Abstract
The histidine kinase-based phosphorelay has emerged as a common strategy among bacteria, fungi, protozoa, and plants for triggering important stress responses and interpreting developmental cues in response to environmental as well as chemical, nutritional, and hormone signals. The absence of this type of signaling mechanism in animals makes the so-called “two-component” pathway an attractive target for development of antimicrobial agents. The best-studied eukaryotic example of a two-component pathway is the SLN1 pathway in Saccharomyces cerevisiae, which responds to turgor and other physical properties associated with the fungal cell wall. One of the two phosphoreceiver proteins known as response regulators in this pathway is Skn7, a highly conserved stress-responsive transcription factor with a subset of activities that are dependent on SLN1 pathway phosphorylation and another subset that are independent. Interest in Skn7as a determinant in fungal virulence stems primarily from its well-established role in the oxidative stress response; however, the involvement of Skn7 in maintenance of cell wall integrity may also be relevant. Since the cell wall is crucial for fungal survival, structural and biosynthetic proteins affecting wall composition and signaling pathways that respond to wall stress are likely to play key roles in virulence. Here we review the molecular and phenotypic characteristics of different fungal Skn7 proteins and consider how each of these properties may contribute to fungal virulence.
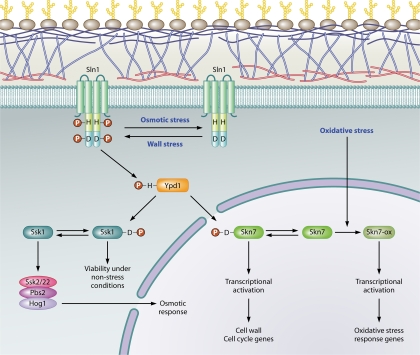
The utilization of histidine kinase-based phosphorelays to trigger stress responses is common in prokaryotes but is also found in free-living eukaryotic microorganisms, such as fungi and Dictyostelium, and in plants (8, 44, 86). In this type of signal transduction pathway, the histidine kinase typically serves a sensory function and responds to specific environmental stimuli by catalyzing autophosphorylation of a conserved histidine residue. In a series of His-to-Asp phosphotransfer steps, the phosphoryl group is ultimately transferred to a conserved aspartate residue in the receiver domain of a response regulator (RR). The SLN1 pathway in Saccharomyces cerevisiae is the best-studied eukaryotic example of a “two-component” signal transduction pathway (where component 1 is a histidine kinase and component 2 is a response regulator). Sln1 is a membrane-associated hybrid histidine kinase whose kinase activity is regulated by turgor and other physical attributes associated with the fungal cell wall (65, 70, 77). The first phosphotransfer step occurs between conserved histidine and aspartate residues within Sln1. The second is between Sln1 and a conserved histidine in the phosphotransfer protein Ypd1, and the final phosphotransfer step is between Ypd1 and the aspartate-containing receiver domains in each of two functionally distinct response regulators, Ssk1 and Skn7 (42, 49, 63) (Fig. 1).
Fig. 1.
The SLN1 phosphorelay in S. cerevisiae. The flow of phosphoryl groups through the SLN1 pathway is shown schematically. Plasma membrane-associated Sln1 kinase is autophosphorylated on the H576 residue under normal growth conditions. Hyperactivation occurs under conditions causing weakening/remodeling of the cell wall (wall stress). Hyperosmolarity and other conditions causing reduced turgor lead to a reduction in Sln1 kinase activity and accumulation of Sln1 in the dephosphorylated form. The phosphoryl group on H576 (H) is transferred to D1144 (D) in the receiver domain of Sln1 and then to H64 of the phosphotransferase, Ypd1, and finally to D554 and D427 in the receiver domains of the cytoplasmic Ssk1 and nuclear Skn7 response regulators, respectively. The unphosphorylated form of Ssk1 interacts with and stimulates the activity of the Ssk2 and Ssk22 MAPKKKs of the HOG1 pathway, while the phosphorylated form of Ssk1 renders it inactive. Phosphorylation of Skn7 leads to activation of a subset of SKN7-dependent genes such as the mannosyltransferase gene, OCH1. However, not all SKN7-dependent genes require aspartyl phosphorylation. For example, SKN7-dependent activation of oxidative stress-responsive genes is independent of the SLN1 pathway.
Skn7 is a stress-responsive transcription factor that is highly conserved among fungi. Skn7 proteins have a uniform architecture, consisting of an N-terminal DNA binding domain similar to that of the heat shock transcription factor (HSF) and a C-terminal receiver domain (5, 6, 33, 53). The receiver domain confers regulation of Skn7 transcriptional activity by His-Asp phosphorelay signaling via phosphorylation of a conserved aspartate in Skn7. This type of regulation distinguishes Skn7 from other eukaryotic transcription factors.
SACCHAROMYCES CEREVISIAE Skn7
Under normal growth conditions, the molecules in the Saccharomyces cerevisiae SLN1 pathway are maintained in the phosphorylated state. Mutations in Sln1, Ypd1, or Ssk1 that prevent phosphorylation are lethal because they lead to constitutive and inappropriate activation of the HOG1 osmotic stress-activated mitogen-activated protein kinase (MAPK) pathway (49) (Fig. 1). Conditions (like osmotic stress) that cause cell shrinkage and loss of turgor reduce Sln1 kinase activity and cause an associated reduction in phosphotransfer to Ypd1, Ssk1, and Skn7 (65). The reduction in kinase and phosphotransfer activity leads to the accumulation of the unphosphorylated form of Ssk1 which interacts with and stimulates the Ssk2 and Ssk22 MAP kinase kinase kinases (MAPKKKs) of the HOG1 pathway (62, 63). In contrast, aspartyl phosphorylation of Skn7 does not affect the osmotic response. Neither deletion nor overexpression of SKN7 has any effect on osmotic stress phenotypes (5, 27).
An increase in Sln1 kinase activity occurs in response to changes affecting the cell wall, such as reduced levels of the glycosylphosphatidylinositol (GPI)-anchored cell wall protein Ccw12 (70) or deletion of the FPS1 glycerol transporter gene (77). The fps1Δ mutation eliminates glycerol efflux and causes intracellular glycerol accumulation (48, 75). Studies of the fps1Δ mutant initially seemed to suggest that increased turgor might stimulate Sln1 kinase activity (77), providing a satisfying counterpoint to the observation that Sln1 kinase activity is reduced under reduced turgor (hyperosmotic) conditions. This model was later modified since increasing turgor by shifting cells to a hypo-osmotic environment did not phenocopy the fps1Δ mutant (70; G. D. Gingerich and J. S. Fassler, unpublished observation), nor was there any evidence that the SLN1-SKN7 pathway could be stimulated by other conditions that activate the protein kinase C (PKC) cell integrity pathway (70). Subsequent studies revealed that the fps1Δ mutation causes alterations in the cell wall, including a reduction in levels of the wall-associated Ccw12 protein (70).
Several observations are consistent with the idea that the cell wall structure is tied to the activity of Sln1 histidine kinase and the SLN1-SKN7 pathway. First, treatment of intact yeast cells with low doses of zymolyase sufficient to alter (including substantial reductions in surface-associated Ccw12 protein) but not remove the cell wall stimulates the Sln1 kinase (70). Second, the SLN1-SKN7 pathway is not only regulated by but also regulates genes involved in cell wall integrity (13, 43), including OCH1, which encodes an α-1,6 mannosyltransferase (59), and NCA3, which encodes a protein that localizes to the cell wall and plays a role in septation (55). Additional evidence tying Skn7 function to the cell wall includes the identification of SKN7 as a dosage suppressor of a mutation in KRE9, which encodes a glycoprotein involved in cell wall β-glucan assembly (6); modest sensitivity of skn7 mutants to hygromycin B (45); identification of the cell wall stress sensor MID2 as a high-copy activator of Skn7 (28); and the synthetic lethality of skn7Δ pkc1Δ double mutants. Taken together, these observations suggest that SKN7 works in parallel with the PKC pathway to coordinate the response to extracellular signals that regulate cell surface integrity (5).
While the role of Skn7 in cell wall signaling and response is dependent on the SLN1 phosphorelay, the involvement of Skn7 in the oxidative stress response is independent of Sln1 and does not require phosphorylation of D427. Although SKN7 null mutants cause severe oxidative stress sensitivity (33), the skn7D427N mutant, which abolishes SLN1-SKN7 signaling, is nonetheless as resistant to oxidative stress as the wild type (43, 52). The oxidative stress phenotype of skn7 null mutants is limited to tert-butyl hydroperoxide and hydrogen peroxide and does not extend to superoxide-generating reagents such as menadione or to the thiol-oxidizing agent, diamide (52; J. S. Fassler and K. E. Mulford, unpublished observations).
The Skn7-mediated oxidative stress response overlaps substantially, if not entirely, with that of Yap1 (23, 39, 52). Yap1 is a basic leucine zipper (bZIP)-type transcription factor whose role in the oxidative stress response is reflected in the oxidant-sensitive phenotype of null mutants (56, 57). Localization of the Yap1 protein is modulated by direct oxidation of cysteine residues (14, 34, 36). In response to hydrogen peroxide exposure, Gpx3-mediated oxidation of Yap1 leads to a Cys303-Cys598 disulfide bond that masks the Yap1 nuclear export signal and allows the protein to accumulate in the nucleus, where it can participate in the activation of OXR genes (15). Skn7 and Yap1 are both involved with activation of many of the key oxidative stress response (OXR) genes, including, for example, TRX2, TSA1, GPX2, AHP1, CCP1, and CTT1 (23, 39, 52, 78).
The promoters of most OXR genes examined thus far contain binding sites for both Skn7 and Yap1 (23, 39, 52), and direct association of each protein with DNA has been observed at many OXR promoters (23, 35, 40, 52, 57, 79, 84). Nonetheless, multiple lines of evidence support an interaction between the two proteins (24; O. Carmel-Harel, K. E. Mulford, and J. S. Fassler, unpublished observation). Following the initial formation of an Skn7-Yap1 complex, Skn7 undergoes serine/threonine phosphorylation, perhaps stabilizing the interacting proteins on the DNA to allow efficient oxidant-stimulated Skn7- and Yap1-dependent activation of OXR genes (24).
The interaction between Skn7 and Yap1 and the ensuing Ser/Thr phosphorylation of Skn7 occur in the context of the receiver domain (24). Hence, both the DNA binding domain and the receiver domain (but not D427) are as important for the Skn7 oxidative stress response as they are for the wall stress response. Of the several mutations identified in the receiver domain that confer an oxidative stress phenotype, at least one, S. cerevisiae skn7-T437A (Scskn7-T437A), has no detectable effect on Sln1-Skn7 signaling (24). Receiver domain mutations specifically affecting the OXR response are defective in the Skn7 interaction with Yap1 and in Ser/Thr phosphorylation of Skn7. Such “split-phenotype” mutations could not be identified following extensive mutagenesis of the DNA binding domain (41). Hence, the ability of Skn7 to mount distinct transcriptional programs in response to different stress conditions is likely to reside in specific interactions of the receiver domain with auxiliary proteins.
In addition to mediating the interaction with Yap1, the Skn7 receiver domain is responsible for the interaction with the Ran binding protein, Mog1 (45). Mog1 is found in association with Skn7 at the SLN1-SKN7-dependent OCH1 promoter but not at the OXR TRX2 promoter. Consistent with a role for Mog1 in SLN1-SKN7-dependent target gene expression, OCH1 expression is reduced in a mog1 deletion mutant. These results suggest that like Yap1, Mog1 might be a stress-specific auxiliary factor (45).
One of the simplest explanations for Skn7 target gene specificity is that partner proteins such as Mog1 and Yap1 compete for an interaction with Skn7. The formation of Skn7-Yap1 complexes following the nuclear localization of Yap1 after oxidant treatment may be at the expense of Skn7-Mog1 complexes. The molecular details of such a model remain to be tested experimentally.
Skn7 IN OTHER MODEL AND PATHOGENIC FUNGI
The presence of response regulator proteins appears to be a universal characteristic of fungal genomes. Most genomes include one SSK1 ortholog and one SKN7 ortholog. Additional classes of genes that encode response regulator (RR) proteins have been reported in a subset of fungi. One class of proteins with RR annotation consist of orthologs of ScRim15, a glucose-repressible serine/threonine protein kinase involved in signal transduction during cell proliferation in response to nutrients and expression of early meiotic genes (81). The “receiver domain” of this protein family aligns well with other receiver domains but is missing key residues, including the phospho-accepting aspartate at conserved positions in the profile hidden Markov model (HMM) (17, 74) of response regulators (PF00072). Some but not all Rim15 proteins contain glutamic acid rather than aspartic acid at the phospho-accepting position, a substitution that might be compatible with constitutive function; others have an alanine. Another class of putative RR protein (AN4134.3/SrrC) reported in Aspergillus nidulans includes a phospho-accepting aspartate but lacks any obvious effector domain (22, 80). Whether this class of protein is capable of undergoing the phosphotransfer reactions, interactions, and conformational changes of typical response regulators awaits further analysis.
SEQUENCE IDENTITY AND DOMAIN ARCHITECTURE IN Skn7 ORTHOLOGS
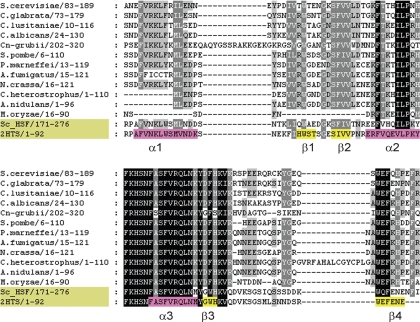
Skn7 orthologs are characterized by two highly conserved domains: an N-terminal HSF-like DNA binding domain and a C-terminal receiver domain which can be easily detected in multiple sequence alignments (Fig. 2 and 3) (38, 60). The HSF domain spans a region of ∼135 residues. Among the fungal orthologs for which tests of Skn7 function have been reported, the HSF domain of Cryptococcus neoformans Skn7 (CnSkn7) is the most distantly related, with 47% identity to the ScSkn7 protein, while the HSF domain of the Candida glabrata protein (CgSkn7) is the most similar, with 71% identity to the ScSkn7 protein (Fig. 2 and Table 1). Within the HSF domain is the highly conserved helix-turn-helix DNA binding motif in which residues of helix 3 known (in heat shock factors and in Skn7 [S137, R140, N143, Y145, and K149]) to be involved in contacting DNA are completely preserved. Likewise, the phenylalanine (F135) and leucine (F142) residues at the edges of helix 2, which are critical in combination for ScSkn7 activity (41), are preserved in most orthologs.
Fig. 2.
Alignment of the HSF-like DNA binding domain in 12 fungal Skn7 orthologs. The residues included in the alignment are indicated to the left. The HSF domain was determined by comparison with the structure of the Kluyveromyces lactis heat shock factor (PDB ID 2HTS). S. cerevisiae Hsf1 is also included in the alignment for comparison. Helices are indicated in pink and β-strands in yellow on the K. lactis sequence. Sequence identifiers were as follows: C. heterostrophus, gi262205082; P. marneffei, gi222160700; C. lusitaniae, gi170877406; A. nidulans, gi259481771; C. neoformans var. grubii, gi54645918; C. glabrata, gi50287867; S. cerevisiae, gi259146967; C. albicans, gi33324593; S. pombe, gi10801610; A. fumigatus, gi14632450; N. crassa, gi94467523; and M. oryzae, gi145616170. The alignment was generated with ClustalW (34) and edited with GeneDoc (56). Shading in the alignment is based on conservation. Black shading indicates invariant residues, and two additional shades of gray represent at least 80% conservation and at least 60% conservation.
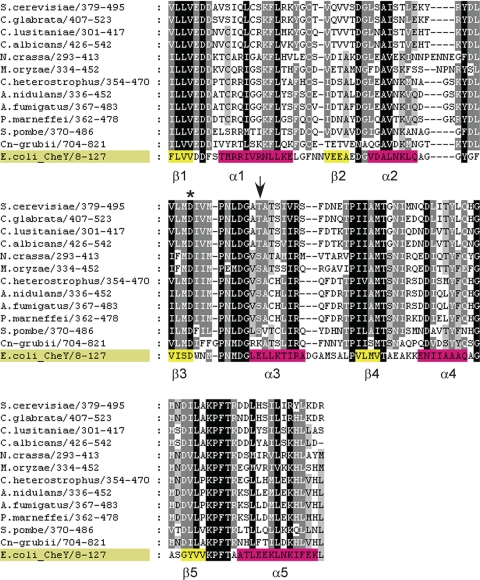
Fig. 3.
Alignment of the response regulator (RR) domain in 12 fungal Skn7 orthologs. The position of the phospho-accepting aspartate is noted with an asterisk. Secondary structure elements are indicated within the E. coli CheY sequence, for which structures have been determined; helices are colored pink and β-sheets yellow. The T437A mutant alleles (see Table 2 and text) are noted with an arrow. The alignment was generated with ClustalW (34) and edited with GeneDoc (56). Shading in the alignment is based on conservation. Black shading indicates invariant residues, and two additional shades of gray represent at least 80% conversion and at least 60% conservation.
Table 1.
Molecular features of fungal Skn7 orthologs
| Organism | Protein | Protein size (aa) | Distance (aa) from N terminus to HSFc | % identityb to HSF domain (135 aa) | Distance (aa) from HSF to RRd | % identity to RR domain (125 aa) | Probability score for CC domaine |
|---|---|---|---|---|---|---|---|
| Animal pathogens | |||||||
| C. glabrata | CgSkn7 | 630 | 73 | 71 | 228 | 92 | 0.85 |
| C. albicans | CaSkn7 | 559 | 24 | 58 | 296 | 75 | 0.999 |
| C. neoformans | CnSkn7 | 1,039/1,040a | 202 | 47 | 384 | 50 | 0.989 |
| P. marneffei | PmSkn7 | 614 | 13 | 57 | 243 | 47 | 0.995 |
| C. lusitaniae | ClSkn7 | 478 | 10 | 61 | 185 | 75 | 0.997 |
| A. fumigatus | AfSkn7 | 622 | 15 | 56 | 246 | 47 | 0.997 |
| Plant pathogens | |||||||
| C. heterostrophus | Dic2/ChSkn7 | 635 | 1f | 52 | 244 | 47 | 0.990 |
| M. oryzae | MoSkn7 | 671 | 9g | 53 | 244 | 46 | 0.53 |
| Model organisms | |||||||
| A. nidulans | SrrA/AnSkn7 | 558 | 1f | 60 | 240 | 47 | 0.895 |
| S. pombe | Prr1/SpSkn7 | 539 | 6 | 60 | 260 | 42 | 0.081 |
| N. crassa | Rrg2/NcSkn7 | 661 | 16 | 58 | 172 | 46 | 0.929 |
| S. cerevisiae | ScSkn7 | 622 | 83 | 100 | 190 | 100 | 0.55 |
There is a slight size difference between the Skn7 proteins in C. neoformans var. grubii and C. neoformans var. neoformans.
Relative to ScSkn7.
HSF, heat shock factor-like DNA binding domain.
RR, response regulator domain.
Highest probability score for a coil-coiled region (CC domain) within 50 bp of the HSF domain (http://www.ch.embnet.org/software/COILS_form.html). Parameters: MTIDK matrix, positions a and d weighted, window width of 21.
Position 1 of the sequence aligns with position 11 in the alignment.
True alignment is between position 23 of the M. oryzae sequence and position 50 (helix α2) of the alignment.
The Skn7 receiver or response regulator (RR) domain is also highly conserved, with orthologs having between 42% and 92% identity to the ScSkn7 protein (Fig. 3 and Table 1). All fungal orthologs have a fully intact domain extending from positions 1 to 112 of the HMM logo (PFAM family Pf00072) (17, 74). Highly conserved residues are universally represented in the amino acid sequence of each species, including the phospho-accepting aspartate residue. Variability in domain length occurs at three positions in the alignment that fall between conserved secondary structural elements in the prototypical Escherichia coli CheY response regulator protein (Protein Data Bank [PDB] identification [ID] 3CHY) (82) and thus are unlikely to perturb the functionality of the domain. In ScSkn7, the T437 residue was found to compromise the interaction of Skn7 with Yap1 in the oxidative stress response without affecting the Sln1-dependent function of Skn7 in wall stress (24). This residue is conserved in all fungal orthologs with either a threonine or serine at this position, with the exception of C. neoformans, which has a lysine (Fig. 3). Interestingly, C. neoformans appears to lack a Yap1 homolog and may therefore lack any selective pressure for a threonine at this position (11, 51, 85).
One additional feature of ScSkn7 is the predicted coiled-coil (CC) domain, located 40 to 80 amino acids (aa) downstream of the HSF domain. The CC domain overlaps the “HR” motif first identified by its similarity to sequences in RhoA effectors ROCK-1 and kinectin proteins (1) and later shown to mediate interactions with some but not all Skn7-interacting proteins, including Rho1 (1) and Mbp1 (4). The probability of coil formation is high in most orthologs (Table 1). One exceptional case is the Schizosaccharomyces pombe Skn7 protein (SpSkn7), in which the probability of coil formation at this position is very low (0.08 compared to 0.55 in S. cerevisiae and 0.99 in several other species) (Table 1). (www.ch.embnet.org/software/COILS_form.html) (47). Whether SpSkn7 utilizes other domains to mediate interactions normally dependent on the CC domain remains to be investigated.
Skn7 proteins vary in size from 478 amino acids in Candida lusitaniae (67) to 1,039 or 1,040 aa in C. neoformans, compared to 620 aa for the ScSkn7 protein, with the main differences occurring at the N terminus (ahead of the HSF domain) and between the HSF/CC and the RR domains. The sizes of the N-terminal domains of ScSkn7 and CgSkn7 (83 and 73 aa, respectively) are longer than the comparable regions in the Skn7 proteins from most fungal pathogens (Aspergillus fumigatus, AfSkn7; C. lusitaniae, ClSkn7; Candida albicans, CaSkn7; Cochliobolus heterostrophus, ChSkn7; Magnaporthe oryzae, MoSkn7; and Penicillium marneffei, PmSkn7), whose sizes range from 1 to 24 aa. This region is longest in CnSkn7 at 202 aa (Table 1). Likewise, the receiver domain is 300 to 400 aa from the N terminus in most species, but 704 aa in C. neoformans. The functional ramifications of these differences in domain spacing and total length of Skn7 proteins are unknown.
The ScSkn7 protein has been localized to the nucleus under a variety of environmental conditions, including osmotic and oxidative and thermal stress, as well as in assorted mutant backgrounds that affect the activity of the SLN1 pathway (45, 46, 64). Likewise, Prr1/SpSkn7 is nuclear in both untreated and oxidant-treated cells. Functional nuclear localization signals (NLSs) have not been experimentally identified. ScSkn7 localization appears to be Ran dependent, since an Skn7-green fluorescent protein (GFP) fusion was cytoplasmically localized in mutants lacking Mog1, a regulator of Ran-mediated nucleocytoplasmic transport (45).
Nuclear localization signal (NLS) prediction algorithms based on an extensive data set of substitution mutants in classical NLS mediated transport in yeast (32) reveal modest (PmSkn7, ClSkn7, SrrA [AnSkn7], CnSkn7, CaSkn7, AfSkn7, and Neurospora crassa Rrg2 [NcSkn7]) or weak or no monopartite signals (ChSkn7, CgSkn7, ScSkn7, SpSkn7, and MoSkn7), and while bipartite signals can be detected in all Skn7 orthologs, the majority are weak (J. S. Fassler, unpublished observations). CaSkn7 is the only protein with a score (bipartite score of 9.3 out of 10 at position 535) indicative of complete nuclear localization by this criterion. Together, the observed constitutive nuclear localization of ScSkn7 and the lack of any strongly predictive classical nuclear localization sequences in the ScSkn7 sequence suggest that fungal Skn7 import into the nucleus is likely to be importin α independent and may instead involve a direct interaction with one of several members of the importin β family, for which nuclear localization signals are not as well characterized.
DISTRIBUTION AND PHYLOGENETIC ANALYSIS OF FUNGAL Skn7 ORTHOLOGS
SKN7 is widely distributed in fungi; examples have been identified in more than 20 Ascomycetes genomes, including endemic fungal pathogens that infect immunocompetent as well as immunocompromised individuals (e.g., Coccidioides immitis, Histoplasma capsulatum, Paracoccidioides brasilliensis, and Blastomyces dermatitidis). The SKN7 gene has also been identified in numerous Basidiomycetes genomes, including the endemic species Cryptococcus gattii. SKN7 orthologs have been identified in Mucormycotina genomes, including Rhizopus oryzae and Phycomyces blakesleeanus but have thus far not been identified in Microsporidia or Chytridiomycota genomes. The absence of SKN7 from these genomes may indicate that the last common ancestor for SKN7 predates the emergence of Dikarya but is not coincident with the earliest events in fungal history (20, 83).
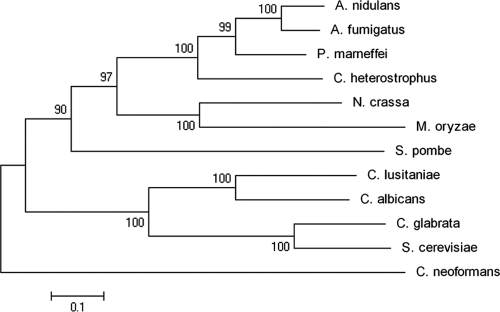
Phylogenetic analysis of Skn7 orthologs from the 12 Ascoymetes and Basidiomycetes taxa discussed in this review is shown in Fig. 4. Inclusion of additional Basidiomycetes taxa did not affect the tree structure. The Skn7 tree is consistent with known species relationships and supports the conclusion that an ancestral SKN7 gene predates the divergence of Ascomycetes and Basidiomycetes. Since trees constructed from domains (data not shown) have the same architecture as trees constructed from the full-length protein, it is likely that the highly conserved HSF-like DNA binding domain and the CheY-like receiver domain have coevolved in the context of an Skn7 protein.
Fig. 4.
Evolutionary relationships of the Skn7 proteins in 12 fungal taxa. Evolutionary history was inferred by the neighbor-joining method (69). The bootstrap consensus tree inferred from 1,000 replicates (18) is taken to represent the evolutionary history of the taxa analyzed. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test is shown next to each branch. Evolutionary distances were computed by the JTT matrix-based method and are shown in the units corresponding to the number of amino acid substitutions per site. All positions containing gaps and missing data were eliminated from the data set. There were a total of 318 positions in the final data set. Phylogenetic analyses were conducted in MEGA4 (76).
PHENOTYPES OF skn7 MUTANTS
A number of fungal SKN7 genes have been characterized by mutational analysis. In nearly all cases, phenotypes were evaluated in deletion or insertion mutants intended to give the null phenotype (Table 2). In P. marneffei, the gene was characterized by complementation of an S. cerevisiae skn7 deletion mutation (7). In C. heterostrophus, analysis of null mutants was supplemented by characterization of point mutants and small deletions isolated in a screen for fungicide resistance (25, 26). In S. pombe and S. cerevisiae, mutations affecting the phospho-accepting aspartate residue have been characterized (5, 24, 27, 33, 42, 58). Analysis of point mutations in SKN7 outside the phospho-accepting aspartate have been reported in S. cerevisiae (24). In the discussion of SKN7 mutant phenotypes below, the S. cerevisiae convention of designating mutations using lowercase italicized letters will be employed. Since the mutations are primarily deletions, the lowercase designation will be taken as shorthand for the deletion mutation, and substitution mutations will be designated by appending the substituted position and the residues involved (e.g., skn7D427N).
Table 2.
Phenotypic features of fungal Skn7 orthologs
| Organism | Gene | Mutation(s) | Phenotypic feature(s)a |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Oxidant sensitivity | Osmotic sensitivity | Fungicide resistance | Thermal sensitivity | Wall stress sensitivity | Other | Virulence | |||
| C. glabrata | CgSKN7 | Deletion | + | NR | NR | NR | − | +/− | |
| C. albicans | CaSKN7 | Deletion | + | + | − | NR | − | Morphogenesis defects | +/− |
| C. neoformans | CnSKN7 | Deletion and insertion | Growth assays and OXR gene expression | − | + | ? | + | Sodium sensitivity, increased melanin, increased mating | + |
| P. marneffei | PmSKN7 | Complementation of the S. cerevisiae gene deletion | + | NR | NR | NR | NR | NR | |
| C. lusitaniae | ClSKN7 | Deletion | + | − | + | − | NR | Pseudohyphal defects | NR |
| A. fumigatus | AfSKN7 | Deletion | + | − | NR | − | + (SDS) | − | |
| C. heterostrophus | dic2/ChSKN7 | Partial and complete deletions and D359N | NR | +/− | + | NR | NR | NR | |
| (Flu and Ipr) | |||||||||
| M. oryzae | MoSKN7 | Deletion | − | +/− | + | NR | − | ||
| A. nidulans | srrA/AnSKN7 | Deletion | + including Men | +/− | + | NR | + (CFW) | Reduced asexual spore formation and conidiospore viability | NA |
| S. pombe | prr1/SpSKN7 | Deletion and D418N | + | − | NR | NR | − | Cold sensitivity, heavy metal sensitivity, and mating sporulation defects | NA |
| N. crassa | rrg2/NcSKN7 | Deletion | Slight | − | NR | − | NR | Conidiation and fertility defects | NA |
| S. cerevisiae | ScSKN7 | Deletion and D427N, D427E, T437A, and HSF domain mutations | HP, TB | − | NR | + | + | NA | |
+, full phenotype observed; +/−, modest phenotype observed; −, no phenotype observed. NR, not reported; NA, not applicable; HP, hydrogen peroxide; TB, tert-butyl hydroperoxide; Men, menadione; Flu, fludioxonil; Ipr, iprodione.
Oxidant sensitivity.
The spectrum of mutant phenotypes attributable to loss of Skn7 function is relatively narrow. Most mutants are sensitive to hydrogen peroxide, t-butyl hydroperoxide, or both, suggesting that Skn7 is nearly universally involved in surviving exposure to certain classes of oxidant. Where tested, most skn7 mutants are not sensitive to menadione, a superoxide-generating compound, or to diamide, which generates oxygen stress conditions by affecting the normal balance between reduced and oxidized glutathione (61, 85). The C. neoformans mutant is distinguished by having a very subtle defect in its response to oxidants that cannot be detected by the usual assay involving a +/− growth phenotype on solid media containing different concentrations of oxidant (3). The mutant does, however, exhibit deficiencies in growth rate after prolonged growth in liquid cultures with added t-butyl hydroperoxide and in oxidant-mediated activation of a subset of oxidative response genes, including TRR1 (thioredoxin reductase) and SOD1 (superoxide dismutase) (11, 85). Consistent with observations in other fungi, even the modest oxidant sensitivity exhibited by the Cnskn7 mutant was specific for certain types of reactive oxygen species (ROS) and did not extend to superoxides nor to reactive nitrogen species (85). The skn7 mutant in the rice blast fungus, M. oryzae, is reportedly insensitive to oxidant; however, the nature of the assay was not described (54). Interestingly, while the A. nidulans srrA mutant shows the expected sensitivity to hydrogen peroxide and t-butyl hydroperoxide, the ssrA sskA double knockout mutant lacking both Ssk1 and Skn7 is also sensitive to high concentrations of menadione, suggesting some participation of at least one Skn7 protein in the response to superoxides (80).
The oxidant sensitive phenotype of S. cerevisiae and S. pombe skn7 mutants is complemented by skn7 alleles containing mutations in the phospho-accepting aspartates (skn7D427N and prr1D418N) (52, 58). This indicates that the oxidative stress function of Skn7 is independent of the SLN1 phosphorelay in at least these two taxa.
In general, the oxidative stress response in S. cerevisiae, C. albicans, and C. glabrata appears to be well conserved and is dependent on a similar subset of transcription factors, including Skn7 and Yap1, as well as Msn2 and Msn4 (12). Wild-type C. glabrata is naturally resistant to higher levels of H2O2 than S. cerevisiae and C. albicans. This form of adaptation is completely abolished in skn7 and yap1 mutants.
Osmotic stress sensitivity and fungicide resistance.
skn7 mutations do not confer a robust osmotic stress phenotype, while mutations in the alternative response regulator, SSK1, are typically strikingly osmosensitive. One exception is the C. albicans ssk1 mutant, which is not osmosensitive but resembles C. albicans skn7 mutants in being sensitive to oxidants (9). Modest osmosensitivity attributable to SKN7 mutations has been noted in C. heterostrophus (25, 26), A. nidulans (22, 80), and M. oryzae (54). In these organisms, the mild osmotic stress phenotype of the skn7Δ mutant is exacerbated in double ssk1 skn7 mutants (26, 54, 80), suggesting that the two response regulators may sometimes have overlapping roles. Cryptococcus skn7 mutants exhibit clear sensitivity to sodium, but not to osmotic conditions per se (3).
Phenylpyrrole fungicides such as fludioxonil activate the stress-activated HOG1 MAPK pathway in filamentous fungi, and null mutations in the HOG1 pathway confer fungicide resistance (30, 31, 87). Mutations in the SKN7 gene also confer partial resistance to fludioxonil (3, 22, 26, 80). In A. nidulans, full fludioxonil resistance was conferred in the absence of both response regulators (sskA and srrA) (80). C. heterostrophus double mutants were likewise resistant to fludioxonil as well as to the dicarboximide fungicide iprodione (25, 26). The observation that the fungicide resistance phenotypes of skn7 and ssk1 mutations are additive indicates that the two pathways work in parallel. Consistent with this conclusion is the observation that although ChSkn7, MoSkn7, and Ssk1 jointly participate in the osmotic stress response and fungicide resistance, only Ssk1 mediates osmolarity and fungicide-induced phosphorylation of the Hog1 ortholog. Neither ChSkn7 nor MoSkn7 has any apparent role in the activation of Hog1 (25, 54). Similarly, both skn7 and ssk1 mutants of C. neoformans confer resistance to fludioxonil, but only the ssk1 mutant exhibits sensitivity to osmotic stress and participates in activation of CnHog1 (3).
Thermal stress.
The presence of an HSF-like DNA binding domain (6), the reported interaction between Skn7 and Hsf1, and the sensitivity of skn7 mutants to acute heat in S. cerevisiae (64) suggest a role for Skn7 orthologs in thermal resistance. However, neither the A. fumigatus (37), C. lusitaniae (67), nor C. neoformans (3, 11) skn7 mutants exhibited this phenotype. A. nidulans srrA conidia proved thermostable, although sskA conidia were sensitive to both thermal stress and cold stress (22). The involvement of other Skn7 proteins in thermal stress has not been reported.
Wall stress.
In S. cerevisiae and S. pombe, the oxidative stress phenotype of skn7 and prr1 mutants is not a consequence of its role in the SLN1-YPD1-SKN7 phosphorelay since mutation of the aspartate residue required for phosphorylation by upstream molecules Sln1 and Ypd1 does not confer oxidative stress sensitivity (52, 58). Whether this is the case for other fungal Skn7 proteins remains to be investigated. In contrast, the role of ScSkn7 in cell wall remodeling is Sln1 (and Skn7 D427) dependent (5, 43, 53, 70). A role for Skn7 orthologs in wall integrity is suggested by the sensitivity of A. fumigatus skn7 mutants to sodium dodecyl sulfate (SDS) (37), and the sensitivity of A. nidulans skn7 mutants to calcofluor white (CFW) (80). In addition, C. neoformans skn7 mutant phenotypes, including flocculation (85), sensitivity to the aminoglycoside antibiotic hygromycin B, and enhanced resistance to the chitin-binding compound Congo Red suggest changes in the cell wall that might include decreased levels of chitin (J. S. Fassler, unpublished). Morphogenetic defects noted in C. albicans (71) and C. lusitaniae (67) might also reflect cell wall remodeling defects in the absence of Skn7; however, direct chemical analysis of purified C. albicans cell walls revealed no differences in the mannosylation or chitin content in wild-type versus skn7 mutant wall fractions (71). The role of aspartyl phosphorylation in the Skn7 wall function in these fungi is unknown since the majority of reports in the literature have utilized deletion mutants.
Melanin production, sexual reproduction, and spore viability.
The C. neoformans skn7 mutant phenotype includes increased melanin production, suggesting a role for CnSkn7 in negative regulation of melanin biosynthetic genes (3). Melanin synthesis is also under the control of the HOG pathway and Ssk1 (3), but the role of Hog1 activation in Skn7 regulation of melanin has not been tested. Melanin production in M. oryzae is likewise under the control of the Ssk1 and Skn7 response regulators, with increased melanin formation in the M. oryzae skn7 mutant due in part to early induction of the transcription factor gene, PIG1, required for hyphal melanization (54).
The C. neoformans skn7 mutant exhibits enhanced mating, as shown by increased filamentation in bilateral matings between MATa skn7 and MATα skn7 mutants as well as in matings between a skn7 strain and a mating-crippled adenylyl cyclase mutant (3). The S. pombe skn7 mutant is also defective in mating and in spore formation (58, 61). The A. nidulans skn7 mutant has normal sexual reproduction but exhibits a striking defect in asexual sporulation and conidiospore viability (22, 80). Interestingly, in the S. pombe prr1D418N mutant sporulation was accelerated and deregulated, proceeding under conditions not normally conducive. These observations suggest that the phosphorylated form of SpPrr1 is required for normal meiotic timing in S. pombe.
TRANSCRIPTIONAL REGULATION OF SKN7 AND SKN7 TARGET GENES
Expression of the SKN7 gene has not been thoroughly characterized. Analysis of publically available transcriptome data for S. cerevisiae suggests that downregulation of SKN7 expression in response to various environmental stresses (oxidative and heat [21], as well as diauxic shift [16]) may be one mechanism for restoring steady-state levels of gene expression as cells recover from stress. A reduction in ScSkn7 protein levels following oxidant treatment of cells has also been observed (X. J. He and J. S. Fassler, unpublished observation). In contrast, expression of the A. nidulans srrA (and sskA) genes was transiently increased in response to oxidative stress (22). The potential for yeast versus mycelial phase-specific regulation of SKN7 was ruled out in P. marneffei (7).
Expression of Skn7 target genes has been examined in S. cerevisiae (24, 42, 43, 52, 64, 79), as well as in other fungi. In-gel catalase activity assays confirmed the effect of the A. nidulans srrA mutation on CatB levels following oxidant treatment (22, 80). Expression of the A. nidulans conidiophore transcription factor gene, brlA, was also reduced in the A. nidulans srrA mutant, perhaps accounting for the observed sporulation defect (80).
Expression of oxidative stress response genes TRX2, CTA1, TRR1, and TSA1 in C. glabrata is clearly Skn7 dependent (68), as is expression of the S. pombe oxidative stress genes ctt1 and trr1. In addition, expression of the S. pombe mating and sporulation genes ste11, mam2, and mei2 is reduced in skn7 null mutants and inappropriately elevated in the skn7D418N mutant (58, 61).
Expression of four C. neoformans oxidative stress response genes was investigated: TRR1 and SOD1 (but not GLR1or TRX2) exhibited Skn7-dependent oxidative stress induction. This is consistent with the observation that Ssk1 rather than Skn7 appears to have a major role in the oxidative stress response in this organism (85). Global expression analysis in Cryptococcus revealed a small set of 86 genes that were significantly regulated in response to Skn7 under normal growth conditions (29). Of these genes, 23 were upregulated and 63 downregulated. Several process-based categories are enriched in the regulated gene set. These include carbohydrate metabolism (6 genes), transport (6 genes), amino acid metabolism (4 genes), and generation of precursor metabolites and energy (3 genes). Whether these genes are directly regulated by Skn7 binding to cis-promoter elements previously characterized in S. cerevisiae (23, 24, 43, 45, 64, 79) remains to be investigated experimentally.
RELATIONSHIP OF Skn7 TO THE Hog1 STRESS-ACTIVATED MAPK PATHWAY
ScSkn7 is nuclear and associates directly with DNA, thus primarily exerting its effects at the level of transcription. ScSsk1, on the other hand, is a cytoplasmic protein and mediates its effects by physically interacting with the MAPK kinase kinases (MAPKKKs) of the HOG1 osmotic response MAPK cascade. This regulatory template appears to be universal for all fungi so far characterized. Although a few Skn7 orthologs are involved in osmotic stress and in fungicide resistance, there are no examples of Skn7 stimulation of Hog1. Likewise, in C. albicans, where oxidative stress is mediated via Ssk1 as well as by Skn7, it is nonetheless the case that Ssk1-dependent oxidative stress is mediated by the Hog1 pathway (9). Hence evidence for cross-communication between the Skn7 response regulator and components of the HOG1 or other MAPK cascades is lacking.
The paradigm for Ssk1 regulation of Hog1 based on work with S. cerevisiae, C. albicans, and S. pombe (reviewed in references 10 and 72) involves activation of the Ssk1-Pbs2-Hog1 MAPK kinase cascade by unphosphorylated Ssk1 and the transient stress-related presence of phosphorylated Hog1, which then translocates into the nucleus to trigger activation of genes that ameliorate the otherwise damaging impact of the stressful environment (50, 66). Unexpected diversity in the relationship between Ssk1 and the Hog1 MAPK cascade is seen in certain C. neoformans strains. CnHog1 is constitutively phosphorylated by Pbs2 MAPKK under normal growth conditions and is rapidly activated by Ssk1-dependent dephosphorylation upon exposure to appropriate stresses (2, 3). Remarkably, the reversal in normal regulation of stress-activated MAPKs relative to other organisms so far characterized is attributable to only two amino acid changes (L248 and M738) in the sequence of the C. neoformans MAPKKK, Ssk2, which may affect the physical interaction of Ssk2 with the upstream Ssk1 response regulator (2).
Skn7 AND ITS RELATIONSHIP TO VIRULENCE
The role of Skn7 in virulence of fungal pathogens appears to be species specific. CnSkn7, CaSkn7, and CgSkn7 play some role in virulence; however, not all Skn7 proteins make a contribution to the pathogenicity of the organism. For example, Skn7 has no impact on the pathogenicity of A. fumigatus (37). Likewise, Moskn7 mutants are fully virulent (54).
Evidence for a role of Skn7 in pathogenicity is most striking in C. neoformans. Survival of tail-vein-infected BALB/c mice increased from 12.5 days (median, wild type) to 25.5 days (skn7 mutant) in experiments employing high inocula and from 22 to 29 days in experiments employing lower inocula. In addition, the fungal burden in brain and lung was 3- to 10-fold higher in wild-type mice than for mice infected with skn7 mutant strains (11). In A/Jcr mice infected by nasal inhalation, median survival of mice infected with the mutant was 47 days, compared to 19 days for infections with wild-type C. neoformans (85).
C. glabrata skn7 mutants are also significantly less virulent than wild-type strains. Mice inoculated with a skn7 mutant strain via lateral tail vein injections showed 2- to 3-fold reductions in fungal burden in kidney, liver, and spleen 7 days after injection compared to wild-type and SKN7 reconstituted strains (68). Virulence of the C. albicans skn7 mutant was also evaluated in a disseminated murine model of candidiasis. In contrast to mice infected with wild-type yeast, which showed signs of morbidity at 3 days, heterozygous and homozygous mutant strains exhibited modest attenuation, with onset of morbidity occurring at 6 to 8 days. In this study, the lack of distinction in viability between heterozygous, homozygous, and reconstituted strains was a source of concern. Furthermore, there were no differences in fungal burden or extent of filamentous growth detected in the kidney at 24 or 48 h postinfection (71).
DISCUSSION
Interest in Skn7 as a virulence determinant primarily stems from its well-established role in the oxidative stress response in S. cerevisiae. Although the majority of Skn7 orthologs in fungal pathogens are known to participate in the oxidative stress response, the role of Skn7 is restricted to certain classes of reactive oxygen species and does not encompass superoxides or reactive nitrogen species. Yeast strains deficient in the response to critical reactive oxygen species are expected to succumb more readily to the reactive oxygen burst generated by immune cells. The premise that the role of Skn7 in C. albicans, C. glabrata, and C. neoformans virulence is related to the oxidative stress function of the protein is weakened by the lack of correlation between the virulence and oxidative stress phenotypes. For example, the C. neoformans skn7 mutant has a very subtle oxidant sensitivity phenotype compared to A. fumigatus (37) (Table 2), yet the CnSkn7 protein is clearly involved in virulence, while the AfSkn7 protein is not (11, 37, 85).
In S. cerevisiae, there is strong evidence for the involvement of Skn7 in cell wall integrity, and the same finding is beginning to emerge in other fungal Skn7 proteins (Table 2). This and other functions of Skn7 (Fig. 5) could ultimately be highly relevant to virulence. In S. cerevisiae, both the oxidative and wall stress functions of Skn7 rely on the DNA binding domain and transcriptional activation capabilities of the protein. Stress-specific activation of Skn7 target genes involves stress-specific posttranslational modifications and protein-protein interactions. For example, the cell wall function of ScSkn7 is dependent on aspartyl phosphorylation of the receiver domain by upstream molecules in the Sln1-dependent His-Asp phosphorelay (70), while the oxidative stress function of ScSkn7 is independent of aspartyl phosphorylation (52) and remains intact even in the skn7D427N mutant. Since wall integrity assays have not been reported for all skn7 mutants and the role of aspartyl phosphorylation in Skn7 function has not been widely assessed, the extent to which a role in wall integrity is an integral feature of all fungal Skn7 orthologs and the nature of its regulation remain unclear.
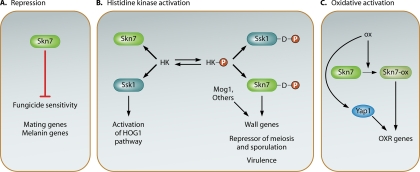
Fig. 5.
The observed spectrum of skn7 phenotypes can be attributed to different forms of the protein. Panel A depicts repression of genes or pathways involved in fungicide sensitivity as well as mating and melanin production explaining the upregulation of melanin and the enhanced mating and fungicide resistance phenotypes observed in certain fungal skn7 null mutants. Panel B depicts regulation of gene expression in response to aspartyl phosphorylation (red P). In S. cerevisiae, substitution mutations that prevent aspartyl phosphorylation of Skn7 by the upstream histidine kinase prevent activation of genes involved in cell wall integrity. S. pombe prr1 null mutants are mating and sporulation defective; however, the nonphosphorylatable prr1D418N mutant exhibits accelerated meiosis and enhanced sporulation, suggesting a role for phosphorylated Skn7 in activating repressors of meiosis and sporulation. Panel C depicts oxidative stress-mediated changes in Skn7 that are independent of aspartyl phosphorylation. In S. cerevisiae, these changes involve serine/threonine phosphorylation as well as interaction with the Yap1 transcription factor.
Since the cell wall is a vital aspect of fungal survival, signaling genes that respond to wall stress are likely an important component of organism's virulence profile. Whether the net effect of such mutations is to compromise survival in the host no doubt depends on complex interactions between the fungal wall molecules and the determinants recognized by the immune system. In S. cerevisiae, a subset of wall-related mutations provoke a “compensatory response” that affords the cell alternative ways to survive in the absence of a normal complement of wall constituents (73). The variable effect of the skn7 mutation on virulence might be a simple reflection of the extent to which loss of Skn7 in a given organism might simultaneously render cells both more (due to loss of the oxidative stress response) and less (due to wall reinforcements) susceptible to host defenses.
An alternate explanation for the absence of correlation between oxidant sensitivity and virulence is suggested by the inverse relationship in C. neoformans. The oxidative stress function of Skn7 might be incompatible with a function in virulence. In organisms like Cryptococcus that lack an ortholog of the Yap1 protein with which Skn7 interacts to mediate the oxidative stress phenotype, Skn7 may have evolved a novel functionality in virulence. Interestingly, the T437 residue in ScSkn7 known to be important for the Skn7-Yap1 interaction is conserved in 17/17 Ascomycetes and several Basidiomycetes genes but is lysine in Crypotcoccus species, including C. neoformans and C. gattii, perhaps reflecting interactions with alternative proteins. At this time, Tremalles mesenterica, a mycoparasitic species from the Tremellales clade, is the only other publically available Basidiomycetes genome with a T/K substitution at this position in Skn7. T. mesenterica lacks a Yap1 homolog but is avirulent in Galleria mellonella infection assays (19). A comparison of SKN7 sequences in the genomes of additional pathogenic Basidiomycetes could be revealing.
ACKNOWLEDGMENTS
We thank Kailash Gulshan, Scott Moye-Rowley (University of Iowa), Maria Cardenas-Corona (Duke University), and John Logsdon (University of Iowa) for critical review of the manuscript and Robert Malone for valuable discussions.
We gratefully acknowledge funding from the NIH (GM59311 to A.H.W., GM68746 to Robert Deschenes and J.S.F., and GM56719 to J.S.F.), the Oklahoma Center for the Advancement of Science and Technology (A.H.W.), and the Center for Biocatalysis and Bioprocessing at the University of Iowa (J.S.F.) for the part of the work described here that is attributable to the authors.
Footnotes
Published ahead of print on 3 December 2010.
REFERENCES
- 1. Alberts A. S., Bouquin N., Johnston L. H., Treisman R. 1998. Analysis of RhoA-binding proteins reveals an interaction domain conserved in heterotrimeric G protein beta subunits and the yeast response regulator protein Skn7. J. Biol. Chem. 273:8616–8622 [DOI] [PubMed] [Google Scholar]
- 2. Bahn Y. S., Geunes-Boyer S., Heitman J. 2007. Ssk2 mitogen-activated protein kinase kinase kinase governs divergent patterns of the stress-activated Hog1 signaling pathway in Cryptococcus neoformans. Eukaryot. Cell 6:2278–2289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bahn Y. S., Kojima K., Cox G. M., Heitman J. 2006. A unique fungal two-component system regulates stress responses, drug sensitivity, sexual development, and virulence of Cryptococcus neoformans. Mol. Biol. Cell 17:3122–3135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bouquin N., Johnson A. L., Morgan B. A., Johnston L. H. 1999. Association of the cell cycle transcription factor Mbp1 with the Skn7 response regulator in budding yeast. Mol. Biol. Cell 10:3389–3400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Brown J. L., Bussey H., Stewart R. C. 1994. Yeast Skn7p functions in a eukaryotic two-component regulatory pathway. EMBO J. 13:5186–5194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Brown J. L., North S., Bussey H. 1993. SKN7, a yeast multicopy suppressor of a mutation affecting cell wall beta-glucan assembly, encodes a product with domains homologous to prokaryotic two-component regulators and to heat shock transcription factors. J. Bacteriol. 175:6908–6915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cao C., Liu W., Li R. 2009. Penicillium marneffei SKN7, a novel gene, could complement the hypersensitivity of S. cerevisiae skn7 disruptant strain to oxidative stress. Mycopathologia 168:23–30 [DOI] [PubMed] [Google Scholar]
- 8. Chang C., Meyerowitz E. M. 1994. Eukaryotes have “two-component” signal transducers. Res. Microbiol. 145:481–486 [DOI] [PubMed] [Google Scholar]
- 9. Chauhan N., et al. 2003. Candida albicans response regulator gene SSK1 regulates a subset of genes whose functions are associated with cell wall biosynthesis and adaptation to oxidative stress. Eukaryot. Cell 2:1018–1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chauhan N., Latge J. P., Calderone R. 2006. Signalling and oxidant adaptation in Candida albicans and Aspergillus fumigatus. Nat. Rev. Microbiol. 4:435–444 [DOI] [PubMed] [Google Scholar]
- 11. Coenjaerts F. E., et al. 2006. The Skn7 response regulator of Cryptococcus neoformans is involved in oxidative stress signalling and augments intracellular survival in endothelium. FEMS Yeast Res. 6:652–661 [DOI] [PubMed] [Google Scholar]
- 12. Cuellar-Cruz M., et al. 2008. High resistance to oxidative stress in the fungal pathogen Candida glabrata is mediated by a single catalase, Cta1p, and is controlled by the transcription factors Yap1p, Skn7p, Msn2p, and Msn4p. Eukaryot. Cell 7:814–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cui Z., Horecka J., Jigami Y. 2002. Cdc4 is involved in the transcriptional control of OCH1, a gene encoding alpha-1,6-mannosyltransferase in Saccharomyces cerevisiae. Yeast 19:69–77 [DOI] [PubMed] [Google Scholar]
- 14. Delaunay A., Isnard A. D., Toledano M. B. 2000. H2O2 sensing through oxidation of the Yap1 transcription factor. EMBO J. 19:5157–5166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Delaunay A., Pflieger D., Barrault M. B., Vinh J., Toledano M. B. 2002. A thiol peroxidase is an H2O2 receptor and redox-transducer in gene activation. Cell 111:471–481 [DOI] [PubMed] [Google Scholar]
- 16. DeRisi J. L., Iyer V. R., Brown P. O. 1997. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science 278:680–686 [DOI] [PubMed] [Google Scholar]
- 17. Eddy S. R. 1998. Profile hidden Markov models. Bioinformatics 14:755–763 [DOI] [PubMed] [Google Scholar]
- 18. Felsenstein J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791 [DOI] [PubMed] [Google Scholar]
- 19. Findley K., et al. 2009. Phylogeny and phenotypic characterization of pathogenic Cryptococcus species and closely related saprobic taxa in the Tremellales. Eukaryot. Cell 8:353–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fitzpatrick D. A., Logue M. E., Stajich J. E., Butler G. 2006. A fungal phylogeny based on 42 complete genomes derived from supertree and combined gene analysis. BMC Evol. Biol. 6:99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gasch A. P., et al. 2000. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell 11:4241–4257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hagiwara D., et al. 2007. The SskA and SrrA response regulators are implicated in oxidative stress responses of hyphae and asexual spores in the phosphorelay signaling network of Aspergillus nidulans. Biosci. Biotechnol. Biochem. 71:1003–1014 [DOI] [PubMed] [Google Scholar]
- 23. He X. J., Fassler J. S. 2005. Identification of novel Yap1p and Skn7p binding sites involved in the oxidative stress response of Saccharomyces cerevisiae. Mol. Microbiol. 58:1454–1467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. He X. J., Mulford K. E., Fassler J. S. 2009. Oxidative stress function of the Saccharomyces cerevisiae Skn7 receiver domain. Eukaryot. Cell 8:768–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Izumitsu K., et al. 2009. Dic2 and Dic3 loci confer osmotic adaptation and fungicidal sensitivity independent of the HOG pathway in Cochliobolus heterostrophus. Mycol. Res. 113:1208–1215 [DOI] [PubMed] [Google Scholar]
- 26. Izumitsu K., Yoshimi A., Tanaka C. 2007. Two-component response regulators Ssk1p and Skn7p additively regulate high-osmolarity adaptation and fungicide sensitivity in Cochliobolus heterostrophus. Eukaryot. Cell 6:171–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ketela T., Brown J. L., Stewart R. C., Bussey H. 1998. Yeast Skn7p activity is modulated by the Sln1p-Ypd1p osmosensor and contributes to regulation of the HOG pathway. Mol. Gen. Genet. 259:372–378 [DOI] [PubMed] [Google Scholar]
- 28. Ketela T., Green R., Bussey H. 1999. Saccharomyces cerevisiae mid2p is a potential cell wall stress sensor and upstream activator of the PKC1-MPK1 cell integrity pathway. J. Bacteriol. 181:3330–3340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ko Y. J., et al. 2009. Remodeling of global transcription patterns of Cryptococcus neoformans genes mediated by the stress-activated HOG signaling pathways. Eukaryot. Cell 8:1197–1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kojima K., Bahn Y. S., Heitman J. 2006. Calcineurin, Mpk1 and Hog1 MAPK pathways independently control fludioxonil antifungal sensitivity in Cryptococcus neoformans. Microbiology 152:591–604 [DOI] [PubMed] [Google Scholar]
- 31. Kojima K., et al. 2004. Fungicide activity through activation of a fungal signalling pathway. Mol. Microbiol. 53:1785–1796 [DOI] [PubMed] [Google Scholar]
- 32. Kosugi S., Hasebe M., Tomita M., Yanagawa H. 2009. Systematic identification of cell cycle-dependent yeast nucleocytoplasmic shuttling proteins by prediction of composite motifs. Proc. Natl. Acad. Sci. U. S. A. 106:10171–10176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Krems B., Charizanis C., Entian K. D. 1996. The response regulator-like protein Pos9/Skn7 of Saccharomyces cerevisiae is involved in oxidative stress resistance. Curr. Genet. 29:327–334 [DOI] [PubMed] [Google Scholar]
- 34. Kuge S., et al. 2001. Regulation of the yeast Yap1p nuclear export signal is mediated by redox signal-induced reversible disulfide bond formation. Mol. Cell. Biol. 21:6139–6150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kuge S., Jones N. 1994. YAP1 dependent activation of TRX2 is essential for the response of Saccharomyces cerevisiae to oxidative stress by hydroperoxides. EMBO J. 13:655–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kuge S., Jones N., Nomoto A. 1997. Regulation of yAP-1 nuclear localization in response to oxidative stress. EMBO J. 16:1710–1720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lamarre C., Ibrahim-Granet O., Du C., Calderone R., Latge J. P. 2007. Characterization of the SKN7 ortholog of Aspergillus fumigatus. Fungal Genet. Biol. 44:682–690 [DOI] [PubMed] [Google Scholar]
- 38. Larkin M. A., et al. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948 [DOI] [PubMed] [Google Scholar]
- 39. Lee J., et al. 1999. Yap1 and Skn7 control two specialized oxidative stress response regulons in yeast. J. Biol. Chem. 274:16040–16046 [DOI] [PubMed] [Google Scholar]
- 40. Lee J., Spector D., Godon C., Labarre J., Toledano M. B. 1999. A new antioxidant with alkyl hydroperoxide defense properties in yeast. J. Biol. Chem. 274:4537–4544 [DOI] [PubMed] [Google Scholar]
- 41. Li S. 2001. Molecular analysis of the Sln1 signaling pathway in the yeast Saccharomyces cerevisiae. Ph.D. thesis. University of Iowa, Iowa City [Google Scholar]
- 42. Li S., et al. 1998. The yeast histidine protein kinase, Sln1p, mediates phosphotransfer to two response regulators, Ssk1p and Skn7p. EMBO J. 17:6952–6962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Li S., et al. 2002. The eukaryotic two-component histidine kinase Sln1p regulates OCH1 via the transcription factor, Skn7p. Mol. Biol. Cell 13:412–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Loomis W. F., Shaulsky G., Wang N. 1997. Histidine kinases in signal transduction pathways of eukaryotes. J. Cell Sci. 110:1141–1145 [DOI] [PubMed] [Google Scholar]
- 45. Lu J. M., Deschenes R. J., Fassler J. S. 2004. Role for the Ran binding protein, Mog1p, in Saccharomyces cerevisiae SLN1-SKN7 signal transduction. Eukaryot. Cell 3:1544–1556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lu J. M., Deschenes R. J., Fassler J. S. 2003. Saccharomyces cerevisiae histidine phosphotransferase Ypd1p shuttles between the nucleus and cytoplasm for SLN1-dependent phosphorylation of Ssk1p and Skn7p. Eukaryot. Cell 2:1304–1314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lupas A., Van Dyke M., Stock J. 1991. Predicting coiled coils from protein sequences. Science 252:1162–1164 [DOI] [PubMed] [Google Scholar]
- 48. Luyten K., et al. 1995. Fps1, a yeast member of the MIP family of channel proteins, is a facilitator for glycerol uptake and efflux and is inactive under osmotic stress. EMBO J. 14:1360–1371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Maeda T., Wurgler-Murphy S. M., Saito H. 1994. A two-component system that regulates an osmosensing MAP kinase cascade in yeast. Nature 369:242–245 [DOI] [PubMed] [Google Scholar]
- 50. Mattison C. P., Ota I. M. 2000. Two protein tyrosine phosphatases, Ptp2 and Ptp3, modulate the subcellular localization of the Hog1 MAP kinase in yeast. Genes Dev. 14:1229–1235 [PMC free article] [PubMed] [Google Scholar]
- 51. Missall T. A., Lodge J. K. 2005. Function of the thioredoxin proteins in Cryptococcus neoformans during stress or virulence and regulation by putative transcriptional modulators. Mol. Microbiol. 57:847–858 [DOI] [PubMed] [Google Scholar]
- 52. Morgan B. A., et al. 1997. The Skn7 response regulator controls gene expression in the oxidative stress response of the budding yeast Saccharomyces cerevisiae. EMBO J. 16:1035–1044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Morgan B. A., Bouquin N., Merrill G. F., Johnston L. H. 1995. A yeast transcription factor bypassing the requirement for SBF and DSC1/MBF in budding yeast has homology to bacterial signal transduction proteins. EMBO J. 14:5679–5689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Motoyama T., et al. 2008. Involvement of putative response regulator genes of the rice blast fungus Magnaporthe oryzae in osmotic stress response, fungicide action, and pathogenicity. Curr. Genet. 54:185–195 [DOI] [PubMed] [Google Scholar]
- 55. Mouassite M., et al. 2000. The 'SUN′ family: yeast SUN4/SCW3 is involved in cell septation. Yeast 16:905–919 [DOI] [PubMed] [Google Scholar]
- 56. Moye-Rowley W. S., Harshman K. D., Parker C. S. 1988. YAP1 encodes a yeast homolog of mammalian transcription factor AP-1. Cold Spring Harbor Symp. Quant. Biol. 53:711–717 [DOI] [PubMed] [Google Scholar]
- 57. Moye-Rowley W. S., Harshman K. D., Parker C. S. 1989. Yeast YAP1 encodes a novel form of the jun family of transcriptional activator proteins. Genes Dev. 3:283–292 [DOI] [PubMed] [Google Scholar]
- 58. Nakamichi N., et al. 2003. Characterization of the Prr1 response regulator with special reference to sexual development in Schizosaccharomyces pombe. Biosci. Biotechnol. Biochem. 67:547–555 [DOI] [PubMed] [Google Scholar]
- 59. Nakayama K., Nagasu T., Shimma Y., Kuromitsu J., Jigami Y. 1992. OCH1 encodes a novel membrane bound mannosyltransferase: outer chain elongation of asparagine-linked oligosaccharides. EMBO J. 11:2511–2519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Nicholas K. B., Nicholas H. B. J., Deerfield D. W. I. 1997. GeneDoc: analysis and visualization of genetic variation. EMBNEW. News 4:14 [Google Scholar]
- 61. Ohmiya R., Yamada H., Kato C., Aiba H., Mizuno T. 2000. The Prr1 response regulator is essential for transcription of ste11+ and for sexual development in fission yeast. Mol. Gen. Genet. 264:441–451 [DOI] [PubMed] [Google Scholar]
- 62. Posas F., Saito H. 1998. Activation of the yeast SSK2 MAP kinase kinase kinase by the SSK1 two-component response regulator. EMBO J. 17:1385–1394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Posas F., et al. 1996. Yeast HOG1 MAP kinase cascade is regulated by a multistep phosphorelay mechanism in the SLN1-YPD1-SSK1 “two-component” osmosensor. Cell 86:865–875 [DOI] [PubMed] [Google Scholar]
- 64. Raitt D. C., et al. 2000. The Skn7 response regulator of Saccharomyces cerevisiae interacts with Hsf1 in vivo and is required for the induction of heat shock genes by oxidative stress. Mol. Biol. Cell 11:2335–2347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Reiser V., Raitt D. C., Saito H. 2003. Yeast osmosensor Sln1 and plant cytokinin receptor Cre1 respond to changes in turgor pressure. J. Cell Biol. 161:1035–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Reiser V., Ruis H., Ammerer G. 1999. Kinase activity-dependent nuclear export opposes stress-induced nuclear accumulation and retention of Hog1 mitogen-activated protein kinase in the budding yeast Saccharomyces cerevisiae. Mol. Biol. Cell 10:1147–1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Ruprich-Robert G., et al. 2008. Contributions of the response regulators Ssk1p and Skn7p in the pseudohyphal development, stress adaptation, and drug sensitivity of the opportunistic yeast Candida lusitaniae. Eukaryot. Cell 7:1071–1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Saijo T., et al. 2010. Skn7p is involved in oxidative stress response and virulence of Candida glabrata. Mycopathologia 169:81–90 [DOI] [PubMed] [Google Scholar]
- 69. Saitou N., Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406–425 [DOI] [PubMed] [Google Scholar]
- 70. Shankarnarayan S., Malone C. L., Deschenes R. J., Fassler J. S. 2008. Modulation of yeast Sln1 kinase activity by the CCW12 cell wall protein. J. Biol. Chem. 283:1962–1973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Singh P., Chauhan N., Ghosh A., Dixon F., Calderone R. 2004. SKN7 of Candida albicans: mutant construction and phenotype analysis. Infect. Immun. 72:2390–2394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Smith D. A., Morgan B. A., Quinn J. 2010. Stress signalling to fungal stress-activated protein kinase pathways. FEMS Microbiol. Lett. 306:1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Smits G. J., Kapteyn J. C., van den Ende H., Klis F. M. 1999. Cell wall dynamics in yeast. Curr. Opin. Microbiol. 2:348–352 [DOI] [PubMed] [Google Scholar]
- 74. Sonnhammer E. L., Eddy S. R., Birney E., Bateman A., Durbin R. 1998. Pfam: multiple sequence alignments and HMM-profiles of protein domains. Nucleic Acids Res. 26:320–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Tamas M. J., et al. 1999. Fps1p controls the accumulation and release of the compatible solute glycerol in yeast osmoregulation. Mol. Microbiol. 31:1087–1104 [DOI] [PubMed] [Google Scholar]
- 76. Tamura K., Dudley J., Nei M., Kumar S. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596–1599 [DOI] [PubMed] [Google Scholar]
- 77. Tao W., Deschenes R. J., Fassler J. S. 1999. Intracellular glycerol levels modulate the activity of Sln1p, a Saccharomyces cerevisiae two-component regulator. J. Biol. Chem. 274:360–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Tsuzi D., Maeta K., Takatsume Y., Izawa S., Inoue Y. 2004. Distinct regulatory mechanism of yeast GPX2 encoding phospholipid hydroperoxide glutathione peroxidase by oxidative stress and a calcineurin/Crz1-mediated Ca2+ signaling pathway. FEBS Lett. 569:301–306 [DOI] [PubMed] [Google Scholar]
- 79. Tsuzi D., Maeta K., Takatsume Y., Izawa S., Inoue Y. 2004. Regulation of the yeast phospholipid hydroperoxide glutathione peroxidase GPX2 by oxidative stress is mediated by Yap1 and Skn7. FEBS Lett. 565:148–154 [DOI] [PubMed] [Google Scholar]
- 80. Vargas-Perez I., Sanchez O., Kawasaki L., Georgellis D., Aguirre J. 2007. Response regulators SrrA and SskA are central components of a phosphorelay system involved in stress signal transduction and asexual sporulation in Aspergillus nidulans. Eukaryot. Cell 6:1570–1583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Vidan S., Mitchell A. P. 1997. Stimulation of yeast meiotic gene expression by the glucose-repressible protein kinase Rim15p. Mol. Cell. Biol. 17:2688–2697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Volz K., Matsumura P. 1991. Crystal structure of Escherichia coli CheY refined at 1.7-A resolution. J. Biol. Chem. 266:15511–15519 [DOI] [PubMed] [Google Scholar]
- 83. Wang H., Xu Z., Gao L., Hao B. 2009. A fungal phylogeny based on 82 complete genomes using the composition vector method. BMC Evol. Biol. 9:195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Wemmie J. A., Wu A. L., Harshman K. D., Parker C. S., Moye-Rowley W. S. 1994. Transcriptional activation mediated by the yeast AP-1 protein is required for normal cadmium tolerance. J. Biol. Chem. 269:14690–14697 [PubMed] [Google Scholar]
- 85. Wormley F. L., Jr., Heinrich G., Miller J. L., Perfect J. R., Cox G. M. 2005. Identification and characterization of an SKN7 homologue in Cryptococcus neoformans. Infect. Immun. 73:5022–5030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Wurgler-Murphy S. M., Saito H. 1997. Two-component signal transducers and MAPK cascades. Trends Biochem. Sci. 22:172–176 [DOI] [PubMed] [Google Scholar]
- 87. Zhang Y., Lamm R., Pillonel C., Lam S., Xu J. R. 2002. Osmoregulation and fungicide resistance: the Neurospora crassa os-2 gene encodes a HOG1 mitogen-activated protein kinase homologue. Appl. Environ. Microbiol. 68:532–538 [DOI] [PMC free article] [PubMed] [Google Scholar]