Abstract
We investigated the spatial pattern of expression of ipdC, a plant inducible gene involved in indoleacetic acid biosynthesis in Erwinia herbicola, among individual cells on plants to gain a better understanding of the role of this phenotype in the epiphytic ecology of bacteria and the factors involved in the regulation of ipdC. Nonpathogenic E. herbicola strain 299R harboring a transcriptional fusion of ipdC to gfp was inoculated onto bean plants, recovered from individual leaves 48 h after inoculation, and subjected to fluorescence in situ hybridization using a 16S rRNA oligonucleotide probe specific to strain 299R. Epifluorescence images captured through a rhodamine filter were used to distinguish the 5carboxytetramethylrhodamine-labeled cells of strain 299R from other leaf microflora. Quantification of the green fluorescence intensity of individual cells by analysis of digital images revealed that about 65% of the 299R cells recovered from bean leaves had higher ipdC expression than in culture. Additionally, 10% of the cells exhibited much higher levels of green fluorescence than the median fluorescence intensity, indicating that they are more heterogeneous with respect to ipdC expression on plants than in culture. Examination of 299R cells in situ on leaf surfaces by confocal laser scanning microscopy after fluorescence in situ hybridization of cells on leaf samples showed that even cells that were in close proximity exhibited dramatically different green fluorescence intensities, and thus, were in a physical or chemical microenvironment that induced differential expression of ipdC.
Erwinia herbicola, a common colonist of aerial plant surfaces, produces the plant hormone indoleacetic acid (IAA) in culture supplemented with tryptophan (1), like many plant-associated bacteria (2, 3). The tat and ipdC genes, encoding a tryptophan aminotransferase and an indolepyruvate decarboxylase, which are involved in the first and second enzymatic steps, respectively, of the IAA biosynthetic pathway in this species, are up-regulated under conditions of low water availability and are plant inducible (unpublished data, ref. 4). Because their expression is controlled by sigma 38 (unpublished data), other environmental conditions also may be involved in their regulation and thus, of IAA synthesis, on plant surfaces. The precise role of IAA production in nonpathogenic bacteria, such as E. herbicola, is still unclear. Previous studies have shown that the synthesis of IAA conferred increased epiphytic fitness to E. herbicola strain 299R on bean plants and pear blossoms (5). In E. herbicola pv. gypsophilae, a pathogen of Gypsophila paniculata that harbors both the IAM and the IpyA pathways, inactivation of IpdC resulted in a 14-fold reduction of its population size during exposure to dry conditions on bean plants (6). We previously have hypothesized that the release of bacterial IAA may increase the leakage of nutrients from plant cells in the vicinity of bacteria that produce it, thus benefiting the bacteria by providing additional substrates for growth (1). In view of the responsiveness of ipdC to osmotic stress and its regulation by sigma 38, IAA production on plant surfaces may be involved in a stress-regulated signaling pathway. A role for IAA production in bacterial signaling has been previously suggested (7). Based on the observations above, we speculate that ipdC will be induced only when E. herbicola is located in microsites on the leaf surface where conditions of low water availability or the presence of appropriate physico/chemical plant signals prevail. In this study, we test this hypothesis by determining the distribution of ipdC expression in individual cells of E. herbicola in culture and on plants.
Most studies of gene regulation in prokaryotic cells address transcriptional activity at the population level. However, the determination of gene expression of individual cells can reveal information about the microenvironment experienced by single cells. For example, quantification of the amount of 12-galactosidase by immunofluorescence in single cells of Ralstonia solanacearum harboring an esp-lacZ fusion and grown in tomato revealed that esp expression was heterogeneous both in culture and in plants (8). Nwoguh et al. (9) have used quantification of 12-galactosidase activity in single cells of nonculturable enteric bacteria to assess their viability after starvation. The green fluorescent protein (GFP), circumvents the use of immunodetection or of a substrate for activity (10). It can be used as a reporter gene along with fluorescence in situ hybridization (FISH), to study the behavior of single cells in situ in complex bacterial communities. For example, the application of FISH to cells harboring a fusion of rrnBP1 to gfp was used to demonstrate the heterogeneous growth activity of Pseudomonas putida cells in developing biofilms (11). In this study, we investigated the distribution of ipdC expression in single cells of E. herbicola on plants using a transcriptional fusion of ipdC to a promoterless gfp, to gain a better understanding of the role of IAA production in the ecology of plant-associated bacteria.
Materials and Methods
Strains, Plasmids, and Growth Media.
E. herbicola (Pantoea agglomerans) strain 299R has been described (1). Plasmid pVMB2 was constructed by insertion of the BamHI fragment of pMB2 (12) containing the ipdC gene and its promoter region into BamHI-restricted pVSP61 (13), a plasmid that is stably maintained in E. herbicola (4). A SacI fragment of pGreenTIR (14) harboring the gene encoding the red-shifted mutant of the GFP (10) and its promoter region then was cloned into the internal SacI site of ipdC on pVMB2 to create pVMG47. To prevent transcription of the ipdC-gfp fusion from the lacZ promoter on pVSP61, a DNA fragment containing three copies of the Ti terminator (15) was ligated into HindIII–BamHI-restricted pVMG47 to create pVMG47T. The control plasmid pKT-bla consists of plasmid pPROBE-KT (pVSP61-based) (16) harboring a constitutively expressed fusion of the promoter of the ampicillin resistance gene (bla) from pBR322 to gfp, and has been described (17). Both pVMG47T and pKT-bla were introduced into strain 299R from Escherichia coli DH5 12 with the helper plasmid pLAFR3 (18) by triparental mating. E. herbicola strain 299R was routinely grown at 28°C in M9 minimal medium containing 0.2% glucose and supplemented with kanamycin at 40 g/ml when appropriate.
Probe Design and Specificity.
A 16S rRNA probe was designed for the specific detection of E. herbicola strain 299R to account for all cells of strain 299R, including those exhibiting undetectable GFP fluorescence, in complex environmental samples. The 16S rDNA in E. herbicola strain 299R was cloned into pCRII-TOPO (Invitrogen) after PCR amplification with primers 27F (AGAGTTTGATCCTGGCTCAG) and 1492R (GGTTACCTTGTTACGACTT) (in the 5′ to 3′ direction). A variable region was sequenced by using primers 805F (ATTAGATACCCTGGTAGTC) and 1100R (AGGGTTGCGCTCGTTG). Probe Eh-299R (5′-GAGTTCCCGCCATCACG-3′) targeted this variable region at target positions 1131 to 1147 (E. coli numbering; ref. 19) and was labeled at the 5′ end with TAMRA (5-carboxytetramethylrhodamine) (Gemini Biotech, Alachua, FL). Probe Eh-299R was tested for specificity with probe match against the rRNA database of the Ribosomal Database Project II (20). The results from probe match revealed that the probe targeted only Thiocapsa rosea, Fluoribacter bozemanae, and Photorhabdus luminescens, which are unlikely to be present in any significant number on bean plants. The specificity of probe Eh-299R also was tested by hybridization to cells of various plant-associated bacterial species. Probe Eh-299R yielded a strong hybridization signal in strain 299R cells, but we failed to detect any signal from Erwinia amylovora, Pseudomonas spp., and Xanthomonas spp. Interestingly, this probe appears also to be rather strain-specific in that only strain 299R and not any of six other strains of E. herbicola that were isolated from various plant species showed a detectable hybridization signal. To further evaluate the specificity of the Eh-299R probe in environmental samples, we probed bacterial cells recovered from leaves sampled from uninoculated bean plants that were incubated under the same conditions as treated plants. Of hundreds of cells recovered from nine different leaves, only a few very faintly fluorescent cells were observed. Cells tested with the general bacterial probe EUB338 as a positive control showed strong hybridization signals. These observations suggested that cells recovered from plants had a sufficiently high ribosome content to hybridize strongly to a 16S rRNA probe and that the Eh-299R probe was specific to E. herbicola strain 299R cells on plants.
The suitability of TAMRA as a fluorescent label in combination with GFP as a fluorescent reporter gene was assessed to ensure that the emission spectrum of TAMRA did not overlap that of the red-shifted GFP. Cells of wild-type strain 299R were cultured in minimal medium and labeled with the TAMRA-Eh-299R probe. No fluorescence above the background was detected with the filter set used to collect GFP fluorescence. Quantitative measurements of the green fluorescence of cells of strain 299R(pVMG47T) grown in minimal medium, in which expression of ipdC-gfp is low, before and after hybridization with the TAMRA-labeled probe, revealed no shift in average green fluorescence intensity of the cells. These observations indicated that the TAMRA label did not interfere with the quantitative measurements of GFP fluorescence.
Plant Inoculation and Bacterial Cell Recovery.
Cells from stationary phase-cultures of strain 299R(pVMG47T) or strain 299R(pKT-bla) were washed twice in potassium phosphate buffer (10 mM, pH 7.0) before being resuspended in distilled water at 2 × 107 cells per ml. Each cell suspension was immediately applied with an atomizer to bean plants (Phaseolus vulgaris cv. Bush Blue Lake 274) (three replicate pots of eight plants per pot) at the first trifoliate leaf stage. The control treatment consisted of spraying the plants with sterile distilled water. The plants then were bagged and incubated in the laboratory at room temperature. Two primary leaves were sampled at random from each pot 48 h after inoculation. Each leaf was washed by repeatedly applying the same 2 ml of distilled water over its entire abaxial and adaxial surfaces with a Pasteur pipette. The resulting suspension of recovered bacterial cells was centrifuged, and the pellet was resuspended in fixation solution.
In Situ Hybridization.
All cells were fixed in 4% paraformaldehyde and stored in 50% ethanol in PBS at −20°C according to a protocol by Amann (21). After fixation, cultured cells and cells recovered from leaf surfaces were spotted onto gelatin-coated slides and dehydrated by successive passage through 50%, 80%, and 98% ethanol. Two to three slides were prepared from each sample of cells. The dehydrated cells were overlayed with 25 hybridization buffer (0.9 M NaCl/100 mM Tris, μl pH 8/0.1% SDS) containing 100 ng of probe and covered with a coverslip. The slides were incubated for at least 4 h at 49°C in a humidity chamber. Excess probe was removed by briefly immersing the slides in water. The slides then were incubated in 50 ml washing buffer (0.9 M NaCl/100 mM Tris, pH 8/0.1% SDS/5 mM EDTA) for 25 min at 49°C, rinsed in water, air-dried, mounted in Prolong (Molecular Probes), and sealed with rubber cement.
When FISH was carried out directly on leaves, individual bean leaves were incubated overnight in a glass Petri dish on filter paper soaked with 10% paraformaldehyde. Small disks of 1 cm in diameter then were cut from leaves with a cork borer and coated with agar to prevent redistribution of the bacterial cells by FISH procedures. Agar coating was performed as follows: each leaf disk was placed into the hole (1.5 cm) of a small square rubber sheet (1 mm thickness) glued to a microscope slide. The disk was covered with 1% agar kept at 40°C and immediately covered with a coverslip to ensure a flat surface. The coverslip was removed after 1 min, and the bacterial cells on the leaf were dehydrated through the agar by successively rinsing the agar surface with 50%, 80%, and 98% ethanol for 3 min each. The agar mount then was equilibrated by incubating it twice for 10 min with hybridization solution and once for 10 min with 30 μl of hybridization solution containing 100 ng of probe Eh-299R, before applying a final 30-μl hybridization solution with 100-ng probe. The agar mount was covered with a coverslip, and FISH was carried out as described above. After FISH, the agar-coated leaf was mounted onto a microscope slide, covered with a coverslip that was glued to the slide with rubber cement, and examined by using confocal laser scanning microscopy (CLSM).
Epifluorescence Microscopy and Image Analysis.
Bacterial cells on slides were visualized by using an Axiophot microscope fitted with a Plan-NEOFLUAR 100×/1.30 oil objective (Zeiss). Bacterial cells were observed with an Endow GFP filter set (Chroma Technology, Brattleboro, VT) for the detection of GFP fluorescence and a standard rhodamine filter set for the detection of TAMRA. The microscope was equipped with a Princeton ST138 cooled charge-coupled device camera (Princeton Instruments, Trenton, NJ). Acquisition of digitized 12-bit images was performed with the MacIntosh version 3.1 of the iplab spectrum software package (Scanalytics, Vienna, VA). Each field of view was first visualized under phase-contrast illumination followed by epifluorescence microscopy with a GFP and rhodamine filter. Exposure time with the GFP filter set was 20 or 5 s for all cells containing ipdC-gfp or bla-gfp fusions, respectively. All cells were exposed for 5 s with the rhodamine filter set. The GFP fluorescence intensity of individual cells of E. herbicola 299R(pVMG47T) or (pKT-bla) was determined with the Macintosh version 3.2 of iplab spectrum as follows: a first binary mask was created from the phase-contrast image to record the profile of each cell within a given field of view, and then overlaid on the corresponding TAMRA fluorescence image to generate a second binary mask formed of strain 299R cells only. This second mask was finally overlayed on the corresponding GFP fluorescence image to determine the intensity of the GFP signal in each cell of strain 299R. Mean pixel intensity per cell was computed from the sum of the intensities of all of the individual pixels averaged over the total number of pixels forming the cell profile.
CLSM.
The spatial distribution of ipdC-gfp expression in E. herbicola on bean leaves was determined by examining the agar-mounted leaf disks after FISH, with a Zeiss LSM150 confocal microscope equipped with argon (488 nm) and krypton (568 nm) lasers. The bacterial cells were visualized on leaves through a 63×/1.2 water objective, and rhodamine and GFP fluorescence was detected with LP 585 and BP 505–530 emission filters, respectively.
Statistical Methods.
Statistical calculations were performed on a total of at least 100 cells per leaf sample with the program statistica (version 5.1) (StatSoft, Tulsa, OK).
Results and Discussion
Frequency Distribution Analysis.
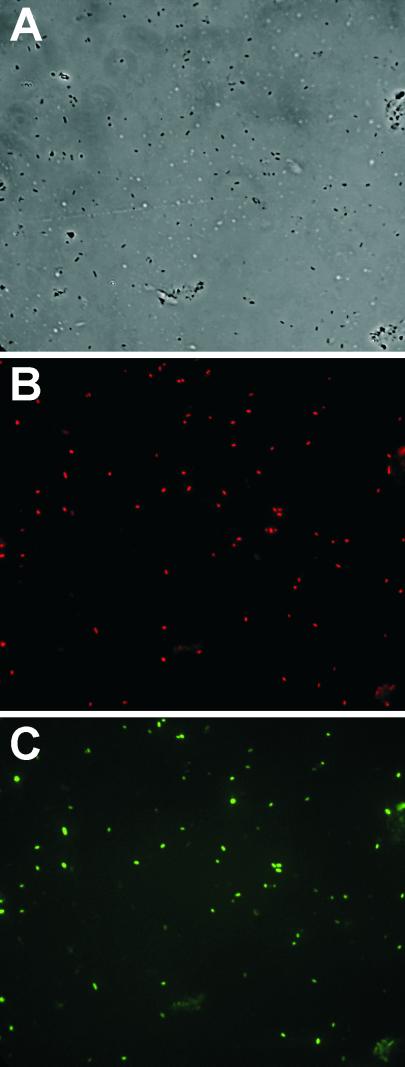
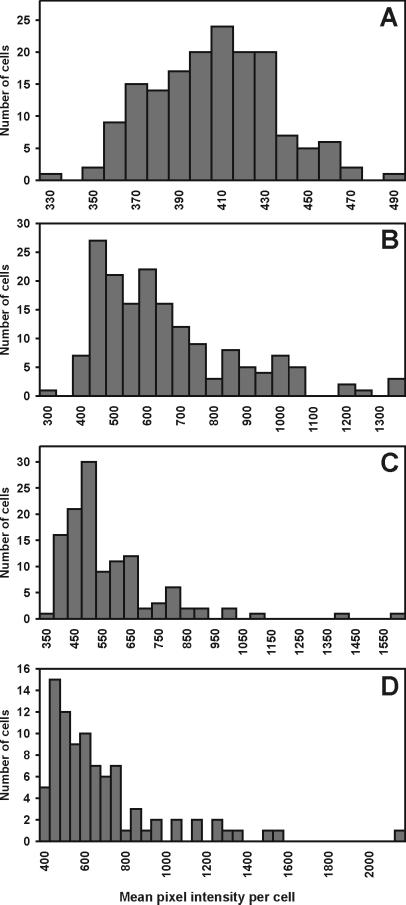
The fluorescence intensity of individual cells of E. herbicola harboring an ipdC-gfp fusion was measured both in culture and after recovery from leaves to assess the extent and uniformity with which the transcription of ipdC was induced on plants. Quantitative determination of ipdC-gfp transcriptional activity in E. herbicola cells grown on bean plant surfaces was enabled by simultaneous detection of targeted cells among the plant bacterial flora by FISH. Analysis of digital images of the green fluorescence of individual cells of strain 299R(pVMG47T) recovered from bean leaves was carried out by estimation of the mean pixel intensity of each TAMRA-labeled cell within a complex population of bacterial cells on microscope slides. As shown in Fig. 1 A and B, strain 299R was a predominant member of the bacterial microflora on bean leaves 2 days after inoculation. Although most strain 299R cells exhibited detectable levels of GFP fluorescence, the population was heterogeneous with respect to ipdC-gfp expression, with some cells showing much higher levels of green fluorescence than others (Fig. 1 B and C). This is further illustrated by comparing the frequency distribution of GFP fluorescence intensity among individual cells of strain 299R(pVMG47T) at the time of inoculation (Fig. 2A) with cells of this strain recovered from leaves 2 days after inoculation (Fig. 2 B–D). Most cells grown in culture exhibited similarly low fluorescence; cell fluorescence was normally distributed (Kolmogorov-Smirnov's d = 0.04, P > 0.2), with an average mean pixel intensity per cell of 401 (Fig. 2A). In contrast, highly right-hand skewed distributions of green fluorescence were observed among cells of strain 299R(pVMG47T) grown on plants (Fig. 2 B–D). In addition to this significant change in distribution of ipdC-gfp activity, the mean fluorescence intensity of most cells grown on leaves increased significantly compared with the average fluorescence observed in culture, whereas a few cells exhibited a much higher fluorescence. For example, at least 65% of the leaf-grown cells had a mean pixel intensity higher than 500 (Fig. 2D), whereas some cells had a mean fluorescence greater than 1,000. Similar distributions were observed for all sampled leaves. The variation in fluorescence intensity of cells was described by the log-normal frequency distribution (Shapiro-Wilk's test on log-transformed mean pixel intensity per cell: w = 0.91, P < 0.001) and suggested that those cells of strain 299R with particularly high levels of ipdC expression experienced physico/chemical conditions in the microenvironment on the plant that were responsible for induction of this gene.
Figure 1.
Micrographs of bacterial cells recovered from bean leaves 2 days after inoculation with E. herbicola 299R(pVMG47T). Only a subset of the mixed population of bacterial leaf colonists shown in the phase-contrast image (A) hybridized to the TAMRA-labeled Eh-299R probe specific for E. herbicola 299R (B). The population of E. herbicola 299R(pVMG47T) cells detected in B was heterogeneous with respect to expression of ipdC-gfp (C). (Magnification: ×10,000).
Figure 2.
Histograms of the distribution of GFP fluorescence intensity among individual cells of E. herbicola 299R(pVMG47T) grown in culture (A) and recovered from three different bean leaves 48 h after inoculation (B–D).
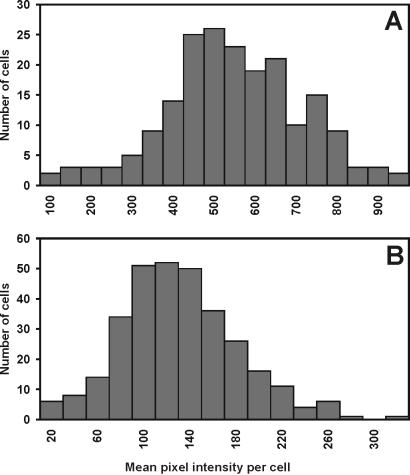
To determine whether the increase in the green fluorescence of strain 299R(pVMG47T) and its heterogeneity on bean leaves reflected true changes in the transcriptional activity of the ipdC promoter, we examined the fluorescence of 299R cells containing a fusion of the promoter of the constitutive ampicillin resistance gene (bla) to a promoterless gfp on pKT-bla, in minimal medium and on bean leaves. This fusion was expected to be constitutively expressed and therefore, insensitive to heterogeneous environments on leaf surfaces. Distribution analysis showed that GFP fluorescence of the control strain 299R(pKT-BLA) was homogenous among cells both on plants and in culture (Fig. 3). Additionally, the average mean fluorescence intensity of cells was significantly lower on plants than in culture. Examination of the variance associated with the mean fluorescence intensity revealed that unlike that of GFP fluorescence of cells containing ipdC-gfp, the variance in fluorescence levels in cells harboring bla-gfp was not higher in cells recovered from plants. This indicated that the increase in both fluorescence intensity and heterogeneity of E. herbicola 299R(pVMG47T) on leaves reflect true induction and variability in ipdC-gfp expression, and not an effect of the plant environment on GFP function per se. The reduced fluorescence of the control strain on plants may be caused by a decreased copy number of the pVSP61-based plasmid or a decreased metabolic rate of cells in that environment. This suggests that the induction of ipdC on leaves is in fact underestimated by this approach.
Figure 3.
Histograms of the distribution of GFP fluorescence intensity among individual cells of E. herbicola 299R(pKT-bla) grown in culture (A) and recovered from a bean leaf 48 h after inoculation (B).
CLSM Observations.
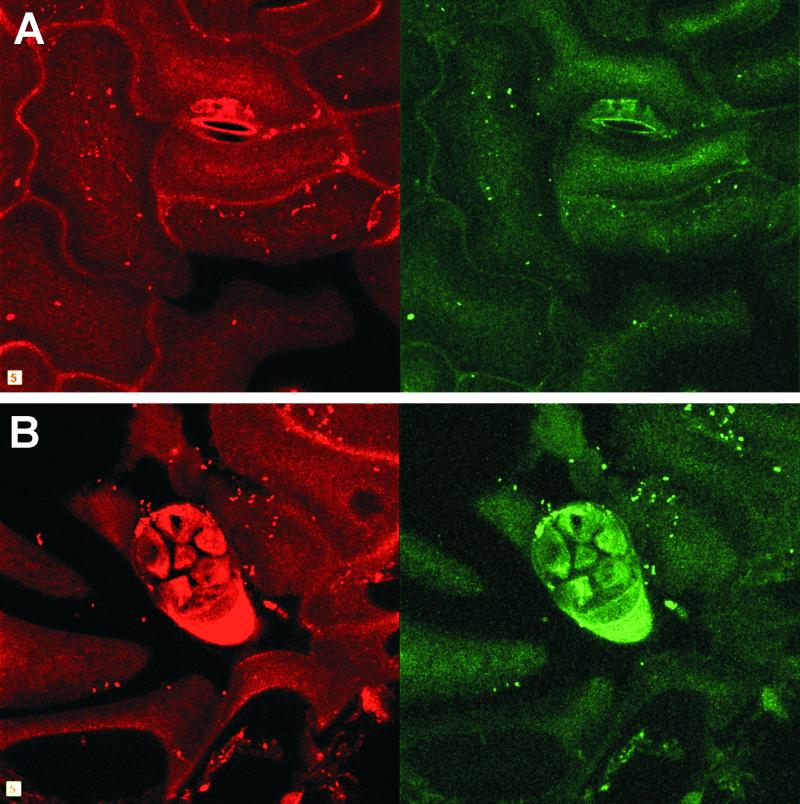
The detection of strain 299R cells with probe Eh-299R in situ on leaves was made possible by the immobilization of bacterial cells on the leaf with agar before FISH. A similar approach using acrylamide was applied successfully to study gene expression in mixed-culture biofilms formed in flow chambers (22). In situ examination of cells of strain 299R(pVMG47T) by CLSM provided insight into the spatial distribution of ipdC expression in the phyllosphere. Although most cells in close proximity exhibited similar levels of GFP fluorescence, some cells having very dim GFP fluorescence often were found near brightly fluorescent cells (Fig. 4). The slightly higher percentage of cells exhibiting low levels of green fluorescence that was observed by CLSM than that observed under the epifluorescence microscope reflects the higher detection limit of green fluorescence due to a lower signal-to-noise ratio in confocal microscopy of plant material.
Figure 4.
Spatial distribution of E. herbicola 299R(pVMG47T) and ipdC-gfp expression on colonized bean leaf adaxial surfaces 2 days after inoculation revealed by CLSM projected z-series. Strain 299R(pVMG47T) cells were detected among the leaf microflora by hybridization to the TAMRA-labeled Eh-299R probe, and imaged in the red channel (Left). The green fluorescence of strain 299R(pVMG47T) cells identified by FISH reflected ipdC-gfp expression and was recorded in the green channel (Right). Large variations in ipdC-gfp expression were detected in different areas of the bean phyllosphere, including regions around stomata (A) and glandular trichomes (B). Note that cells with high GFP fluorescence appear larger than other cells due to the halo effect caused by the high signal gain used to visualize most cells in these studies. Width of square equals 5 μm.
CLSM observations corroborated the quantitative assessment of the distribution of ipdC-gfp activity on leaves and demonstrated that although most 299R cells appeared to be induced and fluorescent, large variations in ipdC-gfp expression occurred, even of closely adjacent cells. The great spatial variation of ipdC expression on leaves, even over small scales, suggests that the leaf surface environment also varies greatly over such small scales. This observation also helps explain our previous report that the coinoculation of an IAA mutant of E. herbicola 299R with its parental strain failed to complement its reduced fitness on beans, even at high ratios of parental to mutant inoculum cells (5). Such a result is also consistent with the suggestion that the release of bacterial IAA has a very localized effect and benefit to the producing cells in the phyllosphere (5).
Although we had previously observed the plant inducibility of ipdC (4), the transcriptional activity of ipdC-inaZ in these studies only reflected an overall increase of ipdC expression among the total population of strain 299R cells recovered from plants and could not address variations in this response. Examination of the distribution of ipdC-gfp transcription in E. herbicola cells in the phyllosphere revealed that although most cells sensed their new habitat on the leaf surface and had increased ipdC expression compared with that in culture, a subpopulation of cells may have been in microenvironments where chemical and/or physical conditions were particularly conducive for expression of ipdC, and presumably for IAA production. Other studies have previously reported the heterogeneous distribution of gene expression in bacterial cells while in their natural habitat (8, 23, 24), supporting our view that cells from a given strain may experience remarkably different environments within a spatially constrained habitat.
Acknowledgments
We thank Norman Pace for advice with FISH and Steve Ruzin and Denise Schichnes of the Biological Imaging Facility of the College of Natural Resources at the University of California at Berkeley for their help with microscopy and image analysis. We are thankful to Robert Mandrell for critical reading of the manuscript. This work was supported by Grant DEB-9615280 from the National Science Foundation.
Abbreviations
- IAA
indoleacetic acid
- FISH
fluorescence in situ hybridization
- GFP
green fluorescent protein
- TAMRA
5-carboxytetramethylrhodamine
- CLSM
confocal laser scanning microscopy
References
- 1.Brandl M, Clark E M, Lindow S E. Can J Micrbiol. 1996;42:586–592. [Google Scholar]
- 2.Lindow S E, Desurmont C, Elkins R, McGourty G, Clark E M, Brandl M T. Phytopathology. 1998;88:1149–1157. doi: 10.1094/PHYTO.1998.88.11.1149. [DOI] [PubMed] [Google Scholar]
- 3.Patten C L, Glick B R. Can J Microbiol. 1996;42:207–220. doi: 10.1139/m96-032. [DOI] [PubMed] [Google Scholar]
- 4.Brandl M T, Lindow S E. Mol Plant-Microbe Interact. 1997;10:499–505. doi: 10.1094/MPMI.1998.11.7.634. [DOI] [PubMed] [Google Scholar]
- 5.Brandl M T, Lindow S E. Appl Environ Microbiol. 1998;64:3256–3263. doi: 10.1128/aem.64.9.3256-3263.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Manulis S, Haviv-Chesner A, Brandl M T, Lindow S E, Barash I. Mol Plant-Microbe Interact. 1998;11:634–642. doi: 10.1094/MPMI.1998.11.7.634. [DOI] [PubMed] [Google Scholar]
- 7.Mazzola M, White F F. J Bacteriol. 1994;176:1372–1382. doi: 10.1128/jb.176.5.1374-1382.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kang Y, Saile E, Schell M A, Denny T P. Appl Environ Microbiol. 1999;65:2356–2362. doi: 10.1128/aem.65.6.2356-2362.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nwoguh C E, Harwood C R, Barer M R. Mol Microbiol. 1995;17:545–554. doi: 10.1111/j.1365-2958.1995.mmi_17030545.x. [DOI] [PubMed] [Google Scholar]
- 10.Chalfie M, Tu Y, Euskirchen G, Ward W W, Prasher D C. Science. 1994;263:802–805. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- 11.Sternberg C, Christensen B B, Johansen T, Toftgaard Nielsen A, Bo Andersen J, Givskov M, Molin S. Appl Environ Microbiol. 1999;65:4108–4117. doi: 10.1128/aem.65.9.4108-4117.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brandl M T, Lindow S E. Appl Environ Microbiol. 1996;62:4121–4128. doi: 10.1128/aem.62.11.4121-4128.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Loper J E, Lindow S E. Appl Environ Microbiol. 1994;60:1934–1941. doi: 10.1128/aem.60.6.1934-1941.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miller W G, Lindow S E. Gene. 1997;191:149–153. doi: 10.1016/s0378-1119(97)00051-6. [DOI] [PubMed] [Google Scholar]
- 15.Brosius J. Gene. 1984;27:161–172. doi: 10.1016/0378-1119(84)90137-9. [DOI] [PubMed] [Google Scholar]
- 16.Miller W G, Leveau J H J, Lindow S E. Mol Plant-Microbe Interact. 2000;13:1243–1250. doi: 10.1094/MPMI.2000.13.11.1243. [DOI] [PubMed] [Google Scholar]
- 17.Miller W G, Brandl M T, Quiñones B, Lindow S E. Appl Environ Microbiol. 2001;67:1308–1317. doi: 10.1128/AEM.67.3.1308-1317.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ditta G, Stanfield S, Corbin D, Helinsli D. Proc Natl Acad Sci USA. 1980;77:7347–7351. doi: 10.1073/pnas.77.12.7347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brosius J, Dull T J, Sleeter D D, Noller H F. J Mol Biol. 1981;148:107–127. doi: 10.1016/0022-2836(81)90508-8. [DOI] [PubMed] [Google Scholar]
- 20.Maidak B L, Cole J R, Parker C T, Garrity G M, Jr, Larsen N, Li B, Lilburn T G, McCaughey M J, Olsen G J, Overbeek R, et al. Nucleic Acid Res. 1999;27:171–173. doi: 10.1093/nar/27.1.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Amann R I. In: Molecular Microbial Ecology Manual. Akkermans A D L, Van Elsas J D, De Bruijn F J, editors. Dordrecht, The Netherlands: Kluwer; 1995. pp. 1–15. [Google Scholar]
- 22.Moller S, Sternberg C, Bo Andersen J, Christensen B B, Ramos J L, Givskov M, Molin S. Appl Environ Microbiol. 1998;64:721–732. doi: 10.1128/aem.64.2.721-732.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Joyner D C, Lindow S E. Microbiol. 2000;146:2435–2445. doi: 10.1099/00221287-146-10-2435. [DOI] [PubMed] [Google Scholar]
- 24.Leveau J H J, Lindow S E. Proc Natl Acad Sci USA. 2001;98:3446–3453. doi: 10.1073/pnas.061629598. [DOI] [PMC free article] [PubMed] [Google Scholar]