Abstract
Comparative genomic studies have identified several Mycobacterium tuberculosis-specific genomic regions of difference (RDs) which are absent in the vaccine strains of Mycobacterium bovis BCG and which may be useful in the specific diagnosis of tuberculosis (TB). In this study, a total of 775 synthetic peptides covering the sequences of 39 open reading frame (ORF) proteins encoded by genes predicted in five RDs of M. tuberculosis, i.e., RD1, RD4, RD5, RD6, and RD7, were tested by enzyme-linked immunosorbent assays for antibody reactivity with sera from HIV-negative pulmonary TB patients (n = 100) and M. bovis BCG-vaccinated healthy subjects (n = 100). The results identified three immunodominant peptides reactive with TB sera, i.e., amino acids (aa) 346 to 370 of RD1ORF Rv3876, aa 241 to 265 of RD6ORF Rv1508c, and aa 325 to 336 of RD6ORF Rv1516c. These peptides had significantly stronger antibody reactivity with sera from TB patients than with sera from healthy subjects (P < 0.05) and significantly higher rates of positivity with TB sera (positives = 66 to 93%) than sera from healthy subjects (positives = 10 to 28%). Antipeptide antibodies were raised in rabbits after immunization with pools of 11 peptides corresponding to each protein. Probing of culture filtrates and whole-cell lysates of M. tuberculosis with antipeptide antibodies suggested the natural expression of Rv1516c in whole-cell lysates of M. tuberculosis. The results suggest the potential of the identified immunodominant RD peptides in the serodiagnosis of TB.
Tuberculosis (TB) is a global infectious disease problem of major concern (34). It is estimated that almost one-third of the world population is infected with Mycobacterium tuberculosis, and 9.3 million new tuberculosis cases with 1.8 million deaths occur annually (34). Among infectious diseases, TB is the second most common killer of adults, after HIV/AIDS, and is among the overall top 10 causes of death in the world. TB was declared a global emergency in 1993 by the World Health Organization (WHO), which was considered to be the first declaration of its kind. In spite of global efforts to control TB in coordination with WHO and governments of various countries, the worldwide burden of TB is worsening, especially among the resource-poor countries of Asia and Africa, primarily due to poverty, malnutrition, immigration, HIV-M. tuberculosis coinfection, and drug resistance (34). In addition to the availability of new drugs and a vaccine to control TB, cost-effective methods/reagents for the specific diagnosis of TB are also required for global control and eradication of TB (11).
The availability of the complete genome sequence of M. tuberculosis H37Rv in 1998 (10), followed by comparative genomics studies of mycobacterial genomes, have identified 11 regions of difference (RDs) specific to M. tuberculosis but deleted/absent in most other mycobacterial organisms, including all of the vaccine strains of M. bovis BCG (7, 14). The major antigens/peptides encoded by the genes present in these RDs may be suitable for specific diagnosis of TB (24). In particular, RD1-encoded antigens have already been shown to have diagnostic potential in T-cell assays (8, 21, 28) and are widely used for diagnosis of active and latent TB, particularly in low-burden and resource-rich countries (24). However, T-cell assays are costly and cumbersome, whereas serological assays to detect antigen-specific antibodies are cost-effective, are easy to perform, and can be applied under situations prevailing in developing countries.
In the past, attempts have been made to determine the seroreactivity of 21 proteins encoded by genes present in RD1, RD2, RD4, RD5, RD6, RD7, RD11, RD14, and RD15, which were obtained as recombinant proteins expressed in Escherichia coli (1, 9, 12, 15, 19, 22, 30). However, these RDs can potentially encode a total of 70 proteins, and only 21 of these were tested for seroreactivity in the studies cited above. This was primarily due to an inability to obtain them as purified recombinant proteins because of the problems associated with DNA amplification, recombinant expression, and purification of mycobacterial proteins expressed in E. coli (1, 2, 6, 30).
To overcome the problems in obtaining full-length pure recombinant proteins of M. tuberculosis RDs, overlapping synthetic peptides are often used in T-cell assays (4, 8, 20, 21, 23, 25). Furthermore, a study with Rv3872 has also shown the potential of synthetic peptides in antibody assays (22). Therefore, in this study, we have used a similar approach to identify the peptides and proteins reactive in antibody assays by using synthetic peptides corresponding to 39 proteins of five M. tuberculosis-specific RDs (RD1, RD4, RD5, RD6, and RD7). A total of 775 peptides were tested for antibody reactivity with sera of TB patients in enzyme-linked immunosorbent assays (ELISAs). In addition, the specificity of the seroreactivity for immunodominant peptides was determined by using sera from BCG-vaccinated healthy subjects, and expression of the respective proteins in M. tuberculosis was studied using antipeptide antibodies raised in rabbits.
MATERIALS AND METHODS
Sera from TB patients and healthy subjects.
Pulmonary TB patients (smear-positive and culture-confirmed cases, n = 100) were recruited from the Chest Diseases Hospital, Kuwait, and healthy subjects (vaccinated with BCG in childhood and confirmed by the presence of a BCG scar, n = 100) were recruited from the Central Blood Bank, Kuwait. All TB patients and healthy subjects were adults, tuberculin skin test positive (induration, ≥10 mm), and negative for human immunodeficiency virus infection. The study was approved by the Ethics Committee of the Faculty of Medicine, Kuwait University, Kuwait. Informed consent was obtained from all subjects. Peripheral blood (5 ml) was collected in plain tubes, and serum samples were separated from clotted blood and kept frozen at −20°C until use.
M. tuberculosis antigens and synthetic peptides.
M. tuberculosis culture filtrate (CF), cell walls (CWs), and whole-cell lysates (WCLs) were provided by K. Dobos, Colorado State University. These preparations were produced as part of NIH, NIAID, contract no. HHSN266200400091C, entitled Tuberculosis Vaccine Testing and Research Materials, which was awarded to Colorado State University. A total of 775 synthetic peptides corresponding to 39 open reading frames (ORFs) of RD1 (12 ORFs, 220 peptides), RD4 (3 ORFs, 80 peptides), RD5 (5 ORFs, 72 peptides), RD6 (11 ORFs, 236 peptides), and RD7 (8 ORFs, 167 peptides) (Table 1) were designed from the amino acid sequences of the proteins predicted from the genome sequence of M. tuberculosis (10, 33). Each peptide was a 25-mer and overlapped with neighboring peptides by 10 amino acids (aa). The peptides were commercially synthesized by Thermo Hybaid GmBH (Ulm, Germany) using 9-fluorenylmethoxy carbonyl chemistry, as described previously (3). The stock concentrations (5 mg/ml) of the peptides were prepared in normal saline (0.9%) by vigorous pipetting, and the working concentrations were prepared by further dilution in phosphate-buffered saline (PBS; pH 7.0), as previously described (26).
TABLE 1.
Synthetic peptides corresponding to ORFs predicted in the RDs
ELISA for detection of antipeptide IgG antibodies in human sera.
The peptides were tested for antibody reactivity by ELISA with pooled sera as well as individual serum samples from TB patients and healthy subjects, according to procedures described previously (5, 12). In brief, wells of 96-well PolySorb plates (Nunc) were coated with 100 μl of 10 μg/ml of each peptide, blocked for 1 h with 100 μl of 5 mg/ml bovine serum albumin (BSA; fraction V [Sigma], pH 8.5), and then incubated for 1 h with diluted human serum (1:100) as the source of primary antibody. Wells without peptide coating and blocked with BSA were used as blanks. The positive controls were CW-coated wells (10 μg/ml) blocked with BSA and incubated with human serum, and negative controls included CW/peptide-coated wells blocked with BSA but without addition of human serum (CW/peptide control) and CW/peptide-noncoated wells blocked with BSA, followed by addition of human serum (serum control). Alkaline phosphatase-conjugated anti-human immunoglobulin G (IgG; 1:1,000; Sigma-Aldrich, Bornem, Belgium) was used as the secondary antibody, and p-nitrophenylphosphatase (pNPP) was used as the substrate for color development. Optical densities (ODs) were recorded at 405 nm. Each sample was tested in duplicate, and the mean OD at 405 nm (OD405) value was calculated. GraphPad Prism software was used for graphical presentation of the ΔOD405 for each serum sample, where ΔOD405 is the OD405 obtained with serum plus peptide minus the OD405 obtained with serum or peptide alone (whichever was higher). The ELISA results were analyzed using Windows software package SPSS, version 17.0 (SPSS, Chicago, IL), to determine median ΔOD405 values for individual peptides with sera from TB patients and healthy subjects and to calculate significant differences (P < 0.05) between the two groups using the Mann-Whitney test.
The ELISA results for a given specimen were considered positive when E/C was ≥2, where E/C is the OD405 obtained with serum plus peptide/OD405 obtained with serum or peptide alone (whichever was higher). E/C values of ≥2 were considered positive (12). The diagnostic value of immunodominant peptides was determined by analyzing the ELISA data in terms of sensitivity, specificity, positive predictive value (PPV), negative predictive value (NPV), and diagnostic accuracy according to standard procedures (32).
Raising of antibodies to RD1ORF Rv3876, RD6ORF Rv1508c, and RD6ORF Rv1516c in rabbits.
Polyclonal antibodies were raised in 4- to 8-month-old male white New Zealand rabbits against pools of 11 overlapping peptides of each protein using procedures described previously (2, 16). The peptides selected for immunization were aa 271 to 295 to aa 421 to 445 of Rv3876, aa 166 to 190 to aa 316 to 340 of Rv1508c, and aa 175 to 199 to aa 325 to 336 of Rv1516c. In brief, each peptide pool (containing individual peptides at 10 μg/ml) was emulsified with an equal volume of incomplete Freund's adjuvant (Sigma) and injected intramuscularly in the right and left thighs. The rabbits were boosted twice at 2-week intervals. The animals were bled from the ear before each immunization and 2 weeks after the last immunization, and sera were used to detect antibodies to peptides using ELISA.
ELISA to detect antibodies in rabbit sera.
Antibody reactivity in rabbit sera against the peptide pools used for immunization, individual peptides constituting each pool, CF, and WCLs were determined by ELISA by testing sera obtained from rabbits before and after immunization, according to procedures described previously (16, 17). In brief, wells of 96-well PolySorb plates (Nunc) were coated with antigens/peptides (10 μg/ml), blocked with the blocking buffer, and incubated with the primary antibody (rabbit sera at 1:100), followed by incubation with secondary antibody (alkaline phosphatase-conjugated anti-rabbit immunoglobulin G) and addition of substrate for color development, as described previously (12). The color intensity was measured by determining the OD405. Antigen/peptide-coated wells in the presence of secondary antibody alone, i.e., without primary antibody, were used as negative controls. The results were expressed in terms of E/C, which is defined above in the section on human sera. E/C values of >2 to 5 were considered weak positive, those between >5 and 10 were considered moderate positive, and those >10 were considered strong positive.
RESULTS
ELISA of RD peptides for antibody reactivity with sera from TB patients and healthy subjects.
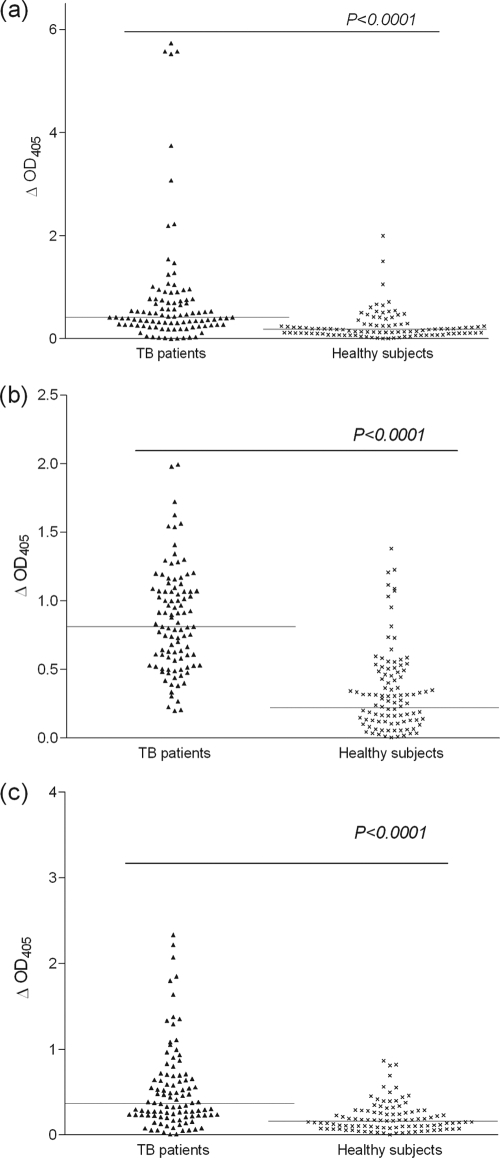
To identify seroreactive peptides of RD ORFs, all 775 peptides were individually tested in ELISAs with a pool of sera obtained from 10 TB patients, and subsequently, the reactive peptides were tested with two additional batches of pooled sera from 10 TB patients in each pool, followed by 30 individual serum samples constituting the pools. These experiments showed that 90 peptides, belonging to 28 of the 39 ORFs of RDs, had ELISA positivity with one or more serum samples, but positivity with >50% of serum samples was observed only with three peptides, i.e., aa 346 to 370 of RD1ORF Rv3876, aa 241 to 265 of RD6ORF Rv1508c, and aa 325 to 336 of RD6ORF Rv1516c (data not shown). The overall ELISA of the immunodominant peptides with sera from 100 TB patients and 100 healthy subjects showed that sera from TB patients exhibited higher ΔOD405 values with peptides of aa 346 to 370 of Rv3876, aa 241 to 265 of Rv1508c, and aa 325 to 336 of Rv1516c (median ΔOD405s = 0.42, 0.81, and 0.37, respectively) than with sera from healthy subjects (median ΔOD405s = 0.18, 0.22, and 0.16, respectively), and the ΔOD405 values for all of these peptides were significantly higher with sera from TB patients than with sera from healthy subjects (P < 0.0001) (Fig. 1 a, b, and c, respectively).
FIG. 1.
ELISA reactivities (ΔOD405 values) of peptide of aa 346 to 370 of RD1ORF Rv3876 (a), peptide of aa 241 to 265 of RD6ORF Rv1508c (b), and peptide of aa 325 to 336 of RD6ORF Rv1516c (c) with sera from 100 TB patients and 100 healthy subjects. Each spot represents the ΔOD405 value obtained with an individual serum sample. The solid lines indicate median ΔOD405 values obtained with sera from TB patients and healthy subjects.
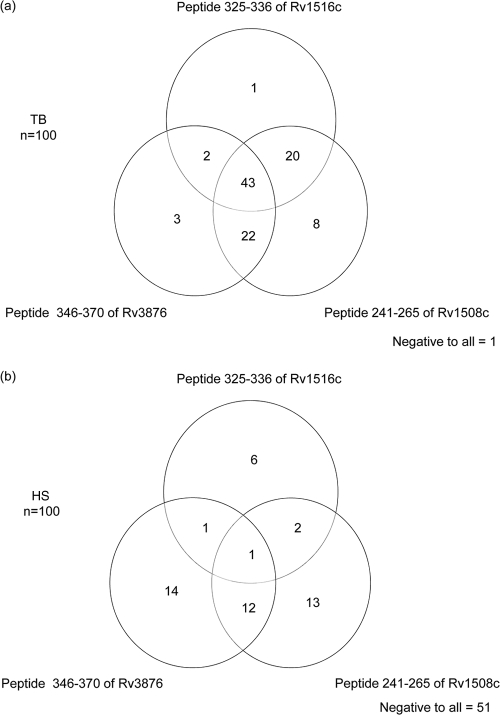
The analyses of ELISA reactivities in terms of percent positives showed that sera from 70%, 93%, and 66% of the TB patients and sera from 28%, 28%, and 10% of the healthy subjects were positive for peptides of aa 346 to 370 of Rv3876, aa 241 to 265 of Rv1508c, and aa 325 to 336 of Rv1516c, respectively (Fig. 2 a, and b, respectively). Furthermore, the results showed that the great majority (87%) of the sera from TB patients and only a small minority (16%) of sera from healthy subjects reacted to two or three peptides (Fig. 2a and b, respectively). Moreover, sera from 12% and 33% TB patients and healthy subjects, respectively, reacted to only one of the peptides, and 1% and 51% of the sera from TB patients and healthy subjects, respectively, did not show ELISA reactivity to any of the peptides (Fig. 2a and b, respectively).
FIG. 2.
Venn diagrams showing seroreactivity of TB patients (TB, n = 100) (a) and (b) healthy subjects (HS, n = 100) with peptides of aa 346 to 370 of RD1ORF Rv3876, aa 241 to 265 of RD6ORF Rv1508c, and aa 325 to 336 of RD6ORF Rv1516c.
To further determine the usefulness of the above peptides in the specific diagnosis of active TB, the results of ELISA reactivity were analyzed in terms of sensitivity, specificity, PPV, NPV, and diagnostic accuracy for both individual peptides and peptides in combination (Table 2). This analysis showed that the peptide of aa 241 to 265 of Rv1508c had the highest sensitivity (93%) and NPV (91%) but the lowest specificity (72%), whereas the peptide of aa 325 to 336 of Rv1516c had the highest specificity (90%) and PPV (89%) but the lowest sensitivity (66%) (Table 2). The combination of peptides (two or more peptides) appeared to give the best results, having the highest diagnostic accuracy (85%), when the results were compared to those obtained with the individual peptides (71%, 82%, and 78% for peptides of aa 346 to 370 of Rv3876, aa 241 to 265 of Rv1508c, and aa 325 to 336 of Rv1516c, respectively) (Table 2).
TABLE 2.
Sensitivities, specificities, PPVs, NPVs, and diagnostic accuracies of peptides of aa 346 to 370 of Rv3876, aa 241 to 265 of Rv1508c, and aa 325 to 336 of Rv1516c individually and in combination
| Peptide | Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) | Diagnostic accuracy (%) |
|---|---|---|---|---|---|
| 346-370 of Rv3876 | 70 | 72 | 72 | 71 | 71 |
| 241-265 of Rv1508c | 93 | 72 | 77 | 91 | 82 |
| 325-336 of Rv1516c | 66 | 90 | 89 | 74 | 78 |
| Two or more peptides | 87 | 84 | 84 | 86 | 85 |
Raising antipeptide antibodies in rabbits and ELISA reactivity of sera to immunizing peptides of Rv3876, Rv1508c, and Rv1516c proteins.
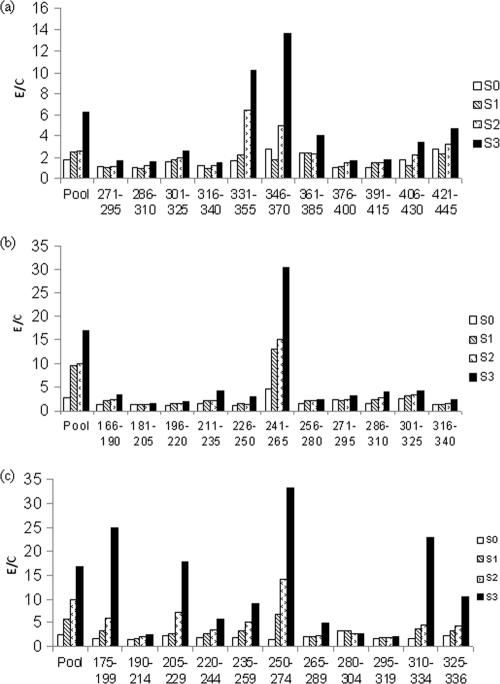
Rabbits were immunized with pools of 11 peptides of the Rv3876, Rv1508c, and Rv1516c proteins, as described in Materials and Methods. To determine if the antibodies were successfully raised against the immunizing peptide pools and to identify the immunogenic peptides in each pool, sera were collected from rabbits before and after each immunization and tested with peptide pools and individual peptides of Rv3876, Rv1508c, and Rv1516c by ELISA. The results showed that moderate to strong titers of antibodies to each peptide pool were successfully raised in rabbits after the third immunization (Fig. 3 a, b, and c). Furthermore, strong antigen-specific antibody reactivities (E/C values ≥ 10) were observed with one or more immunizing peptides of each protein, i.e., peptides of aa 331 to 355 and aa 346 to 370 of Rv3876, the peptide of aa 241 to 265 of Rv1508c, and peptides of aa 175 to 199, aa 205 to 229, aa 250 to 274, aa 310 to 334, and aa 325 to 336 of Rv1516c; moderate reactivities (E/C 5 to < 10) with peptides of aa 220 to 244 and 235 to 239 of Rv1516c; and weak to no reactivity (E/C values < 5.0) to all other peptides (Fig. 3a, b, and c). These results showed that immunization of rabbits with the peptide pools induced strong antibody reactivity to all three peptides that were found to be immunodominant with TB patient sera, i.e., peptides of aa 346 to 370 of Rv3876, aa 241 to 265 of Rv1508c, and aa 325 to 336 of Rv1516c.
FIG. 3.
ELISA reactivity of rabbit sera before (S0) and after the first (S1), second (S2), and third (S3) immunizations with peptide pools and individual peptides of RD1ORF Rv3876 (a), RD6ORF Rv1508c (b), and RD6ORF Rv1516c (c). Rabbits were immunized with pools of peptides, as described in Materials and Methods. Sera were collected before and 2 weeks after each immunization and tested in ELISAs with peptide pools and individual peptides of each pool, as described in Materials and Methods.
Expression of Rv3876, Rv1508c, and Rv1516c proteins in M. tuberculosis.
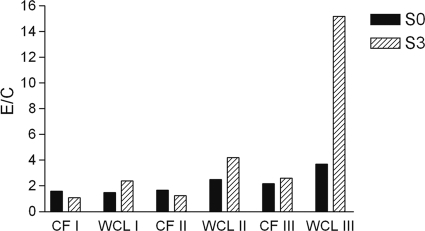
To determine the in vivo expression of Rv3876, Rv1508c, and Rv1516c in M. tuberculosis, CFs rich in secreted proteins and WCLs rich in nonsecreted proteins of M. tuberculosis were tested for antibody reactivity using sera obtained from rabbits before and after the third immunization. The E/C values were similar with CFs using sera from rabbits before and after immunization, whereas WCLs with sera from the rabbit immunized with the peptides of Rv1516c showed increased E/C values (Fig. 4). These results suggest that the Rv1516c protein was expressed in M. tuberculosis as a cytosolic or membrane-associated protein.
FIG. 4.
ELISA reactivity of CF and WCL preparations of M. tuberculosis with sera of rabbits obtained before (S0) and after the third (S3) immunization with peptide pools of RD1ORF Rv3876 (I), RD6ORF Rv1508c (II), and RD6ORF Rv1516c (III). The rabbit sera were tested for ELISA reactivity with CF and WCL preparations of M. tuberculosis using the standard procedures described in Materials and Methods. The results are presented in terms of the E/C, and values >2 were considered positive.
DISCUSSION
In the present study, ELISA was used to identify immunodominant peptides by testing TB patients' sera with 775 synthetic peptides corresponding to ORFs of 39 proteins predicted to be encoded by five M. tuberculosis-specific genomic regions that are deleted in all of the vaccine strains of M. bovis BCG (Table 1) (7). It could be argued that instead of using synthetic peptides, full-length recombinant proteins would have been ideal for antibody testing (27). This is because antibodies recognize epitopes that are mostly conformational and such epitopes are usually present in full-length proteins, whereas synthetic peptides provide linear structures, which may lack conformational epitopes (27). However, M. tuberculosis, being an intracellular pathogen, is exposed to the lytic machinery of macrophages, and, thus, the proteins of the bacteria would be fragmented into small peptides, which in turn can stimulate antibodies that recognize linear epitopes of these proteins (22). Moreover, attempts to obtain full-length RD proteins as recombinant proteins have not always been successful for various reasons, which include the high GC content of mycobacterial DNA; degradation of the expressed proteins by the host proteases; unique codon preferences, being rich in glycine, alanine, proline, and arginine; and nonbinding of the expressed proteins to affinity columns (1, 2, 6, 30). On the other hand, overlapping synthetic peptides covering the sequences of all full-length RD proteins have been successfully synthesized and used in T-cell assays (3). Thus, the use of synthetic peptides provided an opportunity to determine the antibody reactivity of all the RD proteins and identify immunodominant peptides.
The study using ELISAs to detect antibody reactivity with TB patients' sera has identified three immunodominant peptides, i.e., aa 346 to 370 of RD1ORF Rv3876, aa 241 to 265 of RD6ORF Rv1508c, and aa 361 to 372 of RD6ORF Rv1516c (Fig. 1). The antibody reactivity of these peptides was also analyzed by an artificial neural network-based B-cell epitope prediction (ABCpred) server (18), which predicts B-cell epitopes with 66% accuracy (31). The results of our analysis suggested the presence of several high-scoring (above the threshold value of 0.51) epitopes of various lengths in all of the three peptides (data not shown), thus strengthening the observation of their immunodominant recognition by antibodies in human sera.
A search performed using the Basic Local Alignment Search Tool (BLAST) of the National Center for Biotechnology Information (NCBI) to identify homologous sequences showed that all of the immunodominant peptide sequences are 100% conserved in various M. tuberculosis strains (up to 21 laboratory and clinical strains for which the sequences were available in the NCBI database). These findings suggest that differences in infecting M. tuberculosis strains may not affect the antibody reactivity to the identified peptides.
To determine the diagnostic value of the identified immunodominant peptides, the ELISA data were analyzed in terms of sensitivity, specificity, PPV, NPV, and diagnostic accuracy for individual peptides and their combinations. To minimize interplate and day-to-day variations, the ELISA readings were transformed into E/C ratios, and an E/C value of ≥2 was taken as a positive result for the presence of antipeptide antibodies (12). The analysis showed that ELISA positivity for more than one peptide may be more useful in identifying cases with active TB because the best diagnostic accuracy (85%) was observed with peptide combinations (Table 2).
The status of the proteins to which the immunodominant peptides belonged, i.e., the RD1ORF Rv3876, RD6ORF Rv1508c, and RD6ORF Rv1516c proteins, in M. tuberculosis was determined using the Tuberculist server (33), which showed that all of the three proteins are only hypothetical/probable (data not shown), implying that their expression in M. tuberculosis is unknown. Therefore, to ensure that the antibody reactivities to the peptides detected were due to infection with M. tuberculosis, it was essential to confirm their expression in M. tuberculosis. To fulfill this requirement, we have raised peptide-specific antibodies in rabbits and used them as probes to determine natural expression of the respective proteins in M. tuberculosis. As synthetic peptides are smaller and may not be immunogenic by themselves, the peptides of concern have usually been linked to a carrier protein to raise antipeptide antibodies in rabbits (29). However, a recent report has shown that synthetic peptides alone, in the presence of an appropriate adjuvant, could be sufficient to induce antipeptide antibodies in mice (13). The latter approach was used in this study to raise antipeptide antibodies in rabbits. Furthermore, to improve the possibility of raising antipeptide antibodies, peptide pools containing the immunodominant peptides and 10 additional peptides of each protein were used for immunization of rabbits. This was done to facilitate the inclusion of a sufficient number of immunogenic epitopes of each protein in the immunizing peptide pool. In addition, incomplete Freund's adjuvant was used as an adjuvant in each immunization (primary as well as booster) to increase the possibility of raising antibodies against peptides. The results show that antibodies were raised against each peptide pool as well as against some individual peptides in each pool after the third immunization. Moreover, the peptides of aa 346 to 370 of RD1ORF Rv3876, aa 241 to 265 of RD6ORF Rv1508c, and aa 325 to 336 of RD6ORF Rv1516c, which were found to be immunodominant in TB patients and which were predicted to have B-cell epitopes, were among the strongly immunogenic peptides in rabbits as well, thus confirming their immunodominance in two different species.
The expression of the RD1ORF Rv3678, RD6ORF Rv1508c, and RD6ORF Rv1516c proteins in M. tuberculosis was studied using rabbit antisera containing antibodies to peptides by ELISA using two complex preparations of proteins of M. tuberculosis, i.e., culture filtrates and whole-cell lysates. The culture filtrates are rich in secreted proteins, whereas whole-cell lysates are rich in cytosolic and membrane-associated proteins. The ELISA results show that the culture filtrate was consistently negative with antibodies to all three proteins, whereas the whole-cell lysate was positive for reactivity with the rabbit antiserum to Rv1516, thus suggesting that Rv1516c is expressed in M. tuberculosis as either membrane or cytosolic proteins.
In conclusion, the screening of RD peptides for antibody reactivity showed the immunodominance of three peptides, i.e., aa 346 to 370 of RD1ORF Rv3876, aa 241 to 265 of RD6ORF Rv1508c, and aa 325 to 336 of RD6ORF Rv1516c, with sera from TB patients but weak reactivity with sera from BCG-vaccinated healthy subjects. Antipeptide antibodies to the immunodominant peptides were successfully raised in rabbits after immunization with pools of peptides. The probing experiments with rabbit antisera suggested the expression of RD6ORF Rv1516c protein in whole-cell lysates of M. tuberculosis. Our results suggest that the identified seroreactive peptides may be useful in the specific serodiagnosis of TB.
Acknowledgments
This study was funded by Kuwait University, Research Administration Project no. YM08/07.
Footnotes
Published ahead of print on 22 December 2010.
REFERENCES
- 1.Ahmad, S., H. A. Amoudy, J. E. Thole, D. B. Young, and A. S. Mustafa. 1999. Identification of a novel protein antigen encoded by Mycobacterium tuberculosis-specific RD1 region gene. Scand. J. Immunol. 49:515-533. [DOI] [PubMed] [Google Scholar]
- 2.Ahmad, S., S. El-Shazly, A. S. Mustafa, and R. Al-Attiyah. 2005. The six mammalian cell entry proteins (Mce3A-F) encoded by the mce3 operon are expressed during in vitro growth of Mycobacterium tuberculosis. Scand. J. Immunol. 62:16-24. [DOI] [PubMed] [Google Scholar]
- 3.Al-Attiyah, R., and A. S. Mustafa. 2008. Characterization of human cellular immune responses to novel Mycobacterium tuberculosis antigens encoded by genomic regions absent in Mycobacterium bovis BCG. Infect. Immun. 76:4190-4198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Al-Attiyah, R., and A. S. Mustafa. 2010. Characterization of human cellular immune responses to Mycobacterium tuberculosis proteins encoded by genes predicted in RD15 genomic region that is absent in Mycobacterium bovis BCG. FEMS Immunol. Med. Microbiol. 59:177-187. [DOI] [PubMed] [Google Scholar]
- 5.Amoudy, H. A., A. B. H. Al-Asmer, A. T. Abul, and A. S. Mustafa. 1997. Evaluation of complex and defined antigens of Mycobacterium tuberculosis in an IgG specific ELISA for the diagnosis of tuberculosis. Med. Princ. Pract. 6:103-109. [Google Scholar]
- 6.Amoudy, H. A., and A. S. Mustafa. 2008. Amplification of six putative RD1 genes of Mycobacterium tuberculosis for cloning and expression in Escherichia coli and purification of expressed proteins. Med. Princ. Pract. 17:378-384. [DOI] [PubMed] [Google Scholar]
- 7.Behr, M. A., et al. 1999. Comparative genomics of BCG vaccines by whole-genome DNA microarray. Science 284:1520-1523. [DOI] [PubMed] [Google Scholar]
- 8.Brock, I., et al. 2004. Specific T-cell epitopes for immunoassay-based diagnosis of Mycobacterium tuberculosis infection. J. Clin. Microbiol. 42:2379-2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brusasca, P. N., et al. 2001. Immunological characterization of antigens encoded by the RD1 region of the Mycobacterium tuberculosis genome. Scand. J. Immunol. 54:448-452. [DOI] [PubMed] [Google Scholar]
- 10.Cole, S. T., et al. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537-544. [DOI] [PubMed] [Google Scholar]
- 11.Crampin, A. C., J. R. Glynn, and P. E. Fine. 2009. What has Karonga taught us? Tuberculosis studied over three decades. Int. J. Tuberc. Lung Dis. 13:153-164. [PMC free article] [PubMed] [Google Scholar]
- 12.El-Shazly, S., A. S. Mustafa, S. Ahmad, and R. Al-Attiyah. 2007. Utility of three mammalian cell entry proteins of Mycobacterium tuberculosis in serodiagnosis of tuberculosis. Int. J. Tuberc. Lung Dis. 11:676-682. [PubMed] [Google Scholar]
- 13.Felicori, L., et al. 2009. An in vivo protective response against toxic effects of the dermonecrotic protein from Loxosceles intermedia spider venom elicited by synthetic epitopes. Vaccine 27:4201-4208. [DOI] [PubMed] [Google Scholar]
- 14.Gordon, S. V., et al. 1999. Identification of variable regions in the genomes of tubercle bacilli using bacterial artificial chromosome arrays. Mol. Microbiol. 32:643-655. [DOI] [PubMed] [Google Scholar]
- 15.Greenaway, C., et al. 2005. Humoral response to Mycobacterium tuberculosis antigens in patients with tuberculosis in the Gambia. Int. J. Tuberc. Lung Dis. 9:1112-1119. [PubMed] [Google Scholar]
- 16.Hanif, S. N. M., R. Al-Attiyah, and A. S. Mustafa. 2010. Molecular cloning, expression, purification and immunological characterization of three low molecular weight proteins encoded by genes in genomic regions of difference of Mycobacterium tuberculosis. Scand. J. Immunol. 71:353-361. [DOI] [PubMed] [Google Scholar]
- 17.Harboe, M., et al. 2002. Cross-reaction between mammalian cell entry (Mce) proteins of Mycobacterium tuberculosis. Scand. J. Immunol. 56:580-587. [DOI] [PubMed] [Google Scholar]
- 18.IEDB. June 2010, updating date. Epitope prediction and analysis tools. http://tools.immuneepitope.org.
- 19.Koh, K., W. S. E. Soh, and G. T. Seah. 2009. Strong antibody responses to Mycobacterium tuberculosis PE-PGRS62 protein are associated with latent and active tuberculosis. Infect. Immun. 77:3337-3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lalvani, A., et al. 2001. Enumeration of T cells specific for RD1-encoded antigens suggests a high prevalence of latent Mycobacterium tuberculosis infection in healthy urban Indians. J. Infect. Dis. 183:469-477. [DOI] [PubMed] [Google Scholar]
- 21.Liu, X. Q., et al. 2004. Evaluation of T-cell responses to novel RD1- and RD2-encoded Mycobacterium tuberculosis gene products for specific detection of human tuberculosis infection. Infect. Immun. 72:2574-2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mukherjee, P., et al. 2007. The RD1-encoded antigen Rv3872 of Mycobacterium tuberculosis as a potential candidate for serodiagnosis of tuberculosis. Clin. Microbiol. Infect. 13:146-152. [DOI] [PubMed] [Google Scholar]
- 23.Mustafa, A. S. 2005. Recombinant and synthetic peptides to identify Mycobacterium tuberculosis antigens and epitopes of diagnostic and vaccine relevance. Tuberculosis (Edinb.) 85:367-376. [DOI] [PubMed] [Google Scholar]
- 24.Mustafa, A. S. 2010. Cell mediated immunity assays identify proteins of diagnostic and vaccine potential from genomic regions of difference of Mycobacterium tuberculosis. Kuwait Med. J. 42:98-105. [Google Scholar]
- 25.Mustafa, A. S., and F. Shaban. 2010. Mapping of Th1-cell epitope regions of Mycobacterium tuberculosis protein MPT64 (Rv1980c) using synthetic peptides and T-cell lines from M. tuberculosis-infected healthy humans. Med. Princ. Pract. 19:122-128. [DOI] [PubMed] [Google Scholar]
- 26.Mustafa, A. S., R. Al-Attiyah, S. N. Hanif, and F. A. Shaban. 2008. Efficient testing of large pools of Mycobacterium tuberculosis RD1 peptides and identification of major antigens and immunodominant peptides recognized by human Th1 cells. Clin. Vaccine Immunol. 15:916-924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oettinger, T., and A. B. Andersen. 1994. Cloning and B-cell-epitope mapping of MPT64 from Mycobacterium tuberculosis H37Rv. Infect. Immun. 62:2058-2064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Okkels, L. M., et al. 2003. PPE protein (Rv3873) from DNA segment RD1 of Mycobacterium tuberculosis: strong recognition of both specific T-cell epitopes and epitopes conserved within the PPE family. Infect. Immun. 71:6116-6123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Richman, S. J., and R. T. Reese. 1988. Immunologic modeling of a 75-kDa malarial protein with carrier-free synthetic peptides. Proc. Natl. Acad. Sci. U. S. A. 85:1662-1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rosenkrands, I. C., et al. 2008. Identification of Rv0222 from RD4 as a novel serodiagnostic target for tuberculosis. Tuberculosis (Edinb.) 88:335-343. [DOI] [PubMed] [Google Scholar]
- 31.Saha, S., and G. P. S. Raghava. 2006. Prediction of continuous B-cell epitopes in an antigen using recurrent neural network. Proteins 65:40-48. [DOI] [PubMed] [Google Scholar]
- 32.Toman, K. 1981. Sensitivity, specificity and predictive value of diagnostic test. Bull. Int. Union Tuberc. 56:18-28. [PubMed] [Google Scholar]
- 33.Tuberculist. 2008. Tuberculist World-Wide Web server. http://genolist.pasteur.fr/TubercuList/.
- 34.World Health Organization. 2009. Global tuberculosis control. Epidemiology, strategy, financing. WHO report WHO/HTM/2009.411. World Health Organization, Geneva, Switzerland.