Abstract
The phosphate signal transduction (PHO) pathway, which regulates genes in response to phosphate starvation, is well defined in Saccharomyces cerevisiae. We asked whether the PHO pathway was the same in the distantly related fission yeast Schizosaccharomyces pombe. We screened a deletion collection for mutants aberrant in phosphatase activity, which is primarily a consequence of pho1+ transcription. We identified a novel zinc finger-containing protein (encoded by spbc27b12.11c+), which we have named pho7+, that is essential for pho1+ transcriptional induction during phosphate starvation. Few of the S. cerevisiae genes involved in the PHO pathway appear to be involved in the regulation of the phosphate starvation response in S. pombe. Only the most upstream genes in the PHO pathway in S. cerevisiae (ADO1, DDP1, and PPN1) share a similar role in both yeasts. Because ADO1 and DDP1 regulate ATP and IP7 levels, we hypothesize that the ancestor of these yeasts must have sensed similar metabolites in response to phosphate starvation but have evolved distinct mechanisms in parallel to sense these metabolites and induce phosphate starvation genes.
The cellular homeostasis of inorganic phosphate is required for optimal growth and efficient metabolism. The response of the model organism Saccharomyces cerevisiae to extracellular phosphate starvation is well characterized and mediated by the phosphate signal transduction (PHO) pathway (20, 24). To determine whether the PHO pathway is conserved in other Ascomycota fungal species, we screened for PHO pathway mutants in the evolutionarily distantly related Schizosaccharomyces pombe, which last shared a common ancestor with S. cerevisiae more than 1 billion years ago (7).
The PHO pathway in S. cerevisiae often is defined by the regulation of PHO5, which encodes a phosphate starvation-regulated acid phosphatase (17, 20). PHO5 is highly induced during phosphate starvation. ScPho5 activity is detected using a diazo-coupling assay with 1-napthylphosphate (9). Numerous studies have determined that PHO5 transcription is regulated by the specific transcription factors Pho4 and Pho2 and by more general chromatin remodeling complexes, such as SWI/SNF, SAGA, and INO80 (1, 16, 31). Pho4 localization and activity is regulated by a cyclin/cyclin-dependent kinase complex (Pho81/Pho80/Pho85) (11, 12, 25). During high extracellular phosphate conditions, the kinase complex is active and phosphorylates Pho4, leading to nuclear exclusion and little transcription of PHO5 (10, 15). During low extracellular phosphate conditions, Pho81 inhibits the kinase complex through a noncovalent interaction with IP7 (inositol heptakisphosphate) (18, 19). Certain isomers of IP7 increase in abundance in response to phosphate starvation, although how extracellular phosphate concentration leads to these increases is unclear. However, Vip1 is required to phosphorylate IP6 to form 4-PP-IP5 or 6-PP-IP5, and Ddp1 is required for dephosphorylation back to IP6 (19). Increases in IP7 during extracellular phosphate starvation lead to an in vitro loss of the Pho81/Pho80/Pho85 kinase activity through a noncovalent binding of IP7 to Pho81 (18). Unphosphorylated Pho4 is nuclear localized, interacts with Pho2, and cooperatively transcribes PHO5 (15).
In S. pombe, pho1+ is an ortholog of PHO5, and levels of pho1+ transcript increase during phosphate starvation (22, 29, 30). Based on similarity searches, there are no clear orthologs of PHO4, PHO2, or PHO81 in S. pombe, and the closest homologs of PHO80 and PHO85 in S. pombe appear uninvolved in the regulation of acid phosphatase activity (32). Thus, while a similar phosphate starvation response appears in S. pombe relative to S. cerevisiae in which genes that encode structural proteins, such as SpPho1 and SpPho84 (encoding a high-affinity phosphate transporter), are transcriptionally induced during starvation, it is unclear how the starvation response is regulated. To gain insight into the regulation of the PHO pathway in S. pombe, we employed a visual assay of mutants from an S. pombe deletion collection (26) to identify mutants that inappropriately regulate acid phosphatase activity. We identified both uninducible and constitutive mutants, allowing for the description of a genetic pathway regulating pho1+. This genetic pathway includes a putative transcription factor, Pho7, as well as the SWI/SNF chromatin remodeling complex. We conclude from this genetic screen that there is significant flexibility in the regulatory pathway used to sense and respond to phosphate starvation in yeasts.
MATERIALS AND METHODS
Growth conditions and strains.
S. pombe cells were grown in a previously described medium, YES or EMM (5). S. pombe strains used were DP1 (972 h−), DP2 (975 h+), DP3 (ade6-M216 leu1-32 ura4-D18 h+), and DP4 (ade6-M210 his3-D1 leu1-32 ura4-D18 h−), from the laboratory of D. Moazed (Harvard University), and TH4 (an EMS mutant of DP1). DP1 through DP4 are wild-type strains with regard to phosphatase expression, and TH4 constitutively expresses phosphatase. The version 2 deletion collection, purchased from Bioneer (http://pombe.bioneer.co.kr/), was ade6-M216 leu1-32 ura4-D18 h+, and it was manipulated in 384-well format with a Singer robot (14). DP18 (ura4-D18 h+) was used for the plasmid repair of pho7+ and snf5+ through homologous recombination. The strain DP55 is pho1ΔKANMX6 ade6-M210 h+. Screening for uninducible mutants required a modified medium. This medium is 90% SD–10% EMM with no phosphate, which is 0.7 g YNB with no phosphate, 3 g EMM with no phosphate, 4.5 g (NH4)2SO4, 18 g glucose, 0.25 g SP, and 10 mM KH2PO4 (for high-phosphate media only) per 1 liter water (Sunrise Science). The no-phosphate medium 90% SD–0% EMM was used for all phosphate starvation growth conditions. For solid plates, 2% Bacto-agar (Difco) was included.
Visual screen of deletion collection.
A collection of 2,813 deletion strains (14, 26) was arrayed in 384-well format onto solid medium (YES or 90% SD–10% EMM with no phosphate) for ∼6 h. Phosphatase activity was assayed by 1-napthylphosphate and FBSB as described previously (13). Photographs were analyzed utilizing Adobe Photoshop in grayscale using the control strains to determine the cutoff. A list of all mutant strains that were scored as aberrant in either assay relative to the controls is in Table S1 in the supplemental material.
Inactivation of genes in S. pombe.
The deletion of pho1+ (strain DP55) was performed utilizing a fusion PCR protocol with a KANMX6 marker as the core sequence and primers described in Table S3 in the supplemental material (13, 21). The transformation of S. pombe was with lithium acetate and polyethylene glycol 8000 (5). We also deleted pho7+ (strain DP81), asp1+ (strains DP102 and DP103), and snf5+ (strain DP82) from strain 972 using a similar protocol with primers shown in Table S3 in the supplemental material, and we confirmed the phosphatase-uninducible phenotype of pho7Δ and snf5Δ independently.
Secondary screen for linkage of phenotype to gene deletion.
Primers corresponding to each open reading frame (ORF) are listed in Table S3. Genomic DNA preparations followed a modified phenol-chloroform protocol (13). To positively confirm each deletion strain, we required that no amplification product be present when primers corresponding to the deleted gene were used, but for these primers to amplify product from other unrelated mutant DNA. Furthermore, we required that primers not corresponding to the deleted gene would amplify other ORFs to confirm that the DNA preparation was suitable for PCR. To confirm the linkage of the mutant phenotype to the KANMX4 insertion, we backcrossed the library strains to DP1 (h−) on ME solid medium for 3 days and subjected the cross to random spore analysis by the incubation of cells with zymolyase overnight (5). We picked 16 progeny and determined whether the aberrant phosphatase phenotype was linked to the G418-resistant phenotype (KANMX6 confers resistance to G418). This analysis only determines linkage, but this is sufficient to eliminate most phenotypes not caused by the gene deletion. In a few cases, we confirmed our results by tetrad analysis.
PNPP assay.
Strains were grown in 5 ml of YES at 30°C to an optical density at 600 nm (OD600) of 0.15 to 0.3 (approximately 5 h). Cultures were harvested, washed with 90% SD–10% EMM no-phosphate medium, and resuspended at a low density. Cultures were grown in triplicate in 90% SD–10% EMM no-phosphate and high-phosphate media for 16 h at 30°C. Cells were centrifuged, resuspended in water, and assayed at pH 4 as described previously (13) for 10 min. PNPP (p-nitrophenyl phosphate) hydrolysis was measured at OD400. The OD400/OD600 ratio for each reaction was calculated, accounting for dilutions. Student's t tests compared mutant strain phosphatase activity to that of the wild type, and differences were considered significant at P < 0.05.
Time course of pho1+ and pho84+ expression.
Strains were grown in 90% SD–10% EMM liquid medium at 30°C until logarithmic-growth phase (OD600 of 0.2 to 0.5). Cells were pelleted by centrifugation, washed three times, transferred to medium lacking phosphate, and grown at 30°C for 4 h. During the 4 h, cells were harvested at 1-h time points starting immediately after inoculation into medium lacking phosphate. Quantitative reverse-transcription PCR was used to measure the amount of pho84+, pho1+, and act1+ transcript.
RT-qPCR.
For reverse transcription-quantitative PCR (RT-qPCR), mutant strains were grown in 90% SD–10% EMM high-phosphate liquid medium to logarithmic growth phase, washed, inoculated into 90% SD no-phosphate liquid medium, and grown for 4 h. For the analysis of transcript levels in high-phosphate conditions, cells were harvested immediately from logarithmically grown cultures. RNA was purified by an acid phenol protocol (13). The concentration of the RNA was quantified spectrophotometrically. One microgram of RNA was reverse transcribed using an iScript cDNA synthesis kit (Bio-Rad), along with a no-reverse-transcriptase control. The samples were diluted 1:15. Transcripts from 3 μl of the diluted reaction were quantified using a Bio-Rad Chromo-4 with 50 μl Sybr green I reaction mixtures. The amplification protocol was 40 cycles of 94°C for 30 s, 60 to 68°C (depending on primers) for 30 s, 72°C for 45 s, and then 79°C for 10 s, and then a plate read was recorded. A melting curve confirmed the purity of the amplification product, and levels of transcripts were measured relative to a standard curve of genomic DNA. pho1+ and pho84+ transcript amounts were normalized to act1+ transcript, which we confirmed did not change in abundance during phosphate starvation (data not shown). Student's t tests were performed on each of the strains to compare their pho1+ and pho84+ transcription levels to those of the wild type (DP3).
Epistasis.
We exchanged the marker in the constitutive mutants (from KANMX4 to NATMX6) using a PCR product (26) and backcrossed these mutants to confirm that only one NATMX6 insertion was causing the mutant constitutive phenotype. Backcrossed, constitutive, h− mating type strains (ado1Δ, csk1Δ, spcc1393.13Δ, ppn1Δ, and aps1Δ) then were crossed on ME solid medium to the uninducible h+ strains pho7Δ and snf5Δ and the other uninducible strains (data not shown). Several G418R and NATR spores were chosen for each cross, and their mating types were checked by PCR. Additionally, PCR confirmed that the resultant strains had both genes deleted. A phosphatase plate assay was used to identify the phenotype of each double mutant strain, and at least two independent meiotic products with the same genotype were analyzed to confirm the phenotype. To generate plasmids for complementation, we utilized homologous recombination with PCR products of pho7+ and snf5+ to repair a PacI-digested pUR18-YFP plasmid (3). After repair in an S. pombe ura4− strain (DP18), plasmids were rescued into XL1-Blue cells and plasmid DNA was transformed into mutant strains.
Induction of carbon and nitrogen starvation related transcripts.
Strains from the deletion collection that were mutant in pho7+ and snf5+, as well as the wild-type strain (DP3), were grown in EMM plus 3% glucose medium and transferred in triplicate to either EMM 0.1% glucose plus 3% glycerol for 4 h or EMM without nitrogen for 2 h. Cells were harvested, and RNA was subjected to RT-qPCR as previously described. qPCR was used to measure the transcription of fbp1+,which is induced during the shift from glucose to glycerol, and isp6+, which is induced during nitrogen starvation. Both transcripts were normalized to act1+.
RESULTS
Identification of S. pombe mutants aberrant in phosphatase expression.
Given previous studies of pho1+ (30), we predicted that Pho1 is an important acid phosphatase induced during phosphate starvation, and that it is detectable with a diazo coupling reaction (25). We inactivated pho1+ with a KANMX6 cassette and determined the effect of the deletion with an assay on solid medium (Fig. 1A). The lack of phosphatase phenotype (white color) was clearest with a modified S. cerevisiae no-phosphate medium (see Materials and Methods), as there are other putative phosphatases in wild-type S. pombe (35). Preliminarily, we screened with a forward genetic approach and identified a few mutants that aberrantly regulate phosphatase activity (data not shown). We chose a constitutive mutant (TH4) that was a consequence of a single locus based on meiotic analysis (data not shown); we used it and a pho1Δ uninducible strain as controls on either high-phosphate or low-phosphate plates (Fig. 1B).
Fig. 1.
Colorimetric plate phosphatase assays. (A) DP4 and DP3 are wild-type strains; DP55 is a pho1Δ strain, and TH4 is a constitutive mutant. Dark color indicates phosphatase activity. (B) Photograph of a phosphatase plate assay demonstrating the range of phenotypes observed on a sample 384 plate. Coordinate I8 corresponds to the pho1Δ strain in the library. Coordinates A17 and O3 correspond to the wild type and A20 and O5 to the pho1Δ strain we included as controls. The strain at E3 is the pho7Δ strain. (C) Enlarged pictures of mutants. Mutants are shown in the bottom left corner of each tile. Both ado1Δ and aps1Δ mutants are darker in color than surrounding colonies on high-phosphate media, indicating that they are constitutive mutants, and snf5Δ and pho7Δ are lighter than the surrounding strains grown on no-phosphate media, indicating that they are uninducible mutants.
We screened 2,813 S. pombe deletion strains in 384-well format for aberrant phosphatase expression, with the inclusion of controls. The collection was arrayed on YES (high-phosphate) plates and no-phosphate plates, assayed for phosphatase activity using a colorimetric assay (diazo coupling reaction), and then photographed. This assay was repeated on a duplicate array, and photographs were scored blindly for aberrant phenotypes. Approximately 70 strains in total were scored as aberrant in at least one of the screens (see Table S1 in the supplemental material). Strains that appeared either constitutive (8 strains) or uninducible (25 strains) in both screens were chosen for further characterization (see Table S2 in the supplemental material).
To confirm that the mutant phenotypes resulted from the deletion of the predicted gene, we determined whether the ORF was present or absent by amplifying a 300-bp product near the 3′ end of each ORF. We expected that if the appropriate gene was deleted, we would be unable to amplify the ORF but would be able to amplify other ORFs, indicating that a negative result was not a consequence of poor DNA quality. Five of the 33 strains appeared to contain the ORF, suggesting that they were not deleted for the indicated gene; however, it is worth noting that this assay is susceptible to false positives due to contamination or to the partial integration of the KANMX4 cassette. To determine whether the mutant phenotype was linked to the KANMX4 insertion, we crossed each mutant to a wild-type strain (DP1) and assayed the segregation of the mutant phenotype as well as resistance to G418. Nine strains were deleted for the appropriate gene, but the aberrant phosphatase phenotype was not linked to the deletion. Through these two analyses, we identified 11 mutants associated with the uninducible phenotype and 5 mutants with the constitutive phenotype (Table 1). Whereas it appears that there is a high level of false positives, it should be emphasized that our verification procedure was different from the published verification of the collection strains, possibly explaining the discrepancy (14).
Table 1.
List of mutants, their putative functions, and their S. cerevisiae homologs
| S. pombe systematic name | S. pombe standard name | S. cerevisiae ortholog(s) | S. cerevisiae standard name | Product |
|---|---|---|---|---|
| S. pombe systematic name | S. pombe standard name | S. cerevisiae ortholog(s) | S. cerevisiae standard name | Product |
| Uninducible | ||||
| spbc27b12.11c | Pho7 | None apparent | Zinc finger domain transcription factora | |
| spac2f7.08c | Snf5 | YBR289W | Snf5 | SWI/SNF complex subunit Snf5 |
| spbc106.10 | Pka1 | YJL164C, YPL203W, YKL166C | Tpk1 | cyclic AMP-dependent protein kinase catalytic subunit Pka1 |
| spac1071.04c | Spc2 | YML055W | Spc2 | Signal peptidase subunit Spc2a |
| spac13g7.06 | Met16 | YPR167C | Met16 | Phosphoadenosine phosphosulfate reductase |
| spac23h3.13c | Gpa2 | YER020W | Gpa2 | Heterotrimeric G protein alpha-2 subunit Gpa2 |
| spcc757.10 | Vph2 | YKL119C | Vph2 | Endoplasmic reticulum membrane protein involved in assembly of the V-ATPasea |
| spbc3b9.11c | Ctf1 | YGL044C | Rna15 | mRNA cleavage and polyadenylation specificity factor complex subunit Ctf1 |
| spac4c5.02c | Ryh1 | YLR262C | Ypt6 | GTPase Ryh1 |
| spac4f10.04 | YIL153W | Rrd1 | Protein phosphatase type 2A, intrinsic regulatora | |
| spac17h9.04c | YDL167C | Nrp1 | RNA-binding protein | |
| Constitutive | ||||
| spcc338.14 | YJR105W | Ado1 | Adenosine kinasea | |
| spcc1393.13 | YMR027W | DUF89 family protein | ||
| spac1d4.06c | Csk1 | YKL139W | Ctk1 | Cyclin-dependent kinase activating kinase Csk1 |
| spbc713.07c | YDR452W | Ppn1 | Vacuolar polyphosphatasea | |
| spac13g6.14 | Aps1 | YOR163W | Ddp1 | Diadenosine 5′,5′-p1,p6-hexaphosphate hydrolase Aps1 |
Predicted product.
Because some mutants did not exhibit a strong phenotype in the visual phosphatase assay, we quantified phosphatase activity utilizing hydrolysis of p-nitrophenylphosphate with cells grown in high- and low-phosphate conditions (9) (Fig. 2). Based on phosphatase activity, we divided the mutants into three classes: uninducible, constitutive, and hyperinducible. Figure 2 presents only mutants that were statistically different from the wild type and were linked to the deletion, but all of the mutants initially identified are shown in the supplementary information (see Fig. S1 in the supplemental material).
Fig. 2.
Histogram of acid phosphatase activity. Mutants' phosphatase activity was measured using PNPP as a substrate after 16 h of growth in high- and no-phosphate conditions. The strains were ordered by their phosphatase activity during phosphate starvation from least to most activity. All tests were performed in triplicate, and the errors are standard errors. The wild type (DP3) is shown in yellow.
Uninducible mutants.
Eleven uninducible mutants were defined as mutants that did not have wild-type levels of phosphatase activity during phosphate starvation (based on a Student's t test). The uninducible mutants, in general, fall into four classes of genes that, when deleted, express less phosphatase activity than the wild type (Table 1 has a list of mutants and their putative functions). Two mutants affect G protein signaling (pka1+ and gpa2+), two mutants appear to bind RNA (nrp1+ and ctf1+), three mutants may affect protein secretion (spc2+, vph2+, and rhy2+), and three mutants appear to be involved in signaling (a pp2A+ subunit, a zinc finger-containing protein, and snf5+). One additional mutant is defective in PAPS reductase (met16+). A more extensive discussion of how these genes may be involved in signaling phosphate starvation is below. Because essential genes are missing from the collection and because we identified many other mutants that were not linked to a gene deletion, it is likely that other genes required for the increase in phosphatase activity during phosphate starvation remain to be identified.
To identify mutants defective in the phosphate starvation response and not just mutants affecting pho1+ expression and Pho1 secretion, we determined the transcript abundance of other genes during phosphate starvation, such as pho84+, a putative high-affinity phosphate transporter that is regulated by phosphate starvation. We performed a detailed time course of induction to determine the best time to analyze all of the mutants for defects in the transcription of both pho1+ and pho84+ (Fig. 3). We determined that 4 h was an appropriate length of time of phosphate starvation. We examined the expression of these genes in a wild-type strain (DP3) as well as in a strain from the deletion collection harboring a deletion of a gene (which we have named pho7+) that we speculated was important for the phosphate starvation response. Whereas the wild-type strain induced pho1+ and pho84+ ∼5-fold during the period of 4 h, the pho7Δ strain only minimally induced both transcripts, suggesting that pho7+ is required for the increase in the expression of both phosphate starvation-regulated genes (Fig. 3).
Fig. 3.
Time course of transcript levels of pho1+ and pho84+ in a wild-type and a pho7Δ strain. DP3 and the pho7Δ strain were grown as described in Materials and Methods. Errors are standard errors of three separately grown replicates. The ratios that were normalized to 100% are pho1+/act1+ at 1.57 and pho84+/act1+ at 0.71 under phosphate starvation conditions.
To determine whether the mutant strains were affected in their ability to induce the transcription of both pho1+ and pho84+, we grew each mutant strain identified in Table 1 for 4 h in medium lacking phosphate and measured the amount of transcript of the two genes using quantitative PCR of reverse transcribed RNA. We normalized these transcript levels to act1+. Most of the mutants did not significantly differ from the wild type in levels of either transcript; however, because of error and only a 2-fold difference in pho84+ induction, some of these mutants may have subtle defects (Fig. 4).
Fig. 4.
Induction of pho1+ and pho84+ transcription in mutants. Transcript levels were measured by RT-qPCR in 10 uninducible mutant strains after 4 h of phosphate starvation. Errors are standard errors of three separately grown replicates. The wild-type strain (DP3) was used for normalization, and the ratio for pho1+/act1+ is 1.44 and for pho84+/act1+ is 0.41.
Five mutants expressed pho1+ at significantly lower levels than that of the wild type. Three (spc2Δ, gpa2Δ, and ryh1Δ) had significant defects in pho1+ expression but no apparent defects in pho84+ expression, suggesting that they only affect the transcription of pho1+. Interestingly, ryh1+ has been demonstrated previously to affect the glycosylation of Pho1 during secretion, suggesting that there is a feedback mechanism regulating pho1+ transcription (6), and perhaps spc2+ is regulating the same process. Because gpa2+ and pka1+ were identified in our screen and these mutants alter mycelial development during nitrogen starvation, it is tempting to speculate that the PKA pathway is required for the optimal regulation of multiple starvation pathways (4). However, components of the PKA pathway, other than gpa2+ and pka1+, did not appear to have phosphatase phenotypes in our screen.
Importantly, two mutants, snf5Δ and pho7Δ (we have named the latter spbc27b12.11c+ pho7+ because of its apparent role in the PHO pathway), are defective in the induction of both transcripts, suggesting a more general role in regulating the transcription of phosphate starvation genes. Both likely are involved in transcription, as Snf5 is a subunit of the SWI/SNF chromatin remodeling complex (23) and Pho7 contains a zinc finger domain, which often is associated with transcription. Furthermore, SWI/SNF appears to be required for the induction of pho1+, as the deletion of snf5+ in a previous study lowered pho1+ expression ∼8-fold (23).
Constitutive mutants.
Four mutants (ado1Δ, spcc1393.13Δ, csk1Δ, and aps1Δ) have statistically elevated levels of phosphatase activity during high-phosphate growth, and one mutant has a hyperinducible phenotype (ppn1Δ) (Fig. 2). Levels of pho1+ and pho84+ transcript in high-phosphate conditions were quantified using quantitative PCR to confirm the constitutive phosphatase expression (Fig. 5). Interestingly, whereas there is little overlap between the genes identified by the uninducible screens of S. cerevisiae and S. pombe, the aps1Δ, ado1Δ, and ppn1Δ mutants have constitutive or hyperinducible phenotypes in both species (9). The ppn1Δ phenotype observed in Fig. 2 is explained if Ppn1 liberates phosphate from polyphosphate vacuolar stores as it does in S. cerevisiae, leading to a hyperinducible phenotype (33). In high-phosphate conditions, ppn1Δ expresses phosphatase activity similar to that of the wild type because there is sufficient phosphate to repress the signaling pathway. However, during starvation, the ppn1Δ strain is unable to access stores of polyphosphate and consequently starves more quickly than the wild type; thus, this mutant appeared constitutive in the visual assay because it is more likely to induce phosphatase than the wild type. Both ADO1 (encoding adenosine kinase) and DDP1 (encoding an IP7 phosphatase) regulate the PHO pathway upstream in S. cerevisiae, and these gene products regulate IP7 levels and metabolites related to adenosine. Because the constitutive phenotype is common to both species (DDP1 is orthologous to aps1+), it is likely that IP7 levels or adenosine metabolites regulate the same response in S. pombe. To test this idea further, we deleted the VIP1 ortholog in S. pombe (asp1+). We hypothesized that if IP7 levels are critical for signaling phosphate starvation in S. pombe, then the inactivation of the IP6 kinase (asp1+) should cause a defect in phosphatase induction. We did not observe a defect in phosphatase induction during phosphate starvation; however, we did detect a defect in the high-phosphate levels of phosphatase (Fig. 6). Additionally, we examined the strain in the deletion collection and observed a similar phenotype (data not shown). Because our screen did not identify strains that appeared whiter in high-phosphate conditions, we did not initially score it as mutant. These data indicate that IP7 levels are important for phosphatase expression, but they may not be as important as they are in S. cerevisiae.
Fig. 5.
Levels of pho1+ and pho84+ transcript when grown in high-phosphate conditions. Transcript levels were measured by RT-qPCR in constitutive mutant strains and the wild type after 4 h of growth in high-phosphate medium. Errors are standard errors of three separately grown replicates. The wild-type strain (DP3) was used for normalization under high-phosphate conditions, and the ratio of pho1+/act1+ is 0.33 and of pho84+/act1+ is 0.19.
Fig. 6.
asp1+ is required for phosphatase expression in high-phosphate conditions. asp1+ was deleted with a KANMX6 and with a NATMX6 cassette, and deletion was confirmed by PCR. Two deletion strains as well as a G418r and NATr strain that were wild type for asp1+ were replica plated onto high- and no-phosphate agar plates and subjected to a phosphatase plate assay. The lighter color of the mutant strains in high-phosphate medium is reproducible in multiple isolates.
The mutants that constitutively express phosphatase in S. pombe and do not appear to have the same phenotype in S. cerevisiae provide clues as to how the starvation signal is processed in S. pombe. For example, the strong constitutive phenotype of the csk1Δ strain suggests that this kinase represses the pathway, possibly by phosphorylating a downstream component of the pathway. Additionally, the conserved gene spcc1393.13+ may be involved in repression, although its function is unknown.
Epistasis.
To determine possible genetic interactions and infer pathway order, we analyzed the phenotypes of double mutants by crossing constitutive and uninducible mutants to one another. First, we exchanged the marker in the constitutive mutant (from KANMX4 to NATMX6), backcrossed these mutants to confirm that only one NATMX6 insertion was causing the mutant constitutive phenotype, and isolated the mutant in the mating type (h−) opposite that of the deletion collection. We initially performed this analysis with 10 different uninducible mutants and the constitutive or hyperinducible mutants aps1Δ, csk1Δ, spcc1393.13Δ, ppn1Δ, and ado1Δ (data not shown). Only two uninducible mutants (pho7Δ and snf5Δ) were capable of suppressing the phenotypes of any of the constitutive mutants; however, all of the uninducible mutants were capable of suppressing the ppn1Δ strain, which is consistent with the hypothesis that Ppn1 plays a role in altering kinetics but not actual signaling (Fig. 7). We conclude that Pho7 and Snf5 regulate the transcription of pho1+, and based on these epistasis experiments, they act downstream of all of the gene products mutated in the constitutive mutants.
Fig. 7.
Epistatic tests. Double mutant strains were arrayed on solid medium with and without phosphate and subjected to a phosphatase assay. Asterisks indicate wild-type strains; both the high- and no-phosphate photographs are of identical strains.
We discovered during the analysis of the snf5Δ strains that an additional locus is important for the uninducible phenotype of the snf5Δ strain. We subjected the snf5Δ strain to an extensive backcross (>200 random spores) and determined that only half of the strains that were G418r had the uninducible phenotype, but that all of the uninducible mutants were G418r, suggesting that snf5+ is necessary but not sufficient for the uninducible phenotype. We analyzed numerous progeny during the epistasis experiment that were NATr G418r. When the four constitutive mutants were crossed to the snf5Δ strain, we were able to isolate uninducible double mutants, suggesting that when the uninducible genotype was combined with the constitutive mutant the uninducible phenotype was epistatic (Fig. 7). Additionally, when we deleted snf5+ in a wild-type (DP1) background, there was no obvious phenotype related to phosphatase activity (data not shown). The additional locus required to observe the uninducible phenotype is unknown at this time.
To confirm that pho7+ and snf5+ are critical for the uninducible phenotype, we backcrossed pho7Δ and snf5Δ mutant strains to a ura4− strain and constructed ura4+ plasmids containing genomic copies of these genes. We determined that only a pho7+ plasmid complemented the pho7Δ mutant and only a snf5+ plasmid complemented the snf5Δ mutant (data not shown). This suggests that the additional locus in the snf5Δ mutant is not pho7+. These results also further validate that these two genes were properly identified in the deletion collection.
Pho7 regulates phosphate starvation genes but not glucose or nitrogen starvation genes.
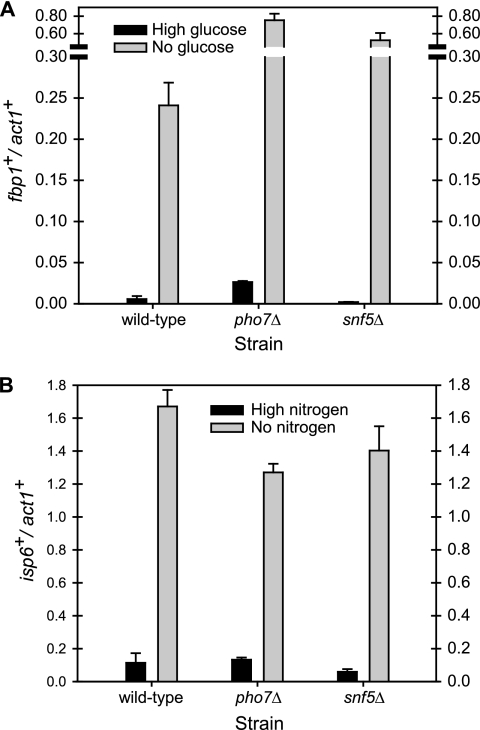
To determine whether pho7+ and snf5+ are regulators of multiple environmental stresses, we examined the expression of genes that are upregulated during nitrogen and carbon starvation (Fig. 8). If Pho7 is a phosphate starvation-regulated transcription factor, then a pho7Δ strain should be unaffected in the transcription of genes unrelated to phosphate starvation. We examined the transcription of fbp1+, which is induced during the shift from glucose to glycerol, and isp6+, which is induced during nitrogen starvation (8, 28). Wild-type and mutant strains pho7Δ and snf5Δ induced these transcripts appropriately, suggesting that the mutants are specific to phosphate starvation.
Fig. 8.
pho7Δ and snf5Δ are not defective in the transcriptional induction of carbon- and nitrogen-regulated transcripts. Strains from the deletion collection that were mutant in the two genes and wild type (DP3) were grown in EMM plus 3% glucose medium and transferred in triplicate to either EMM 0.1% glucose plus 3% glycerol for 4 h or EMM with nitrogen for 2 h. Cells were harvested and RNA subjected to RT-qPCR as previously described.
DISCUSSION
Utilizing a collection of S. pombe deletion strains, we were able to identify a rudimentary PHO pathway in an archiascomycete distantly related to S. cerevisiae, and we conclude that there are significant differences between the two PHO pathways (Fig. 9). Approximately 40% of our mutants were false positives, in that the phenotype did not correlate with the deleted gene, underscoring the need for secondary validation. In most cases, the appropriate gene was deleted, but the PHO pathway-related phenotype was not linked to the gene deletion. Regardless, the deletion collection greatly facilitated the identification of a novel signal transduction pathway.
Fig. 9.
Model of PHO pathway in two yeast species. See the introduction for a discussion of the PHO pathway in S. cerevisiae. The proposed PHO pathway in S. pombe shares at least four genes in common with S. cerevisiae, and they are indicated by the blue circles. We hypothesize that IP7 levels increase in response to phosphate starvation in a manner very similar to that of S. cerevisiae; these increased levels activate directly or indirectly the Pho7 protein (yellow circle). There are other genes involved, but their predicted role in the pathway is unclear, and they are indicated by empty circles. spcc1393.13+ and csk1+ must act upstream of the transcription factors based on epistasis experiments, but whether they regulate IP7 levels is unknown. Additionally, gpa2+ and pka1+ do not suppress the constitutive phenotypes, so they could be acting separately from the pathway, influencing IP7 levels, regulating the activation of Pho7 or Snf5 in a redundant manner, or regulating the secretion of Pho1. The genes in green circles are hypothesized to be regulating the secretion of Pho1.
The PHO pathway in S. pombe has novel regulation relative to that of S. cerevisiae. Pho7 likely is a transcription factor that is epistatic to the constitutive mutants, which is consistent with its role in transcription. It is interesting that performing a BLAST search with Pho7 uncovers a possible ortholog only in Schizosaccharomyces japonicas (E value, 3.8 × 10−14), with other species possessing zinc finger-containing domains but not obvious orthologs. This suggests that other yeasts have different transcription factors that are responsive to phosphate starvation. This is the third type of transcription factor identified that regulates phosphate starvation responses, with a basic helix-loop-helix factor in many Ascomycetes typified by ScPho4 (2), a myb-like factor in plants typified by Psr1 in Arabidopsis thaliana and Chlamydomonas reinhardtii (27, 34), and now a zinc finger factor in Archiascomycetes. It seems likely that there is dramatic flexibility in the acquisition of a phosphate-responsive transcription factor in evolution.
On the other hand, there are common components of the PHO pathway between S. cerevisiae and S. pombe, and these components either arose through convergent evolution or were shared by the common ancestor of these yeasts. The conservation of the acid phosphatase and high-affinity phosphate transporters in many species, including many non-yeast species, suggests that the common ancestor of many eukaryotes contained these genes. Furthermore, the similar phenotypes of the aps1Δ, ado1Δ, and ppn1Δ mutants in both yeasts suggest that IP7, adenosine, and polyphosphate metabolism influenced the phosphate starvation response in the ancestor. Finally, mutations in SWI/SNF in both species affect the transcriptional induction of genes during phosphate starvation, and thus the involvement of chromatin remodeling in this pathway may have been an ancestral trait.
We have determined that there are common components as well as novel components of the PHO pathway in S. pombe relative to that of S. cerevisiae. The putative orthologs of PHO80 and PHO85 in S. pombe do not have any obvious phenotype in our screen (data not shown), and there is not a PHO4 ortholog. Thus, S. pombe developed a regulated transcriptional pathway parallel to S. cerevisiae using pho7+ and other genes. We postulate that the two yeasts have generated a very similar transcriptional output through two distinct mechanisms. While these signaling pathways did not arise through convergent evolution, because some signaling components were shared in the common ancestor, we believe they represent an example of parallel evolution.
By beginning to define the PHO pathway in a yeast distantly related to S. cerevisiae, we can speculate an evolutionary scheme for the acquisition of a phosphate starvation response. It seems likely that the earliest organisms had systems to acquire low concentrations of inorganic phosphate (high-affinity phosphate transporters) and to access organic phosphate sources (phosphatases). It would be advantageous to produce only these systems during inorganic phosphate starvation because of energy constraints; therefore, the ability to sense phosphate starvation and to accumulate these systems would be selected. Because the two yeasts have the same phenotype in response to the deletion of aps1+, yeasts (and possibly other eukaryotes) may accumulate IP7 in response to phosphate starvation. Alterations of ATP charge may impact the levels of IP7, although the mechanism underlying the phenotype of the ado1Δ strain still is unclear. We hypothesize that many different protein-based mechanisms to sense IP7 levels arose. In S. cerevisiae, Pho81 binds IP7 and eventually regulates the transcription factor Pho4. In S. pombe, we postulate that IP7 regulates Pho7 either directly or indirectly, possibly through the Csk1 kinase. Our work suggests that the ancestral state of metabolic by-products (in this case, IP7) of phosphate starvation are present in all unicellular eukaryotes, and different early species utilized many different pathways to sense the metabolic by-product.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Department of Biology, the College of Arts and Sciences, and the Center for Undergraduate Research and Fellowships at Villanova University and by a grant from the National Science Foundation (RUI-MCB-0747799). Additional support was from the Larry L. Hillblom Foundation (to D.M.C.) and the Howard Hughes Medical Institute (to J.S.W.).
We thank Heather Lander and Eric Christenson for the development of tools used in this study.
Footnotes
Supplemental material for this article may be found at http://ec.asm.org/.
Published ahead of print on 17 December 2010.
REFERENCES
- 1. Barbaric S., Munsterkotter M., Goding C., Horz W. 1998. Cooperative Pho2-Pho4 interactions at the PHO5 promoter are critical for binding of Pho4 to UASp1 and for efficient transactivation by Pho4 at UASp2. Mol. Cell. Biol. 18:2629–2639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Barbarić S., Munsterkotter M., Svaren J., Horz W. 1996. The homeodomain protein Pho2 and the basic-helix-loop-helix protein Pho4 bind DNA cooperatively at the yeast PHO5 promoter. Nucleic Acids Res. 24:4479–4486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Barbet N., Muriel W. J., Carr A. M. 1992. Versatile shuttle vectors and genomic libraries for use with Schizosaccharomyces pombe. Gene 114:59–66 [DOI] [PubMed] [Google Scholar]
- 4. Dodgson J., et al. 2009. Functional genomics of adhesion, invasion, and mycelial formation in Schizosaccharomyces pombe. Eukaryot. Cell 8:1298–1306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Forsburg S. L., Rhind N. 2006. Basic methods for fission yeast. Yeast 23:173–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. He Y., et al. 2006. Genetic and functional interaction between Ryh1 and Ypt3: two Rab GTPases that function in S. pombe secretory pathway. Genes Cells 11:207–221 [DOI] [PubMed] [Google Scholar]
- 7. Hedges S. B. 2002. The origin and evolution of model organisms. Nat. Rev. Genet. 3:838–849 [DOI] [PubMed] [Google Scholar]
- 8. Hoffman C. S., Winston F. 1990. Isolation and characterization of mutants constitutive for expression of the fbp1 gene of Schizosaccharomyces pombe. Genetics 124:807–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Huang S., O'Shea E. K. 2005. A systematic high-throughput screen of a yeast deletion collection for mutants defective in PHO5 regulation. Genetics 169:1859–1871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jeffery D. A., Springer M., King D. S., O'Shea E. K. 2001. Multi-site phosphorylation of pho4 by the cyclin-CDK pho80-pho85 is semi-processive with site preference. J. Mol. Biol. 306:997–1010 [DOI] [PubMed] [Google Scholar]
- 11. Kaffman A., Rank N. M., O'Neill E. M., Huang L. S., O'Shea E. K. 1998. The receptor Msn5 exports the phosphorylated transcription factor Pho4 out of the nucleus. Nature 396:482–486 [DOI] [PubMed] [Google Scholar]
- 12. Kaffman A., Rank N. M., O'Shea E. K. 1998. Phosphorylation regulates association of the transcription factor Pho4 with its import receptor Pse1/Kap121. Genes Dev. 12:2673–2683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kerwin C. L., Wykoff D. D. 2009. Candida glabrata PHO4 is necessary and sufficient for Pho2-independent transcription of phosphate starvation genes. Genetics 182:471–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kim D. U., et al. 2010. Analysis of a genome-wide set of gene deletions in the fission yeast Schizosaccharomyces pombe. Nat. Biotechnol. 28:617–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Komeili A., O'Shea E. K. 1999. Roles of phosphorylation sites in regulating activity of the transcription factor Pho4. Science 284:977–980 [DOI] [PubMed] [Google Scholar]
- 16. Lam F. H., Steger D. J., O'Shea E. K. 2008. Chromatin decouples promoter threshold from dynamic range. Nature 453:246–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lau W. W., Schneider K. R., O'Shea E. K. 1998. A genetic study of signaling processes for repression of PHO5 transcription in Saccharomyces cerevisiae. Genetics 150:1349–1359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lee Y. S., Huang K., Quiocho F. A., O'Shea E. K. 2008. Molecular basis of cyclin-CDK-CKI regulation by reversible binding of an inositol pyrophosphate. Nat. Chem. Biol. 4:25–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lee Y. S., Mulugu S., York J. D., O'Shea E. K. 2007. Regulation of a cyclin-CDK-CDK inhibitor complex by inositol pyrophosphates. Science 316:109–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lenburg M. E., O'Shea E. K. 1996. Signaling phosphate starvation. Trends Biochem. Sci. 21:383–387 [PubMed] [Google Scholar]
- 21. Longtine M. S., et al. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14:953–961 [DOI] [PubMed] [Google Scholar]
- 22. Maundrell K., Nurse P., Schonholzer F., Schweingruber M. E. 1985. Cloning and characterization of two genes restoring acid phosphatase activity in pho1− mutants of Schizosaccharomyces pombe. Gene 39:223–230 [DOI] [PubMed] [Google Scholar]
- 23. Monahan B. J., et al. 2008. Fission yeast SWI/SNF and RSC complexes show compositional and functional differences from budding yeast. Nat. Struct. Mol. Biol. 15:873–880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mouillon J. M., Persson B. L. 2006. New aspects on phosphate sensing and signalling in Saccharomyces cerevisiae. FEMS Yeast Res. 6:171–176 [DOI] [PubMed] [Google Scholar]
- 25. O'Neill E. M., Kaffman A., Jolly E. R., O'Shea E. K. 1996. Regulation of PHO4 nuclear localization by the PHO80-PHO85 cyclin-CDK complex. Science 271:209–212 [DOI] [PubMed] [Google Scholar]
- 26. Roguev A., et al. 2008. Conservation and rewiring of functional modules revealed by an epistasis map in fission yeast. Science 322:405–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rubio V., et al. 2001. A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae. Genes Dev. 15:2122–2133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sato S., Suzuki H., Widyastuti U., Hotta Y., Tabata S. 1994. Identification and characterization of genes induced during sexual differentiation in Schizosaccharomyces pombe. Curr. Genet. 26:31–37 [DOI] [PubMed] [Google Scholar]
- 29. Schwaninger R., Dumermuth E., Schweingruber M. E. 1990. Effects of seven different mutations in the pho1 gene on enzymatic activity, glycosylation and secretion of acid phosphatase in Schizosaccharomyces pombe. Mol. Gen. Genet. 221:403–410 [DOI] [PubMed] [Google Scholar]
- 30. Schweingruber M. E., Edenharter E., Zurlinden A., Stockmaier K. M. 1992. Regulation of pho1-encoded acid phosphatase of Schizosaccharomyces pombe by adenine and phosphate. Curr. Genet. 22:289–292 [DOI] [PubMed] [Google Scholar]
- 31. Steger D. J., Haswell E. S., Miller A. L., Wente S. R., O'Shea E. K. 2003. Regulation of chromatin remodeling by inositol polyphosphates. Science 299:114–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tanaka K., Okayama H. 2000. A pcl-like cyclin activates the Res2p-Cdc10p cell cycle “start” transcriptional factor complex in fission yeast. Mol. Biol. Cell 11:2845–2862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Thomas M. R., O'Shea E. K. 2005. An intracellular phosphate buffer filters transient fluctuations in extracellular phosphate levels. Proc. Natl. Acad. Sci. U. S. A. 102:9565–9570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wykoff D. D., Grossman A. R., Weeks D. P., Usuda H., Shimogawara K. 1999. Psr1, a nuclear localized protein that regulates phosphorus metabolism in Chlamydomonas. Proc. Natl. Acad. Sci. U. S. A. 96:15336–15341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yang J. W., Dhamija S. S., Schweingruber M. E. 1991. Characterisation of the specific p-nitrophenylphosphatase gene and protein of Schizosaccharomyces pombe. Eur. J. Biochem. 198:493–497 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.