and microsphere immunoassay (MIA) (Luminex) using recombinant GP5 and membrane (M) proteins and the nucleocapsid (N) protein (16-19, 25-27, 29-31, 33, 38, 50, 58). Immunoblotting studies confirmed that infected horses respond to a number of viral structural proteins and that sera from horses other than carrier stallions most consistently recognized the conserved carboxy-terminal region of the M protein (33). However, little is known about the equine humoral immune response to the nonstructural proteins of EAV. The nsps of EAV are the first viral proteins synthesized in cells infected with EAV and are essential to the viral replication cycle (47, 57). Thus, it is reasonable to predict that the antibodies directed against nsps might appear early in the course of EAV infection based on their levels of abundance and immunogenicity. Therefore, it is hypothesized that a distinct antibody response, similar to the immune response to structural proteins of EAV, may be generated to some of the nsps. The current study aimed to determine the humoral immune response of EAV-infected horses to each of the nsps encoded by the ORF 1a and ORF 1b region of the viral genome (the proteolytic cleavage products derived from pp1a and pp1ab). Specifically, in the present study, the recombinant nsps expressed in mammalian cells and E. coli were utilized in combined immunoprecipitation and Western immunoblotting analyses to establish the specificity of the antibody responses of EAV-infected or vaccinated horses to the nsps of EAV.
MATERIALS AND METHODS
Cells.
High-passage (passage 399 [P399] to P409) rabbit kidney 13 (KY RK-13) and baby hamster kidney 21 (BHK-21 [ATCC CCL-10]; P61 to P80) cells were cultured and maintained in Eagle's minimum essential medium (EMEM) (Mediatech, Herndon, VA) supplemented with 10% fetal calf serum (FCS) (HyClone, Logan, UT), 100 IU/ml penicillin, 100 μg/ml streptomycin, 1 μg/ml amphotericin B, and 0.06% sodium bicarbonate at 37°C.
Antibodies.
Monoclonal antibodies specific for nsp1 of EAV (12A4) have been previously described (58). Similarly, monospecific polyclonal rabbit antisera recognizing EAV nsp2 (48), nsp3 (rabbit 98.E3 [43]), nsp4 (a 1:1 mix of anti-nsp4M and anti-nsp4C [48]), nsp7 and nsp8 (48), and nsp10 (56) have been previously described. In addition, we used previously unpublished antisera against nsp9 and nsp11, both of which were raised by immunizing rabbits with full-length expression products purified from Escherichia coli (J. C. Zevenhoven, D. D. Nedialkova, and E. J. Snijder, unpublished data). Commercially available anti-FLAG (Agilent Technologies, Santa Clara, CA) and anti-His (Invitrogen, Carlsbad, CA) monoclonal antibodies were used to detect FLAG- and His-tagged fusion proteins in Western immunoblotting analyses, respectively.
Equine sera.
Sera from 11 horses that were seropositive for antibodies to EAV by virus neutralization assay were used to characterize the equine humoral immune response to the EAV nsps (Table 1). The panel consisted of three serum samples from horses that were experimentally inoculated with the virulent Bucyrus (VB) strain or the KY77 and KY84 strains of EAV, four serum samples from stallions confirmed to be persistently infected carriers of EAV (stallions D, E, G, and R) (11, 28, 42), and four serum samples from horses vaccinated with the modified live-virus vaccine strain of EAV (ARVAC; Fort Dodge Animal Health Laboratories [now Pfizer Animal Health Inc., New York, NY]). Two equine serum samples negative for neutralizing antibodies to EAV were included as controls.
TABLE 1.
Serologic responses of horses to EAV nsps following experimental infection with VB, KY77, and KY84 strains of EAV, persistent infection, and vaccination
| Horse no. | Breeda | Status/time | VN titer | Protein immunoprecipitationb |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| nsp1 | nsp2 | nsp3 | nsp4 | nsp5 | nsp7 | nsp8 | nsp9 | nsp10 | nsp11 | nsp12 | ||||
| Experimentally infected horses | ||||||||||||||
| 1462 (VB) | TB | 2 mo postinfection | ≥1:1,024 | − | + | − | +w | + | − | − | − | − | − | + |
| 77E853 (KY77) | TB | 2 mo postinfection | 1:1,024 | − | + | − | + | + | − | − | − | − | − | +* |
| 198/199 (KY84) | TB | 42 days postinfection | 1:1,024 | − | + | − | + | + | − | − | − | − | − | +w |
| Persistently infected stallions | ||||||||||||||
| D | TB | Carrier | 1:512 | − | − | − | +w | +w | − | − | − | − | − | +w |
| E | TB | Carrier | 1:256 | − | + | − | + | + | − | − | − | − | − | + |
| R | DWB | Carrier | ≥1:512 | − | + | − | + | +w | − | − | − | − | − | +w |
| G | STB | Carrier | 1:256 | − | + | − | + | − | − | − | − | − | − | +w |
| Vaccinated horses | ||||||||||||||
| SR-10258 (no. 800) | TB | ≥1:512 | − | +w | − | +w | − | − | − | − | − | − | + | |
| 94-593 | TB | 128 days postvaccinion | 1:512 | − | +w | − | +w | − | − | − | − | − | − | + |
| 508 | STB | 8 mo postvaccinion | 1:512 | − | +w | − | +w | − | − | − | − | − | − | + |
| 478 | STB | 8 mo postvaccinion | 1:128 | − | +w | − | +w | − | − | − | − | − | − | + |
TB, Thoroughbred; DWB, Dutch Warmblood; STB, Standardbred.
−, not immunoprecipitated; +, immunoprecipitated; +w,immunoprecipitated protein signal was weaker than those of other positive sera in Western immunoblotting analysis; +*, proteins expressed in both mammalian and bacterial cells were immunoprecipitated with equine sera.
Virus neutralization (VN) test.
The neutralizing antibody titers of the test sera were determined as described by the World Animal Health Organization (OIE) and Senne et al. (39, 44). Briefly, serial 2-fold dilutions of each sample from 1:4 to 1:512 were made in MEM (Invitrogen, Carlsbad, CA) containing 10% guinea pig complement (Rockland Immunochemicals, Gilbertsville, PA). Each serum sample was tested in duplicate in 96-well plates. An equal volume of a virus dilution containing an estimated 200 50% tissue culture infective doses (TCID50) of the modified live-virus vaccine strain of EAV (ARVAC; Fort Dodge Animal Health) was added to each well, except the serum controls. The plates were shaken to ensure mixing of the well contents and then incubated for 1 h at 37°C. A suspension of high-passage (P399 to P409) RK-13 cells was added to each well in a volume double that of the serum-virus mixtures, and the plates were incubated for 72 h at 37°C until viral cytopathic effect had fully developed in the virus control wells. The titer of a sample was recorded as the reciprocal of the highest serum dilution that provided at least 50% neutralization of the reference virus.
Construction of plasmids for expression of recombinant EAV nsp1 to nsp12 in mammalian cells and E. coli.
The 12 nonstructural proteins (nsp1 to nsp12) of EAV were PCR amplified from the EAV recombinant virulent Bucyrus strain (rVBS) full-length infectious cDNA clone-containing plasmid (pEAVrVBS; GenBank accession number DQ846751 [10]) using the primer pairs listed in Table 2 . The PCR amplification was performed with high-fidelity Pfu DNA polymerase enzyme (Agilent, Santa Clara, CA) according to the manufacturer's protocol. The nsp5, nsp6, nsp10, and nsp12 coding regions were amplified using reverse primers with the FLAG tag coding sequence followed by a downstream stop codon. Subsequently, the individual PCR products were gel purified, digested with restriction enzymes EcoRI and XhoI, and cloned into the pCAGGS eukaryotic expression vector (generously donated by Brenda Hogue, Arizona State University, Tempe, AZ) (37). To remove the ORF 1a/ORF 1b ribosomal frameshift site in the nsp9 coding sequence and allow full-length nsp9 expression, mutations were introduced into the pCAGGS-nsp9 construct with site-directed PCR mutagenesis using the Quick Change II XL Site-Directed Mutagenesis kit (Agilent, Santa Clara, CA) following the manufacturer's instructions. Specifically, the ORF 1a/ORF 1b ribosomal frameshift site had been removed by mutating nucleotide A-5404 to C and inserting a C between G-5399 and T-5400 (55) (numbered according to GenBank accession number DQ846751). Plasmids containing individual nsp coding sequences were transformed into E. coli (DH5α) and grown at 37°C overnight. Plasmids were purified from overnight cultures of E. coli using the QIAprep Spin Miniprep plasmid extraction kit (Qiagen, Valencia, CA). Following purification, individual plasmids were identified and characterized by restriction enzyme analysis for correct orientation of the insert. The nucleotide identity of each construct was confirmed by automatic BigDye terminator cycle sequencing (Eurofins MWG-Operon, Huntsville, AL). The plasmids containing individual nsps (nsp1 to nsp12) were identified as pCAGGS-nsp1 to pCAGGS-nsp12, respectively.
TABLE 2.
Primers used for PCR amplification of individual nsps for cloning into pCAGGS vector
| ORF | Protein (aaa length) | Nucleotide location in the genomeb | Directionc | Primer sequence (5′-3′)d | Recombinant plasmid | Predicted molecular mass (with modifications) (kDa) |
|---|---|---|---|---|---|---|
| 1ab | nsp1 (260) | 255-1004 | F | ttcGAATTCaccATGGCAACCTTCTCCGCTACTGG | pCAGGS-nsp1 | 28.6 (28.60) |
| R | ttcCTCGAGttaGCCGTAGTTGCCAGCAGGCAA | |||||
| nsp2 (571) | 1005-2717 | F | ttcGAATTCaccATGGGCTACAATCCACCAGGGGAC | pCAGGS-nsp2 | 61.4 (61.50) | |
| R | ttcCTCGAGttaACCTATCAGCCGGAACCCCGGA | |||||
| nsp3 (233) | 2718-3416 | F | ttcGAATTCaccATGGGATGGATTTATGGGATATGC | pCAGGS-nsp3 | 25 (25.1) | |
| R | ttcCTCGAGttaTTCAAACACCATCCCGCCCTC | |||||
| nsp4 (204) | 3417-4028 | F | ttcGAATTCaccATGGGGCTATTCAGGTCACCGAAGG | pCAGGS-nsp4 | 21 (21.2) | |
| R | ttcCTCGAGttaCTCTCTATTGGATAAGCCATC | |||||
| nsp5 (162) | 4029-4514 | F | ttcGAATTCaccATGAGCAGCCTTTCTGGACCTCAG | pCAGGS-nsp5-FLAG | 18.1 (19.30) | |
| R | ttcCTCGAGttaCTTATCGTCGTCATCCTTGTAATCCTCCAGGAAGTATTTCATCATG | |||||
| nsp6 (22) | 4515-4580 | F | ttcGAATTCaccATGGGAGGAGTGAAAGAGAGTGTCACC | pCAGGS-nsp6-FLAG | 2.3 (3.40) | |
| R | ttcCTCGAGttaCTTATCGTCGTCATCCTTGTAATCCTCCTGGGTAATTGGTTTGC | |||||
| nsp7 (225) | 4581-5255 | F | ttcGAATTCaccATGAGTCTCACTGCAACATTAGC | pCAGGS-nsp7 | 25.2 (25.40) | |
| R | ttcCTCGAGttaTTCATAGCTCCCCTTGCCCAGC | |||||
| nsp8 (50) | 5256-5405 | F | ttcGAATTCaccATGGGCCTAGATCAGGACAAAGTG | pCAGGS-nsp8 | 5.5 (5.60) | |
| R | ttcCTCGAGtcaGTTTAACTGATTCACTGCCTC | |||||
| nsp9 (693) | 5256-7333 | F | ttcGAATTCaccATGGGCCTAGATCAGGACAAAGTG | pCAGGS-nsp9 | 76.8 (76.90) | |
| R | ttcCTCGAGttaCTCATACTGCTTGGTGCGGAAG | |||||
| nsp10 (467) | 7334-8734 | F | ttcGAATTCaccATGAGTGCCGTGTGCACAGTTTGTGG | pCAGGS-nsp10-FLAG | 50.5 (51.60) | |
| R | ttcCTCGAGttaCTTATCGTCGTCATCCTTGTAATCTTGCTTTTCCCAGCCACAG | |||||
| nsp11 (219) | 8735-9391 | F | ttcGAATTCaccATGTCCAACAAAATTTCGTGCCTC | pCAGGS-nsp11 | 24.2 (24.30) | |
| R | ttcCTCGAGttaCTCTTGGACATAAAAGGTCGC | |||||
| nsp12 (119) | 9392-9748 | F | ttcGAATTCaccATGGGTGTTGATGCAGTTACATCAGC | pCAGGS-nsp12-FLAG | 12.5 (13.60) | |
| R | ttcCTCGAGttaCTTATCGTCGTCATCCTTGTAATCCACGGGCCCAATGACTGAACC |
aa, amino acids.
Nucleotide positions for the primers are based on GenBank accession number DQ846751.
F, forward; R, reverse.
The restriction enzyme sites used for cloning are underlined (EcoRI, GAATTC; XhoI, CTCGAG). “acc” and “tta” were inserted as the Kozak consensus sequence and the stop codon in the forward and reverse primers, respectively. The C-terminal FLAG tags in the nsp5, nsp6, nsp10, and nsp12 reverse primers are indicated in boldface.
For generating bacterial expression plasmids for EAV nsp1 to nsp12, each nsp coding sequence was amplified from pEAVrVBS using the primers listed in Table 3. Subsequently, PCR products were gel purified, digested with EcoRI and XhoI, and cloned into the pQE-TriSystem His·Strep 2 vector (Qiagen, Valencia, CA), which has promoters for expression in E. coli, insect cells, and mammalian cells, allowing expression of His·Strep-tagged proteins from a single vector. The constructs containing nsp1 to nsp12 were designated pQE-rVBSnsp1 through pQE-rVBSnsp12. Following transformation and purification, individual plasmids were identified and characterized by restriction enzyme analysis for correct orientation of the insert. The nucleotide identity of each construct was confirmed by automatic BigDye terminator cycle sequencing (Eurofins MWG-Operon, Huntsville, AL).
TABLE 3.
Primers used for PCR amplification of individual nsps for cloning into pQE-TriSystem His·Strep 2 vector
| ORF | Protein (aaa length) | Nucleotide location in the genomeb | Directionc | Primer sequence (5′-3′)d | Recombinant plasmid | Predicted molecular mass (with modifications) (kDa) |
|---|---|---|---|---|---|---|
| 1ab | nsp1 (260) | 225-1004 | F | ttcGAATTCtATGGCAACCTTCTCCGCTACTGG | pQE-rVBSnsp1 | 28.6 (32.50) |
| R | ttcCTCGAGGCCGTAGTTGCCAGCAGG | |||||
| nsp3 (233) | 2718-3416 | F | ttcGAATTCtGGATGGATTTATGGGATATGC | pQE-rVBSnsp3 | 25 (28.8) | |
| R | ttcCTCGAGTTCAAACACCATCCCGCCCTC | |||||
| nsp4 (204) | 3417-4028 | F | ttcGAATTCtGGGCTATTCAGGTCACCGAAGG | pQE-rVBSnsp4 | 21 (24.9) | |
| R | ttcCTCGAGCTCTCTATTGGATAAGCCATC | |||||
| nsp6 (22) | 4515-4580 | F | ttcGAATTCtGGAGGAGTGAAAGAGAGTGTCACC | pQE-rVBSnsp6 | 2.3 (6.10) | |
| R | ttcCTCGAGCTCCTGGGTAATTGGTTTGC | |||||
| nsp7 (225) | 4581-5255 | F | ttcGAATTCtAGTCTCACTGCAACATTAGC | pQE-rVBSnsp7 | 25.2 (29.10) | |
| R | ttcCTCGAGTTCATAGCTCCCCTTGCCCAGC | |||||
| nsp8 (50) | 5256-5405 | F | ttcGAATTCtGGCCTAGATCAGGACAAAGTG | pQE-rVBSnsp8 | 5.5 (9.60) | |
| R | ttcCTCGAGGTTTAACTGATTCACTGCCTC | |||||
| nsp10 (467) | 7334-8734 | F | ttcGAATTCtAGTGCCGTGTGCACAGTTTGTGG | pQE-rVBSnsp10 | 50.5 (54.40) | |
| R | ttcCTCGAGTTGCTTTTCCCAGCCACAG | |||||
| nsp12 (119) | 9392-9748 | F | ttcGAATTCtGGTGTTGATGCAGTTACATCAGC | pQE-rVBSnsp12 | 12.5 (16.40) | |
| R | ttcCTCGAGCACGGGCCCAATGACTGAACC |
aa, amino acids.
Nucleotide positions for the primers are based on GenBank accession number DQ846751.
F, forward; R, reverse.
The restriction enzyme sites used for cloning are underlined (EcoRI, GAATTC; XhoI, CTCGAG). A single nucleotide “t” was added after the EcoRI restriction enzyme site in the forward primer to allow in-frame expression of recombinant protein.
Expression of recombinant nsp1 to nsp12 in mammalian cells.
Expression of recombinant EAV nsps was performed in BHK-21 cells transfected with individual plasmids containing nsp1 to nsp12 coding regions using Fugene HD (Promega) according to the manufacturer's instructions. Briefly, BHK-21 cells were plated the day before transfection at a density of 5 × 105 cells into a 100-mm cell culture dish in 17 ml of complete growth medium. A total of 19 μg of each plasmid was mixed with Fugene HD reagent (Promega), and the complex was incubated for 10 min at room temperature (RT). Subsequently, each mixture was added onto confluent monolayers of BHK-21 cells and incubated at 37°C in a 5% CO2 incubator. At 24 h posttransfection, the cells were lysed in NP-40 cell lysis buffer (50 mM Tris [pH 7.4], 250 mM NaCl, 5 mM EDTA, 50 mM NaF, 1 mM Na3VO4, 1% NP-40, and 0.02% NaN3) supplemented with 1× proteinase inhibitor cocktail (Pierce) and 1 mM phenylmethanesulfonyl fluoride (PMSF). Insoluble materials were removed by centrifugation at 4°C for 10 min at 13,000 × g in a microcentrifuge. Cleared cell lysates were stored at −80°C for further use. The expression and validity of each recombinant protein were confirmed by indirect immunofluorescence and Western immunoblotting analyses.
Expression and purification of recombinant EAV nsp1 to nsp12 expressed in bacterial cells.
Plasmids pQE-rVBSnsp1, -nsp3, -nsp4, -nsp6, -nsp7, -nsp8, -nsp10, and -nsp12 were transformed in an expression strain of E. coli, M15[pREP4], and a single colony was used to inoculate 10 ml of Luria-Bertani (LB) medium containing both ampicillin (100 μg/ml) and kanamycin (25 μg/ml). The culture was grown overnight, diluted 1:20 in fresh LB medium with ampicillin and kanamycin, and subsequently grown at 37°C until an optical density at 600 nm (OD600) of 0.6 was reached. Recombinant protein expression was induced by addition of 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG). Bacterial cells were harvested 4 h after induction by centrifugation at 4,000 × g for 20 min at 4°C and stored at −20°C. Individual recombinant fusion proteins containing 8× His tag were purified by the Ni-nitrilotriacetic acid (NTA) (Qiagen, Valencia, CA) agarose affinity isolation procedure. Briefly, induced bacterial cell pellets were resuspended in buffer B (100 mM NaH2PO4, 10 mM Tris base, and 8 M urea, pH 8.0) and incubated for 1 h at RT with gentle mixing. The lysates were clarified at 10,000 × g for 30 min at RT. The supernatant containing recombinant protein was decanted into a 50% Ni-NTA slurry and rotated gently for 1 h at RT. After binding, the agarose containing column was washed twice with buffer C (100 mM NaH2PO4, 10 mM Tris base, and 8 M urea, pH 6.3). Elution was performed in 0.5-ml fractions of buffer D (100 mM NaH2PO4, 10 mM Tris base, and 8 M urea, pH 5.9) four times, followed by elution in 0.5-ml fractions of buffer E (100 mM NaH2PO4, 10 mM Tris base, and 8 M urea, pH 4.5) four times. Fractions containing the same recombinant protein were pooled and stored at −80°C for further use.
Immunofluorescence assays.
For the indirect immunofluorescence assay (IFA), BHK-21 cells grown on glass coverslips in 24-well plates were transfected with 0.55 μg of each nsp expression plasmid (nsp1 to nsp12) using Fugene HD (Promega, Madison, WI) according to the manufacturer's instructions. At 18 h posttransfection, cells were fixed in 4% paraformaldehyde in phosphate-buffered saline (PBS) (pH 7.4) for 30 min at RT and washed three times with PBS (pH 7.4) containing 10 mM glycine (glycine-PBS). After permeabilization with 0.2% Triton X-100 in PBS (pH 7.4) for 10 min, coverslips were incubated with the appropriate nsp-specific monoclonal (MAb) or polyclonal antibody or with anti-FLAG tag MAb (1:200) in PBS containing 5% fetal bovine serum (FBS). After three 10 mM glycine-PBS washes, coverslips were incubated with fluorescein isothiocyanate (FITC)-conjugated goat anti-mouse or anti-rabbit IgG (Southern Biotech, Birmingham, AL) for 1 h at RT. The coverslips were washed and mounted in 4′,6′-diamidino-2-phenylindole (DAPI)-containing aqueous mounting medium (Vector Laboratories, Burlingame, CA) and observed under an inverted fluorescence microscope.
Western immunoblotting.
The solubilized proteins were mixed with 5× reducing sample buffer containing 100 mM dithiothreitol (DTT) (Pierce, Rockford, IL) and incubated for 10 min at RT. Samples were resolved in an SDS-12% polyacrylamide gel. The gel was transferred to a polyvinylidene difluoride (PVDF) membrane (Bio-Rad, Hercules, CA). Blocking was performed in 5% skim milk powder dissolved in TBS-T (10 mM Tris-HCl [pH 7.6], 150 mM NaCl, 0.05% Tween 20). The blots were incubated with primary antibody for 1 h at RT, followed by biotinylated goat anti-mouse or anti-rabbit antibodies (Invitrogen, Carlsbad, CA) for 1 h. Subsequently, the blots were incubated with streptavidin-horseradish peroxidase tertiary antibody (Invitrogen, Carlsbad, CA) and detected using an enhanced chemiluminescence (ECL) reaction (Amersham, Piscataway, NJ).
Immunoprecipitation.
Immunoprecipitation of individual EAV nsps was performed using a Dynabeads Protein G (Invitrogen, Carlsbad, CA) immunoprecipitation kit following the manufacturer's instructions. Briefly, 10 μl of equine antiserum was added to the Dynabeads Protein G and incubated with rotation for 10 min at RT, allowing antibody binding to the beads via the Fc region. The bead-bound antibody complex was washed with binding buffer. EAV nsp-containing cell lysate was mixed with the bead-bound antiserum complex and incubated for 30 min at RT with constant rotation. Then, the bead-antibody-antigen complex was washed 3 times with washing buffer. The immunoprecipitated target antigen was eluted in elution buffer and mixed with 5× reducing sample buffer containing DTT (Pierce, Rockford, IL) and heated for 5 min at 70°C. Subsequently, denatured samples were resolved by SDS-12% PAGE and subjected to Western immunoblotting analysis as described previously.
RESULTS
Characterization of EAV nonstructural proteins expressed in mammalian cells.
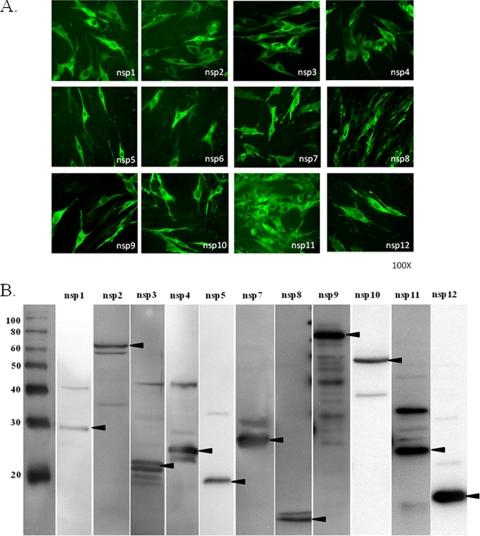
cDNA fragments corresponding to 12 nonstructural proteins (nsp1 to nsp12) of the VB strain of EAV were cloned and expressed in BHK-21 cells. Due to lack of protein-specific antisera, recombinant proteins nsp5, nsp6, nsp10, and nsp12 were expressed as C-terminal FLAG-tagged fusion proteins from each construct. The expression and validity of each recombinant protein was confirmed by IFA and Western immunoblotting analyses with protein-specific rabbit antisera and monoclonal anti-FLAG antibody (Fig. 1 A and B). All 12 proteins, except for nsp6 (a very short peptide of 22 amino acids), which was expressed in only a few cells, were expressed at high levels and detected by IFA (Fig. 1A). Consistent with the IFA result, nsp6 could not be detected in the Western immunoblotting assay, indicating that expression of this protein is insufficient, or it may be lost during SDS-PAGE and Western immunoblotting due to its extremely small size (3.4 kDa after modification). As shown in Fig. 1B, all of the recombinant proteins migrated according to their predicted sizes, listed in Table 2.
FIG. 1.
(A) Expression of EAV nonstructural proteins in BHK-21 cells as detected by immunofluorescence assay. For this purpose, BHK-21 cells were transfected with plasmids encoding individual nsps and examined at 24 h posttransfection by IFA. Protein-specific monoclonal antibody 12A4 (nsp1), protein-specific rabbit anti-peptide sera (nsp2, nsp3, nsp4, nsp7 and nsp8, nsp9, and nsp11), or anti-FLAG monoclonal antibody (nsp5, nsp6, nsp10, and nsp12) and FITC-conjugated goat anti-mouse or rabbit IgG (H+L) were used for detection of each protein. The recombinant proteins are identified at the bottoms of the images. (B) Western immunoblotting analyses of recombinant EAV nsps expressed in mammalian cells, performed with protein-specific monoclonal antibodies as specified for panel A. The molecular size markers (in kilodaltons) are shown on the left, and the nsps are identified above the lanes. The arrowheads indicate the position of each recombinant protein.
Characterization of EAV nonstructural proteins expressed in E. coli.
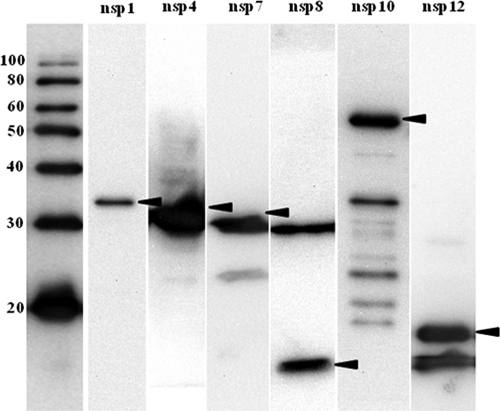
As an alternative to the use of a mammalian expression system, we employed a bacterial expression system with a C-terminally 8× His-tagged vector as a secondary/purification strategy. This system is more convenient for production of larger amounts of protein, which could be used as antigen in future diagnostic tests. Segments of the EAV genome containing the sequences of nsp1, nsp3, nsp4, nsp6, nsp7, nsp8, nsp10, and nsp12 were successfully inserted into the pQE-TriSystem His·Strep 2 expression vector. When E. coli cells were transformed with each plasmid expressing individual nsp and induced with IPTG, bands migrating at positions corresponding to nsp1, nsp4, nsp7, nsp8, nsp10, and nsp12 with molecular masses of approximately 32 kDa, 26 kDa, 29 kDa, 9 kDa, 55 kDa, and 16 kDa, respectively, as indicated in Table 3, were observed on Western blots (Fig. 2). Although the predicted molecular mass of nsp4 is 21 kDa, nsp4 was detected as an approximately 30-kDa product, as documented previously (48). Nsp1 and nsp12 were expressed at high concentrations in both soluble and insoluble fractions of cell lysates. In contrast, larger amounts of nsp3, nsp4, nsp6, nsp7, nsp8, and nsp10 could be recovered from insoluble fractions of cell lysates compared to soluble fractions. Therefore, proteins were purified under denaturing conditions using an immobilized-metal affinity chromatography procedure with the Ni-NTA resin column. Although expression of nsp3 and nsp6 proteins was confirmed in a Western immunoblotting analysis before purification, these proteins could not be recovered after purification using Ni-NTA resin columns. Accordingly, bacterially expressed nsp3 and nsp6 could not be further evaluated in the study. Furthermore, nsp2, nsp5, nsp9, and nsp11 were toxic and could not be expressed in E. coli M15(pREP4) using the pQE-TriSystem His·Strep 2 vector.
FIG. 2.
Western immunoblot analyses of recombinant nsp4, nsp7, nsp8, nsp9, nsp10, and nsp12 of EAV expressed in E. coli. The samples were total lysates from IPTG-induced cultures of E. coli M15(pREP4) cells transformed with pQE-TriSystem His·Strep 2. Total bacterial cell lysate was bound to an Ni-NTA resin column. The bound individual recombinant proteins were eluted in elution buffer and subjected to Western immunoblotting analysis. Recombinant proteins were detected using anti-His monoclonal antibody. The molecular size markers (in kilodaltons) are shown on the left, and the nsps are identified above the lanes. The arrowheads indicate the positions of the proteins.
Immunoprecipitation of EAV nsp1 to nsp12 expressed in mammalian cells with equine serum.
To determine the equine antibody response to nonstructural proteins of EAV, recombinant proteins nsp1 to nsp12 expressed in mammalian cells were subjected to immunoprecipitation using 11 serum samples containing antibodies to EAV. The VN antibody titers of these serum samples varied between 1:128 and ≥1:1,024 (Table 1).
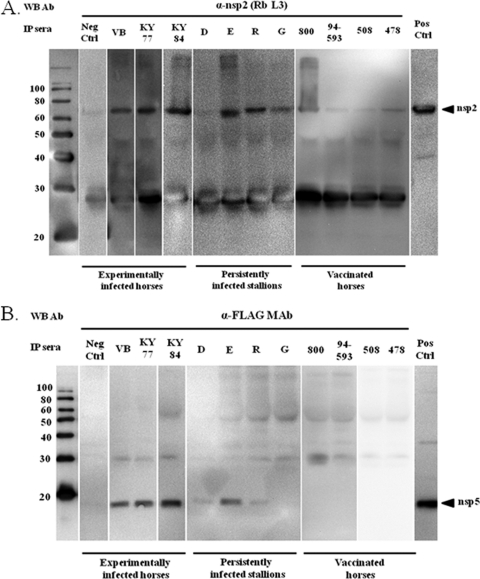
Individual recombinant nsps expressed in mammalian cells were immunoprecipitated using well-characterized equine serum samples from experimentally and persistently infected horses plus sera from horses vaccinated with the commercial modified live-virus vaccine (ARVAC; Fort Dodge Animal Health Laboratories [now Pfizer Animal Health Inc., New York, NY]). Briefly, total solubilized proteins derived from nsp-transfected (nsp1 to nsp12) or pCAGGS-empty vector-transfected BHK-21 cells were immunoprecipitated with equine sera, and the immunoprecipitated proteins were subjected to SDS-polyacrylamide gel electrophoresis. As a control, two equine serum samples negative for EAV antibodies were included in these assays. Individual nsps in the immunoprecipitates were then identified with an nsp-specific monoclonal antibody (nsp1), a specific rabbit antiserum (nsp2, nsp3, nsp4, nsp7, nsp8, nsp9, and nsp11), or an anti-FLAG monoclonal antibody (nsp5, nsp6, nsp10, and nsp12) by Western immunoblotting analysis. All three sera from horses that were previously experimentally infected with EAV strains VB, KY77, and KY84 precipitated recombinant nsp2 expressed in mammalian cells. Also, three out of four serum samples from persistently infected horses recognized nsp2, although serum from stallion G gave a weak precipitation reaction compared to the others. In contrast, all four serum samples from vaccinated horses weakly immunoprecipitated nsp2, as detected by Western immunoblotting assay (Fig. 3 A). Similarly, all three sera from horses experimentally infected with different EAV strains reacted strongly with recombinant nsp5 expressed in mammalian cells. Interestingly, sera from persistently infected horses did not consistently immunoprecipitate nsp5. Sera from stallion E gave a strong immunoprecipitation reaction with nsp5 (comparable to sera from experimentally infected horses), while sera from stallions D and R reacted very weakly and serum from stallion G did not react at all. In contrast, none of the sera from vaccinated horses were able to immunoprecipitate nsp5 (Fig. 3B). All of the sera from experimentally infected horses gave a very weak immunoprecipitation reaction with nsp12 (Fig. 4). Equine serum from the horse experimentally inoculated with the KY77 strain strongly immunoprecipitated nsp12 compared to sera from two other horses experimentally inoculated with the VB and KY84 strains of EAV. None of the sera from persistently infected stallions and vaccinated horses recognized nsp12 expressed in mammalian cells. Interestingly, none of the 11 equine serum samples evaluated in this study were able to immunoprecipitate nsp1, nsp3, nsp4, nsp7, nsp8, nsp9, nsp10, and nsp11 expressed in mammalian cells (Table 1). Based on these results using recombinant proteins expressed in mammalian cells, only nsp2 and nsp5 were consistently recognized by equine antiserum. As expected, none of the proteins were immunoprecipitated by the two negative-control sera, confirming the specificity of EAV nsp recognition. Furthermore, none of the equine sera reacted with the solubilized cellular proteins from pCAGGS-empty-vector-transfected BHK-21 cells (data not shown).
FIG. 3.
Immunoprecipitation analyses of nsp2 (A) and nsp5 (B) with sera from experimentally and persistently infected horses and vaccinated horses (ARVAC; Pfizer Animal Health Inc., New York, NY). Nsp2 and nsp5 expressed in mammalian cells were immunoprecipitated (IP) using equine sera and detected by Western immunoblotting (WB) analysis with an anti-nsp2 rabbit serum and the anti-FLAG tag monoclonal antibody, respectively. The molecular size markers (in kilodaltons) are shown on the left, and the serum samples used for immunoprecipitation are identified above the lanes. The extreme right-hand lanes contain corresponding recombinant proteins expressed in mammalian cells as a control. Neg Ctrl, negative control; Pos Ctrl, positive control.
FIG. 4.
Immunoprecipitation analysis of nsp12 with sera from experimentally and persistently infected horses and vaccinated horses. Nsp12 expressed in mammalian cells was immunoprecipitated using equine sera and detected by Western blot analysis with an anti-FLAG tag monoclonal antibody. For molecular size markers and controls, see the legend to Fig. 3.
Immunoprecipitation of EAV nonstructural proteins expressed in E. coli with equine serum.
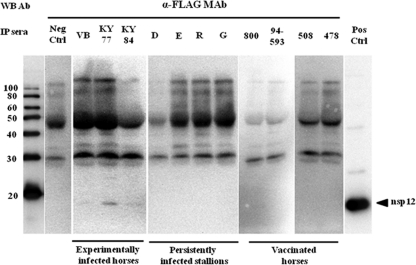
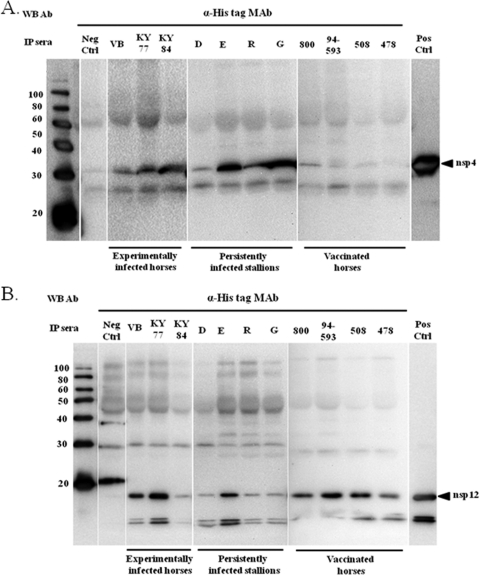
In order to verify whether equine serum from acutely (experimentally) infected, persistently infected, and vaccinated horses can react with bacterially expressed antigens similar to antigens expressed in mammalian cells, purified nsp1, nsp3, nsp4, nsp7, nsp8, nsp10, and nsp12 proteins were subjected to immunoprecipitation analyses with equine antisera. Consistent with the immunoprecipitation results using antigens expressed in mammalian cells, none of the tested sera recognized nsp1, nsp3, nsp7, nsp8, and nsp10 antigens. Interestingly, sera from horses used in this study immunoprecipitated nsp4 and nsp12 expressed in bacterial cells, although they did not previously immunoprecipitate the same proteins expressed in mammalian cells (Fig. 5 A and B). With the exception of serum from one persistently infected stallion, other sera from persistently infected stallions and all three experimentally infected horses strongly immunoprecipitated nsp4 expressed in E. coli. A serum sample from the persistently infected stallion D gave a weak immunoprecipitation reaction compared to other samples (Fig. 5A). Interestingly, the serum samples from vaccinated horses were only able to weakly immunoprecipitate nsp4. Previously, most tested sera did not react or reacted weakly with nsp12 expressed in mammalian cells. In contrast, most sera reacted strongly with bacterially expressed nsp12 antigen (Fig. 5B). Sera from VB- and KY77-inoculated horses and stallion E reacted strongly with the bacterially expressed nsp12 antigen, while the rest of the sera (a KY84-inoculated horse and stallions D, R, and G) reacted only weakly with this antigen in immunoprecipitation assays. Additionally, all four sera from vaccinated horses were also able to strongly immunoprecipitate nsp12. Based on these results, horses consistently mount an immune response to EAV nsp4 and nsp12. Equine antisera from experimentally infected and vaccinated horses, as well as persistently infected stallions, were unable to immunoprecipitate nsp1, nsp7, nsp8, and nsp10 expressed in E. coli. As expected, none of the proteins were immunoprecipitated with negative-control sera, confirming the specificity of our approach.
FIG. 5.
Immunoprecipitation analyses of nsp4 (A) and nsp12 (B) with sera from acutely and persistently infected horses and vaccinated horses (ARVAC; Pfizer). Nsp4 and nsp12 expressed in E. coli were immunoprecipitated using equine sera and detected by Western immunoblot analysis with the anti-His monoclonal antibody. For molecular size markers and controls, see the legend to Fig. 3.
DISCUSSION
In this study, we generated recombinant EAV nsp1 to nsp12 using mammalian and E. coli expression systems. Most of the proteins migrated at the predicted molecular size, as indicated in Tables 2 and 3. However, several nsps expressed in mammalian cells showed nonspecific protein bands on the membrane that were also recognized by protein-specific rabbit antisera, e.g., nsp2, nsp3, nsp4, nsp9, and nsp11 (Fig. 1B). This observation was considered to be only background bands or possible instability/degradation of expressed proteins. Previous studies have shown that nsp2, nsp3, and nsp5 contain predicted membrane-spanning domains that may serve to anchor nsps to the modified intracellular membranes with which the viral replication complex is presumed to be associated (56). Incomplete disassociation of these nsps from cellular proteins during SDS-PAGE could have caused some of these extra bands with higher molecular masses. Consistent with previous reports, the recombinant nsp4 (predicted as 21 kDa and 25 kDa with modifications in mammalian and bacterial expression systems, respectively) migrated as a protein of approximately 30 kDa, a phenomenon described as aberrant migration of this protein during SDS-PAGE (49, 53). Additionally, the two bands of nsp4 visible by Western immunoblotting analysis using nsp4-specific rabbit antisera have been reported previously (53).
The difficulties encountered during expression of recombinant nsp2, nsp5, nsp9, and nsp11 and purification of nsp3 and nsp6 in E. coli were likely related to the inherent toxicity, codon usage differences between bacterial and mammalian cells, and/or misfolding of these proteins when expressed from the pQE-TriSystem vector. Although expression of nsp3 and nsp6 was confirmed in pilot experiments, these proteins could not be recovered by Ni-NTA purification. It is plausible that misfolding of the proteins masked their 8× His tag and prevented binding to the Ni-NTA resin. Specifically, nsp2 and nsp5, which showed high immunoreactivity against equine sera when expressed in mammalian cells, may need to be expressed using a different expression vector and E. coli strain to generate recombinant proteins that could be used in diagnostic assays in the future.
Previous studies on characterization of the equine humoral immune response to EAV focused mainly on detection of antibodies to major viral structural proteins, especially GP5 and the M and N proteins (15, 16, 18, 19, 26, 32, 33, 38, 58). To further characterize the host immune response to EAV proteins, individual recombinant nsps expressed in both mammalian and bacterial cells were immunoprecipitated with EAV antibody-positive and -negative sera and identified with individual protein-specific monoclonal antibodies or protein-specific rabbit antisera by Western immunoblotting analyses. These viral proteins synthesized during an early phase of infection in host cells are degraded in the cytoplasm by the proteasome, and the resulting peptides are translocated into the endoplasmic reticulum, where they are loaded onto major histocompatibility complex (MHC) class molecules (1). It has been suggested that during antigen processing, antigenic sites buried in native form, such as hydrophobic regions, are exposed to the immune system, making them more immunogenic (2, 3). The data showed that nsp2, nsp4, nsp5, and nsp12 are the most immunogenic of the 12 nsps (Table 1) and that horses tend to mount an immune response to these viral proteins. All three sera from experimentally infected horses consistently recognized nsp2, nsp4, nsp5, and nsp12. In contrast, the antibody responses to nsp2, nsp4, and nsp12 in persistently infected stallions were somewhat variable. In addition, nsp5 was the only protein that consistently reacted with sera from carrier stallions. These data suggested that the serologic response of horses against nsps that clear the virus following acute infection might be different from that of persistently infected horses. It is possible that horses mount a strong immune response to viral nsps in the acute phase (when virus replication is active) that remains until the late convalescent phase but decreases over time. In the presence of high-titer neutralizing antibodies in the serum during persistent infection, EAV is harbored in the ampulla of the reproductive tract of the stallion (52). Therefore, it could be postulated that virus replicates in immunologically privileged cells in the stallion's reproductive tract, preventing (or reducing) the exposure of nsps to the host immune system. However, some of the nsps may still be able to induce a limited immune response during prolonged persistent infection in the reproductive tract of the stallion. Moreover, all nsps may not be equally immunogenic, and this may also vary among different strains of EAV. This could explain the difference between the serologic response to nsps in vaccinated animals and that in the horses experimentally inoculated with more virulent field strains (VB, KY77, and KY84 strains). The response of horses vaccinated with the MLV vaccine strain (ARVAC) was also different from that of experimentally and persistently infected horses. Sera from vaccinated horses recognized nsp2 and nsp12, while nsp4 and nsp5 reacted very weakly or did not react at all. Furthermore, different horses may also respond differently to EAV nsps during infection. It has been shown previously that immune responses to EAV structural proteins differ with the infecting virus strain and the duration of infection (33). The M protein was most consistently recognized by various serum samples, whereas the responses to the N and GP5 proteins were variable (26, 33).
Interestingly, the equine humoral immune responses to nsp4 and nsp12 differed depending on whether the protein was expressed in mammalian or bacterial cells. Most sera, except for those from vaccinated horses, strongly reacted with nsp4 expressed in bacteria. However, none of the sera immunoprecipitated nsp4 when it was expressed in mammalian cells. Similarly, the equine sera from experimentally infected horses or persistently infected stallions either were only weakly immunoprecipitated or did not precipitate the nsp12 expressed in mammalian cells while they strongly reacted with and immunoprecipitated the nsp12 expressed in E. coli. The reason for this difference in recognition of the same antigen expressed in different expression systems might be due to the failure of antigenic epitopes to be presented in their native form to be recognized by the equine antibodies. It is also possible that nsp4 and nsp12 proteins, when expressed in a mammalian expression system, are not processed properly or are misfolded; this may lead to obscuring both the conformational and linear epitopes recognized by equine antibodies. However, the proteins expressed in E. coli are denatured during the purification process, and this may well expose the linear epitopes recognized by equine antibodies and facilitate immunoprecipitation of both nsp4 and nsp12 proteins.
In PRRSV infection in pigs, nsp1, nsp2, and nsp7 were found to be the most immunogenic nsps of the virus (12, 40, 41). Subsequently, PRRSV nsp1α/β and nsp7, and also nsp2, have been explored as potential antigens for the development of serological diagnostic assays. However, our data strongly suggest that horses do not mount a strong humoral antibody response to EAV nsp1 and nsp7 compared to pigs infected with PRRSV. In our hands, it appears that EAV nsp1 is toxic in both mammalian and bacterial cells, as expression of the protein was not highly efficient compared to other nsps. Therefore, the suboptimal antigen concentration provided for the immunoprecipitation assay could have adversely affected the result. As in PRRSV, there is significant variation in EAV nsp2 (5, 62). However, unlike the PRRSV nsp2, until now, no B-cell or T-cell epitopes have been identified in this protein in the case of EAV (20, 41, 60).
The VN test is the OIE (World Animal Health Organization)-prescribed regulatory test for serologic detection of EAV infection of the horse (39). However, the assay is expensive, labor-intensive, and time-consuming to perform. The results of the assay tend to vary among laboratories when insufficient attention is paid to standardization of both test reagents and procedures. To address this problem, we previously developed an MIA using recombinant structural proteins of EAV. However, the sensitivity and specificity of the assay were 92.6% and 92.9%, respectively, compared to the VN test (25). In the current study, we identified the key EAV nsps recognized by horses during acute and persistent infection, as well as following vaccination. Thus, the recombinant nsps (nsp2, nsp4, nsp5, and nsp12) identified in this study, in combination with the structural proteins, could be used in a new MIA to further improve the specificity and sensitivity of the assay. Moreover, nsp5 could be a potential candidate to be used as an antigen in a companion differential diagnostic test to distinguish horses vaccinated with the current modified live-virus vaccine (ARVAC) from naturally infected horses. Once we develop this new MIA, it will be used to further characterize the antibody response to EAV by testing sequential serum samples from experimentally inoculated and vaccinated horses (10, 11, 42, 61). We also plan to evaluate this MIA with a large panel of equine sera determined to be positive and negative for EAV antibody by VN test.
In summary, data presented in this study showed that the equine humoral immune response to EAV nsps is directed against nsp2, nsp4, nsp5, and nsp12. None of the horse sera recognized nsp1, nsp3, and nsp6 through nsp11 when recombinant antigens were used in immunoprecipitation assays. Among these proteins, nsp2 and nsp12 were consistently recognized by either experimentally and persistently infected horses or vaccinated horses. In contrast, nsp4 and nsp5 were recognized efficiently only by sera from experimentally and persistently infected horses, but not by those from vaccinated horses. Therefore, this study suggests that these antigens can be used for serodiagnostic testing of EAV infection. Incorporation of the immunogenic nsps in a new MIA, along with structural proteins as antigens, might greatly improve the sensitivity and specificity of the previously developed MIA. An improved MIA is already under development in our laboratory. Thus, a multiplex MIA including the most immunogenic structural and nonstructural proteins of EAV could become a potential alternative to the standard VN test.
Acknowledgments
This study was supported by funds from the Frederick Van Lennep Endowment and the Kentucky Agricultural Experiment Station, College of Agriculture, University of Kentucky. Yun Young Go is the recipient of a Geoffrey C. Hughes Foundation graduate fellowship.
We thank Brenda Hogue, Arizona State University, Tempe, AZ, for providing the pCAGGS expression vector and Fred Wassenaar, Jessika Zevenhoven, and Danny Nedialkova of the Department of Medical Microbiology, Center of Infectious Diseases, Leiden University Medical Center, Leiden, Netherlands, for assistance in EAV nsp antiserum production. We also thank Kathleen M. Shuck for critical reading of the manuscript.
Footnotes
Published ahead of print on 8 December 2010.
REFERENCES
- 1.Abbas, A. K., and A. H. Lichtman. 2000. Cellular and molecular immunology. Saunders, Philadelphia, PA.
- 2.Allen, P. M., D. J. Strydom, and E. R. Unanue. 1984. Processing of lysozyme by macrophages: identification of the determinant recognized by two T-cell hybridomas. Proc. Natl. Acad. Sci. U. S. A. 81:2489-2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allen, P. M., and E. R. Unanue. 1984. Differential requirements for antigen processing by macrophages for lysozyme-specific T cell hybridomas. J. Immunol. 132:1077-1079. [PubMed] [Google Scholar]
- 4.Balasuriya, U. B., et al. 2004. Characterization of the neutralization determinants of equine arteritis virus using recombinant chimeric viruses and site-specific mutagenesis of an infectious cDNA clone. Virology 321:235-246. [DOI] [PubMed] [Google Scholar]
- 5.Balasuriya, U. B., et al. 2004. Genetic characterization of equine arteritis virus during persistent infection of stallions. J. Gen. Virol. 85:379-390. [DOI] [PubMed] [Google Scholar]
- 6.Balasuriya, U. B., and N. J. MacLachlan. 2004. The immune response to equine arteritis virus: potential lessons for other arteriviruses. Vet. Immunol. Immunopathol. 102:107-129. [DOI] [PubMed] [Google Scholar]
- 7.Balasuriya, U. B., N. J. Maclachlan, A. A. De Vries, P. V. Rossitto, and P. J. Rottier. 1995. Identification of a neutralization site in the major envelope glycoprotein (GL) of equine arteritis virus. Virology 207:518-527. [DOI] [PubMed] [Google Scholar]
- 8.Balasuriya, U. B., et al. 1997. Neutralization determinants of laboratory strains and field isolates of equine arteritis virus: identification of four neutralization sites in the amino-terminal ectodomain of the G(L) envelope glycoprotein. Virology 232:114-128. [DOI] [PubMed] [Google Scholar]
- 9.Balasuriya, U. B., P. V. Rossitto, C. D. DeMaula, and N. J. MacLachlan. 1993. A 29K envelope glycoprotein of equine arteritis virus expresses neutralization determinants recognized by murine monoclonal antibodies. J. Gen. Virol. 74:2525-2529. [DOI] [PubMed] [Google Scholar]
- 10.Balasuriya, U. B., et al. 2007. Development and characterization of an infectious cDNA clone of the virulent Bucyrus strain of Equine arteritis virus. J. Gen. Virol. 88:918-924. [DOI] [PubMed] [Google Scholar]
- 11.Balasuriya, U. B., et al. 1999. Equine arteritis virus derived from an infectious cDNA clone is attenuated and genetically stable in infected stallions. Virology 260:201-208. [DOI] [PubMed] [Google Scholar]
- 12.Brown, E., et al. 2009. Antibody response to porcine reproductive and respiratory syndrome virus (PRRSV) nonstructural proteins and implications for diagnostic detection and differentiation of PRRSV types I and II. Clin. Vaccine Immunol. 16:628-635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bryans, J. T., M. E. Crowe, E. R. Doll, and W. H. McCollum. 1957. Isolation of a filterable agent causing arteritis of horses and abortion by mares; its differentiation from the equine abortion (influenza) virus. Cornell Vet. 47:3-41. [PubMed] [Google Scholar]
- 14.Cavanagh, D. 1997. Nidovirales: a new order comprising Coronaviridae and Arteriviridae. Arch. Virol. 142:629-633. [PubMed] [Google Scholar]
- 15.Chirnside, E. D., A. A. de Vries, J. A. Mumford, and P. J. Rottier. 1995. Equine arteritis virus-neutralizing antibody in the horse is induced by a determinant on the large envelope glycoprotein GL. J. Gen. Virol. 76:1989-1998. [DOI] [PubMed] [Google Scholar]
- 16.Chirnside, E. D., P. M. Francis, A. A. de Vries, R. Sinclair, and J. A. Mumford. 1995. Development and evaluation of an ELISA using recombinant fusion protein to detect the presence of host antibody to equine arteritis virus. J. Virol. Methods 54:1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chirnside, E. D., P. M. Francis, and J. A. Mumford. 1995. Expression cloning and antigenic analysis of the nucleocapsid protein of equine arteritis virus. Virus Res. 39:277-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cho, H. J., et al. 2000. Detection of antibodies to equine arteritis virus by a monoclonal antibody-based blocking ELISA. Can. J. Vet. Res. 64:38-43. [PMC free article] [PubMed] [Google Scholar]
- 19.Cook, R. F., S. J. Gann, and J. A. Mumford. 1989. The effects of vaccination with tissue culture-derived viral vaccines on detection of antibodies to equine arteritis virus by enzyme-linked immunosorbent assay (ELISA). Vet. Microbiol. 20:181-189. [DOI] [PubMed] [Google Scholar]
- 20.de Lima, M., A. K. Pattnaik, E. F. Flores, and F. A. Osorio. 2006. Serologic marker candidates identified among B-cell linear epitopes of Nsp2 and structural proteins of a North American strain of porcine reproductive and respiratory syndrome virus. Virology 353:410-421. [DOI] [PubMed] [Google Scholar]
- 21.den Boon, J. A., et al. 1991. Equine arteritis virus is not a togavirus but belongs to the coronaviruslike superfamily. J. Virol. 65:2910-2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Deregt, D., A. A. de Vries, M. J. Raamsman, L. D. Elmgren, and P. J. Rottier. 1994. Monoclonal antibodies to equine arteritis virus proteins identify the GL protein as a target for virus neutralization. J. Gen. Virol. 75:2439-2444. [DOI] [PubMed] [Google Scholar]
- 23.Doll, E. R., J. T. Bryans, J. C. Wilson, and W. H. McCollum. 1968. Immunization against equine viral arteritis using modified live virus propagated in cell cultures of rabbit kidney. Cornell Vet. 48:497-524. [PubMed] [Google Scholar]
- 24.Glaser, A. L., A. A. de Vries, and E. J. Dubovi. 1995. Comparison of equine arteritis virus isolates using neutralizing monoclonal antibodies and identification of sequence changes in GL associated with neutralization resistance. J. Gen. Virol. 76:2223-2233. [DOI] [PubMed] [Google Scholar]
- 25.Go, Y. Y., et al. 2008. Development of a fluorescent-microsphere immunoassay for detection of antibodies specific to equine arteritis virus and comparison with the virus neutralization test. Clin. Vaccine Immunol. 15:76-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hedges, J. F., et al. 1998. Detection of antibodies to equine arteritis virus by enzyme linked immunosorbant assays utilizing G(L), M and N proteins expressed from recombinant baculoviruses. J. Virol. Methods 76:127-137. [DOI] [PubMed] [Google Scholar]
- 27.Hedges, J. F., U. B. Balasuriya, and N. J. MacLachlan. 1999. The open reading frame 3 of equine arteritis virus encodes an immunogenic glycosylated, integral membrane protein. Virology 264:92-98. [DOI] [PubMed] [Google Scholar]
- 28.Hedges, J. F., U. B. Balasuriya, P. J. Timoney, W. H. McCollum, and N. J. MacLachlan. 1999. Genetic divergence with emergence of novel phenotypic variants of equine arteritis virus during persistent infection of stallions. J. Virol. 73:3672-3681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jeronimo, C., and D. Archambault. 2002. Importance of M-protein C terminus as substrate antigen for serodetection of equine arteritis virus infection. Clin. Diagn. Lab Immunol. 9:698-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kheyar, A., et al. 1997. Expression cloning and humoral immune response to the nucleocapsid and membrane proteins of equine arteritis virus. Clin. Diagn. Lab Immunol. 4:648-652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kondo, T., Y. Fukunaga, K. Sekiguchi, T. Sugiura, and H. Imagawa. 1998. Enzyme-linked immunosorbent assay for serological survey of equine arteritis virus in racehorses. J. Vet. Med. Sci. 60:1043-1045. [DOI] [PubMed] [Google Scholar]
- 32.Kondo, T., S. Sugita, Y. Fukunaga, and H. Imagawa. 1998. Identification of the major epitope in the GL protein of equine arteritis virus (EAV) recognized by antibody in EAV-infected horses using synthetic peptides. J. Equine Sci. 9:19-23. [Google Scholar]
- 33.MacLachlan, N. J., et al. 1998. Serologic response of horses to the structural proteins of equine arteritis virus. J. Vet. Diagn. Invest. 10:229-236. [DOI] [PubMed] [Google Scholar]
- 34.McCollum, W. H. 1986. Responses of horses vaccinated with avirulent modified-live equine arteritis virus propagated in the E. Derm (NBL-6) cell line to nasal inoculation with virulent virus. Am. J. Vet. Res. 47:1931-1934. [PubMed] [Google Scholar]
- 35.McCollum, W. H. 1970. Vaccination for equine viral arteritis, p. 143-151. In J. T. Bryans and H. Gerber (ed.), Proc. 2nd Int. Conf. Equine Infect. Dis., S. Karger AG, Basel, Switzerland.
- 36.Molenkamp, R., et al. 2000. The arterivirus replicase is the only viral protein required for genome replication and subgenomic mRNA transcription. J. Gen. Virol. 81:2491-2496. [DOI] [PubMed] [Google Scholar]
- 37.Niwa, H., K. Yamamura, and J. Miyazaki. 1991. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 108:193-199. [DOI] [PubMed] [Google Scholar]
- 38.Nugent, J., et al. 2000. Development and evaluation of ELISA procedures to detect antibodies against the major envelope protein (G(L)) of equine arteritis virus. J. Virol. Methods 90:167-183. [DOI] [PubMed] [Google Scholar]
- 39.Office International des Epizooties. 2004. OIE manual of diagnostic tests and vaccines for terrestrial animals, 5th ed., vol. 2. Office International des Epizooties, Paris, France.
- 40.Oleksiewicz, M. B., A. Botner, and P. Normann. 2001. Semen from boars infected with porcine reproductive and respiratory syndrome virus (PRRSV) contains antibodies against structural as well as nonstructural viral proteins. Vet. Microbiol. 81:109-125. [DOI] [PubMed] [Google Scholar]
- 41.Oleksiewicz, M. B., A. Botner, P. Toft, P. Normann, and T. Storgaard. 2001. Epitope mapping porcine reproductive and respiratory syndrome virus by phage display: the nsp2 fragment of the replicase polyprotein contains a cluster of B-cell epitopes. J. Virol. 75:3277-3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Patton, J. F., et al. 1999. Phylogenetic characterization of a highly attenuated strain of equine arteritis virus from the semen of a persistently infected standardbred stallion. Arch. Virol. 144:817-827. [DOI] [PubMed] [Google Scholar]
- 43.Pedersen, K. W., Y. van der Meer, N. Roos, and E. J. Snijder. 1999. Open reading frame 1a-encoded subunits of the arterivirus replicase induce endoplasmic reticulum-derived double-membrane vesicles which carry the viral replication complex. J. Virol. 73:2016-2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Senne, D. A., J. E. Pearson, and E. A. Carbrey. 1985. Equine viral arteritis: a standard procedure for the virus neutralization test and comparison of results of a proficiency test performed at five laboratories, p. 29-34. Proc. 89th Annu. Meet. United States Anim. Health Assoc. United States Animal Health Association, Richmond, VA.
- 45.Snijder, E. J. 2001. Arteriviruses, p. 1205-1220. In D. M. Knipe and P. M. Howley (ed.), Fields virology. Lippincott Williams and Wilkins, Philadelphia, PA.
- 46.Snijder, E. J., and J. J. Meulenberg. 1998. The molecular biology of arteriviruses. J. Gen. Virol. 79:961-979. [DOI] [PubMed] [Google Scholar]
- 47.Snijder, E. J., and W. J. Spaan. 2006. Arteriviruses, p. 1337-1355. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 5th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
- 48.Snijder, E. J., A. L. Wassenaar, and W. J. Spaan. 1994. Proteolytic processing of the replicase ORF1a protein of equine arteritis virus. J. Virol. 68:5755-5764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Snijder, E. J., A. L. Wassenaar, L. C. van Dinten, W. J. Spaan, and A. E. Gorbalenya. 1996. The arterivirus nsp4 protease is the prototype of a novel group of chymotrypsin-like enzymes, the 3C-like serine proteases. J. Biol. Chem. 271:4864-4871. [DOI] [PubMed] [Google Scholar]
- 50.Starik, E., A. Ginter, and P. Coppe. 2001. ELISA and direct immunofluorescence test to detect equine arteritis virus (EAV) using a monoclonal antibody directed to the EAV-N protein. J. Vet. Med. B Infect. Dis. Vet. Public Health 48:1-9. [DOI] [PubMed] [Google Scholar]
- 51.Timoney, P. J., and W. H. McCollum. 1993. Equine viral arteritis. Vet. Clin. North Am. Equine Pract. 9:295-309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Timoney, P. J., et al. 1987. The carrier state in equine arteritis virus infection in the stallion with specific emphasis on the venereal mode of virus transmission. J. Reprod. Fertil. Suppl. 35:95-102. [PubMed] [Google Scholar]
- 53.van Aken, D., et al. 2006. Expression, purification, and in vitro activity of an arterivirus main proteinase. Virus Res. 120:97-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.van Aken, D., J. Zevenhoven-Dobbe, A. E. Gorbalenya, and E. J. Snijder. 2006. Proteolytic maturation of replicase polyprotein pp1a by the nsp4 main proteinase is essential for equine arteritis virus replication and includes internal cleavage of nsp7. J. Gen. Virol. 87:3473-3482. [DOI] [PubMed] [Google Scholar]
- 55.van Dinten, L. C., S. Rensen, A. E. Gorbalenya, and E. J. Snijder. 1999. Proteolytic processing of the open reading frame 1b-encoded part of arterivirus replicase is mediated by nsp4 serine protease and is essential for virus replication. J. Virol. 73:2027-2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.van Dinten, L. C., A. L. Wassenaar, A. E. Gorbalenya, W. J. Spaan, and E. J. Snijder. 1996. Processing of the equine arteritis virus replicase ORF1b protein: identification of cleavage products containing the putative viral polymerase and helicase domains. J. Virol. 70:6625-6633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Van Hemert, M. J., and E. J. Snijder. 2008. The arterivirus replicase, p. 83-101. In S. Perlman, T. Gallagher, and E. J. Snijder (ed.), Nidoviruses. American Society for Microbiology, Washington, DC.
- 58.Wagner, H. M., U. B. Balasuriya, and N. James MacLachlan. 2003. The serologic response of horses to equine arteritis virus as determined by competitive enzyme-linked immunosorbent assays (c-ELISAs) to structural and non-structural viral proteins. Comp. Immunol. Microbiol. Infect. Dis. 26:251-260. [DOI] [PubMed] [Google Scholar]
- 59.Weiland, E., et al. 2000. Monoclonal antibodies directed against conserved epitopes on the nucleocapsid protein and the major envelope glycoprotein of equine arteritis virus. J. Clin. Microbiol. 38:2065-2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yan, Y., et al. 2007. Monoclonal antibody and porcine antisera recognized B-cell epitopes of Nsp2 protein of a Chinese strain of porcine reproductive and respiratory syndrome virus. Virus Res. 126:207-215. [DOI] [PubMed] [Google Scholar]
- 61.Zhang, J., et al. 2008. Amino acid substitutions in the structural or nonstructural proteins of a vaccine strain of equine arteritis virus are associated with its attenuation. Virology 378:355-362. [DOI] [PubMed] [Google Scholar]
- 62.Zhang, J., et al. 2010. Molecular epidemiology and genetic characterization of equine arteritis virus isolates associated with the 2006-2007 multi-state disease occurrence in the USA. J. Gen. Virol. 91:2286-2301. [DOI] [PubMed] [Google Scholar]
- 63.Ziebuhr, J., E. J. Snijder, and A. E. Gorbalenya. 2000. Virus-encoded proteinases and proteolytic processing in the Nidovirales. J. Gen. Virol. 81:853-879. [DOI] [PubMed] [Google Scholar]