Abstract
Human β-defensins 2 and 3 are small cationic peptides with antimicrobial activity against the fungal pathogen Candida albicans. We found that hog1 and pbs2 mutants were hypersensitive to treatment with these peptides, pointing to a role of the high-osmolarity glycerol (HOG) pathway in the response to defensin-induced cell injury.
Candida albicans is a member of the human flora, colonizing the gastrointestinal and genitourinary tracts and the skin, where it dwells as a commensal. However, if given the opportunity by the human host, C. albicans can cause superficial, mucosal, or even life-threatening deep-tissue infections, which highlights the need for rapid and effective antifungal therapies. In recent years, there has been an increasing interest in the development of antimicrobial peptides as an alternative therapy for microbial infections, mainly due to their rapid and broad-spectrum activities (6, 19).
Human β-defensins 2 and 3 (hBD-2 and hBD-3) display potent in vitro activity against C. albicans (11, 10, 21), but their antifungal mechanism is poorly understood. They do not seem to cause gross membrane disruption in C. albicans (21), and a moderate membrane permeabilization has been reported only for hBD-3 (21, 12).
C. albicans has been shown to initiate an osmotic-stress response to histatin 5, a salivary antimicrobial peptide, via activation of the high-osmolarity glycerol (HOG) mitogen-activated protein kinase (MAPK) pathway (20). Histatins and β-defensins have different structural properties and roles in human immunity (2). Thus, it was interesting to investigate if the C. albicans MAP kinases, and Hog1 in particular, were involved in the regulation of a response to human β-defensins. We exploited insertion mutant libraries developed in our laboratory (8, 13, 14, 16, 3) to investigate the genetic mechanism underlying the action of hBD-2 on C. albicans cells.
We developed a high-throughput assay to test the activities of β-defensins on C. albicans with the aid of a microplate reader, a tool known to quantify fungal growth inhibition (5). Inocula were prepared by allowing cells to grow overnight in synthetic complete (SC) medium at 30°C (4). Cells were inoculated into 3 ml of SC medium to an optical density at 600 nm (OD600) of 0.25, grown to an OD600 of approximately 1, harvested by centrifugation, washed twice in 10 mM sodium phosphate buffer (NaPB; Na2HPO4-NaH2PO4, pH 7.4), and resuspended in the same buffer. Ten microliters of the cell suspension (OD600 = 1) was added to 130 μl of hBD-2 solution in NaPB and also to the same volume of buffer only. Cells were incubated at 37°C for 1 h with shaking, and three 40-μl aliquots of each incubation mixture were loaded into a 96-well plate containing 60 μl of yeast extract-peptone-dextrose (YPD) (1× final concentration). Appropriate blanks were also included. The microplate was incubated at 30°C with orbital shaking for 24 h in a microplate reader (Tecan Infinite M100), which recorded the OD600 at 1-h intervals. The OD600 value for each strain at a given time point was calculated as the average of triplicate OD600 values, after blank subtraction. Growth curves were generated for each mutant strain and compared to that of the reference strain.
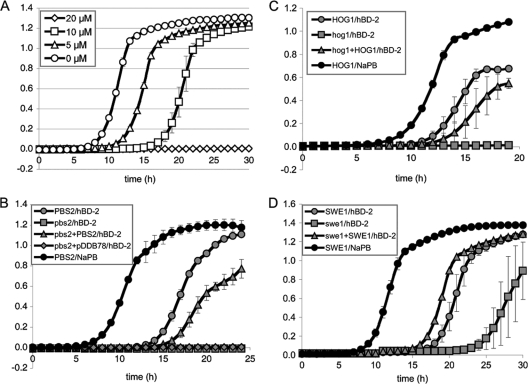
We first tested the effects of various concentrations of hBD-2 on our reference strain, DAY286. C. albicans cells were sensitive to hBD-2 under the conditions of our assay in a dose-dependent manner (Fig. 1A). Increasing concentrations of hBD-2 resulted in increasingly longer lag phases when the reference strain was grown in liquid YPD after treatment with the peptide. In this way, cells treated with NaPB alone entered exponential phase at approximately 7 h, while cells treated with 5 μM and 10 μM hBD-2 entered exponential phase at 10 and 16 h, respectively. An hBD-2 concentration of 20 μM resulted in complete lethality in the time frame of our assay, as no growth was observed during the 30 h of our assay.
Fig. 1.
(A) hBD-2 at the indicated concentrations inhibits the growth of reference strain DAY286, grown on rich medium, under our assay conditions as determined with a microplate reader. The y axis measures OD600 values. Error bars represent the standard deviations of results from three wells. (B to D) pbs2 (B), hog1 (C), and swe1 (D) mutant cells are hypersensitive to hBD-2. Error bars represent the ranges between results for at least two isolates of each mutant.
For the purpose of our screen, we chose an hBD-2 concentration of 6 μM to allow the identification of both hypersensitive and resistant mutants. We performed the screen as follows. One isolate of each mutant strain was treated with 6 μM hBD-2, and its growth curve in YPD was compared to that of the reference strain. If the mutant strain entered the exponential growth phase at approximately the same time as the reference strain, it was considered to display a wild-type phenotype for hBD-2 treatment (this is denoted by “wt” in Table S1 of the supplemental material). If the mutant strain entered exponential phase earlier or later than the reference strain, then at least one more independent isolate was tested. When treatment with hBD-2 resulted in all the independent isolates of the mutant strain entering logarithmic phase at least 4 h later than the reference strain, the mutant was considered hypersensitive (“HS” in Table S1 of the supplemental material). If the independent isolates entered logarithmic phase at least 4 h before the reference strain, then the mutant was considered resistant. If the mutant strain displayed a growth defect on YPD medium after treatment with NaPB alone, then the inhibition of growth could not be attributed to the hBD-2 treatment (these strains are denoted by “GD” for growth defect in Table S1 of the supplemental material).
Our screen of 244 mutants for transcription factors, kinases, and kinase-associated proteins resulted in the identification of three strains that were hypersensitive to hBD-2 (see Table S1 in the supplemental material). Two of these strains carried insertions in the kinase genes PBS2 and SWE1. The third strain has a homozygous null mutation for the gene encoding the MAP kinase Hog1. To our surprise, we found no mutants with a resistance phenotype.
To confirm our observations and rule out the possibility that the hypersensitivity of these mutants was due to undetected insertions in other loci, we repeated our assay with the corresponding complemented strains, and we also tested hog1 and pbs2 homozygous-deletion mutants (see the supplemental material for details on strains). Again, treatment of the pbs2, hog1, and swe1 mutant cells with 6 μM hBD-2 caused a strong inhibition of growth in comparison to the growth of the reference strain (Fig. 1B, C, and D). This was specific to the treatment with hBD-2, as reintroduction of one copy of the PBS2, HOG1, or SWE1 gene resulted in a response to hBD-2 similar to that of the reference strain. The effect of hBD-2 on pbs2 and hog1 cells was more pronounced than for swe1 cells, since the swe1 mutant resumed growth after approximately 22 h in YPD, but the pbs2 and hog1 mutants displayed no growth within the 30-hour incubation of our assay (data not shown).
We then focused on the hog1 and pbs2 mutants, since their phenotypes were more pronounced. The hypersensitivity of pbs2 and hog1 mutant cells to hBD-2 was also evident in a spot dilution assay. Cells were grown, washed, and resuspended as described for the liquid assay. Cells were subsequently incubated with different concentrations of hBD-2 in NaPB buffer in a final volume of 40 μl (OD600 = 0.5) at 37°C for 1 h, serially diluted with NaPB, and spotted onto YPD-agar plates. Plates were photographed after 2 to 3 days of growth at 30°C. Incubation with the peptide inhibited the growth of the reference strain, partially at a concentration of 6 μM and completely at 12 μM (Fig. 2A). The inhibition of growth after incubation with 6 μM hBD-2 was more pronounced for the pbs2 and hog1 mutants. As before, reintroduction of one copy of the PBS2 or HOG1 gene conferred on the mutant wild-type sensitivity to hBD-2, confirming that the inhibition of growth is specific to the hBD-2 treatment. All together, our results point to a central role of the Hog1 pathway in the response to hBD-2.
Fig. 2.
pbs2 and hog1 cells are hypersensitive to hBD-2 (A) and hBD-3 (B).
To investigate the role of the Hog1 pathway in a more general response to β-defensins, we tested the effect of hBD-3 on pbs2 and hog1 mutant cells by a spot dilution assay. Treatment with 1.5 μM hBD-3 caused a partial inhibition of the growth of the reference strain (Fig. 2B). The inhibition of growth was almost complete in the case of pbs2 and hog1 mutant cells treated with the peptide, and the corresponding complemented strains showed the same growth as the reference strain. The pbs2 and hog1 mutants were also hypersensitive to 1.5 μM hBD-3 in our liquid assay (data not shown). These results indicate that the Hog1 pathway is involved in the response to both hBD-2 and hBD-3 and suggest a general response mechanism for defensins.
Both Pbs2 and Hog1 are members of the HOG pathway, and they have been previously shown to physically interact in Saccharomyces cerevisiae (15) and in C. albicans (7) by coimmunoprecipitation. In view of our results, we were interested in investigating if the exposure of C. albicans cells to β-defensins elicited the interaction between Pbs2 and Hog1. The vesicle-capture interaction (VCI) assay recently developed by Boysen et al. (4) allows the detection of protein-protein interactions in live C. albicans cells. This novel technique exploits the sequestering of an Snf7-fused protein of interest to an endosome-derived class E compartment by the ESCRT (endosomal sorting complex required for transport) pathway, which then recruits a green fluorescent protein (GFP)-fused interacting protein. Thus, a positive interaction yields fluorescent endosomes and class E compartments (4). To assay Pbs2-Hog1 interaction, a C. albicans vps4Δ/vps4Δ strain carrying both a Pbs2-GFP fusion and a Hog1-Vps32 fusion was treated with 6 μM hBD-2, 1.5 μM hBD-3, or buffer (NaPB), and the cells were inspected for the formation of protein-protein interaction foci with a Nikon Eclipse E800 fluorescence microscope (Melville, NY) and a 100× NA 1.4 objective. Untreated cells displayed the basal level of interaction observed previously (Fig. 3) (4). Treatment with hBD-2 and hBD-3 caused the interaction of Hog1 and Pbs2, as evidenced from the presence of bright foci in the cells treated with the peptides, in contrast to what occurred in the cells treated with buffer (Fig. 3). The formation of foci resulting from the interaction of Hog1 and Pbs2 was observed rapidly after the addition of the peptides, and it was sustained over the 1-h period of our experiment (data not shown). The control cells exhibited a uniform distribution of fluorescence, with the occasional formation of small foci, possibly due to the upregulation of PBS2-GFP expression after the salt treatment. These results suggest that β-defensins elicit the interaction of Pbs2 and Hog1 in C. albicans.
Fig. 3.
Hog1 and Pbs2 interaction is augmented in a VCI assay after treatment with hBD-2 and hBD-3.
We provide the first evidence that C. albicans responds to hBD-2 and hBD-3 via the HOG pathway. Osmotic, oxidative, and heavy-metal stresses are known to activate the Hog1 MAP kinase, which in turn triggers a transcriptional response aimed to rescue the cells from the source of injury (9). Pbs2 has been previously shown to be required for osmotic- and oxidative-stress regulation of Hog1p localization and activity (1, 17). Our VCI results suggest that the injury imposed on C. albicans cells by human defensins appears to share common features with osmotic and/or oxidative stress. The serine-threonine protein kinase Rck2 and the transcription factor Sko1 have been previously implicated in the Hog1-dependent response to osmotic stress (9, 16). However, sko1 and rck2 mutant cells did not display an hBD-2 phenotype under the conditions of our assay (see Table S1 in the supplemental material), indicating that they do not play a role in the Hog1-mediated response to hBD-2.
Treatment with histatin 5, another antimicrobial peptide, has been recently shown to cause activation of Hog1, as well as the core and osmotic-stress transcriptional responses (20). Taken together, this and our results suggest that the Hog1 kinase plays a key role in the response to antimicrobial peptides. This is an interesting observation in light of the different structures and properties of the defensin and histatin molecules. Activation of the HOG pathway may have evolved in C. albicans as an adaptive response to the relatively constant exposure to antimicrobial peptides with which the fungus has to cope as a commensal of humans. Further investigations will be required to evaluate the contribution of the Hog1-mediated osmotic- and oxidative-stress responses to the rescue of cells from β-defensin activity.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Joelle E. Gabay for her advice at the onset of this project and to Ryan Subaran for providing insertion mutants. We thank members of our laboratory for advice and discussion.
This work was supported by NIH grant 7R01AI057804 to A.P.M., NIH fellowship F32AI071439 to J.R.B., and a National University of Ireland Travelling Studentship to S.F.
Footnotes
Supplemental material for this article may be found at http://ec.asm.org/.
Published ahead of print on 3 December 2010.
REFERENCES
- 1. Arana D. M., Nombela C., Alonso-Monge R., Pla J. 2005. The Pbs2 MAP kinase kinase is essential for the oxidative-stress response in the fungal pathogen Candida albicans. Microbiology 151:1033–1049 [DOI] [PubMed] [Google Scholar]
- 2. Bals R. 2000. Epithelial antimicrobial peptides in host defense against infection. Respir. Res. 1:141–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Blankenship J. R., Fanning S., Hamaker J. J., Mitchell A. P. 2010. An extensive circuitry for cell wall regulation in Candida albicans. PLoS Pathog. 6:e1000752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Boysen J. H., Fanning S., Newberg J., Murphy R. F., Mitchell A. P. 2009. Detection of protein-protein interactions through vesicle targeting. Genetics 182:33–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Broekaert W. F., Terras F. R. G., Cammue B. P. A., Vanderleyden J. 1990. An automated quantitative assay for fungal growth inhibition. FEMS Microbiol. Lett. 69:55–59 [Google Scholar]
- 6. Bulet P., Stocklin R., Menin L. 2004. Anti-microbial peptides: from invertebrates to vertebrates. Immunol. Rev. 198:169–184 [DOI] [PubMed] [Google Scholar]
- 7. Cheetham J., et al. 2007. A single MAPKKK regulates the Hog1 MAPK pathway in the pathogenic fungus Candida albicans. Mol. Biol. Cell 18:4603–4614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Davis D. A., Bruno V. M., Loza L., Filler S. G., Mitchell A. P. 2002. Candida albicans Mds3p, a conserved regulator of pH responses and virulence identified through insertional mutagenesis. Genetics 162:1573–1581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Enjalbert B., et al. 2006. Role of the Hog1 stress-activated protein kinase in the global transcriptional response to stress in the fungal pathogen Candida albicans. Mol. Biol. Cell 17:1018–1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Feng Z., et al. 2005. Human beta-defensins: differential activity against candidal species and regulation by Candida albicans. J. Dent. Res. 84:445–450 [DOI] [PubMed] [Google Scholar]
- 11. Joly S., Maze C., McCray P. B., Jr., Guthmiller J. M. 2004. Human beta-defensins 2 and 3 demonstrate strain-selective activity against oral microorganisms. J. Clin. Microbiol. 42:1024–1029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Krishnakumari V., Rangaraj N., Nagaraj R. 2009. Antifungal activities of human beta-defensins HBD-1 to HBD-3 and their C-terminal analogs Phd1 to Phd3. Antimicrob. Agents Chemother. 53:256–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nobile C. J., Mitchell A. P. 2005. Regulation of cell-surface genes and biofilm formation by the C. albicans transcription factor Bcr1p. Curr. Biol. 15:1150–1155 [DOI] [PubMed] [Google Scholar]
- 14. Norice C. T., Smith F. J., Jr., Solis N., Filler S. G., Mitchell A. P. 2007. Requirement for Candida albicans Sun41 in biofilm formation and virulence. Eukaryot. Cell 6:2046–2055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Posas F., Saito H. 1997. Osmotic activation of the HOG MAPK pathway via Ste11p MAPKKK: scaffold role of Pbs2p MAPKK. Science 276:1702–1705 [DOI] [PubMed] [Google Scholar]
- 16. Rauceo J. M., et al. 2008. Regulation of the Candida albicans cell wall damage response by transcription factor Sko1 and PAS kinase Psk1. Mol. Biol. Cell 19:2741–2751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Smith D. A., Nicholls S., Morgan B. A., Brown A. J., Quinn J. 2004. A conserved stress-activated protein kinase regulates a core stress response in the human pathogen Candida albicans. Mol. Biol. Cell 15:4179–4190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reference deleted.
- 19. Thevissen K., Kristensen H. H., Thomma B. P., Cammue B. P., Francois I. E. 2007. Therapeutic potential of antifungal plant and insect defensins. Drug Discov. Today 12:966–971 [DOI] [PubMed] [Google Scholar]
- 20. Vylkova S., Jang W. S., Li W., Nayyar N., Edgerton M. 2007. Histatin 5 initiates osmotic stress response in Candida albicans via activation of the Hog1 mitogen-activated protein kinase pathway. Eukaryot. Cell 6:1876–1888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Vylkova S., Nayyar N., Li W., Edgerton M. 2007. Human β-defensins kill Candida albicans in an energy-dependent and salt-sensitive manner without causing membrane disruption. Antimicrob. Agents Chemother. 51:154–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.