Abstract
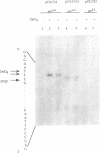
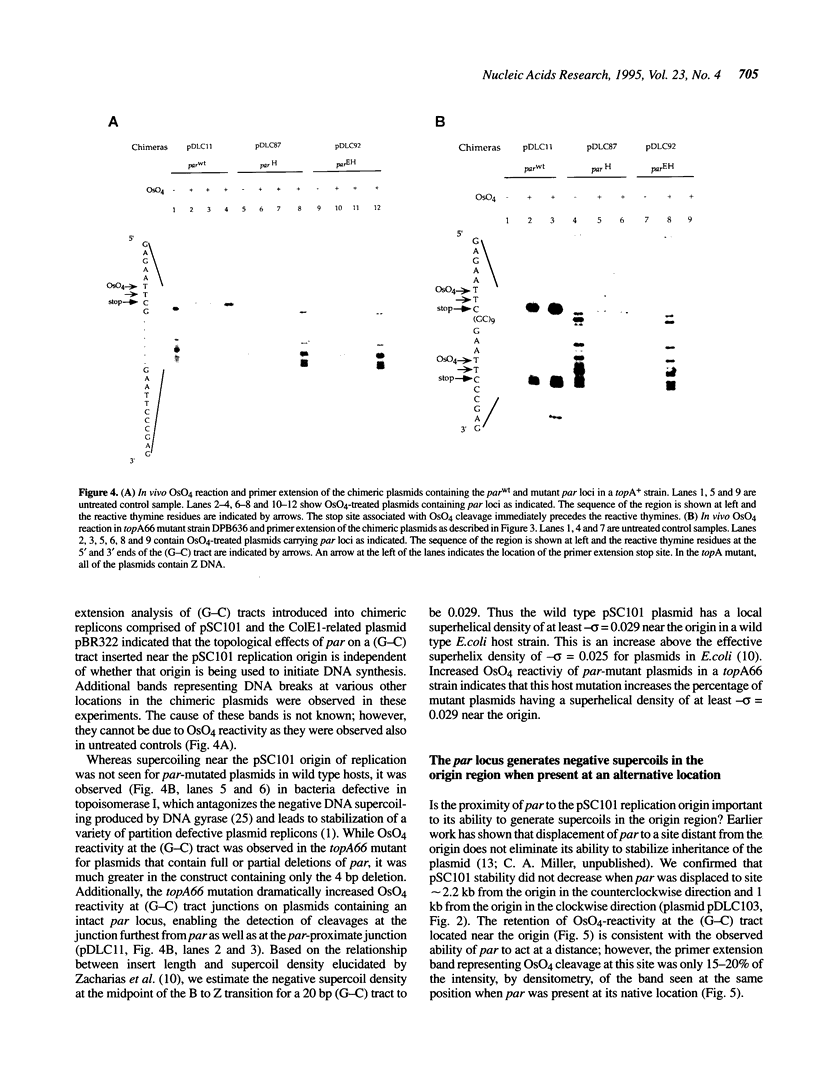
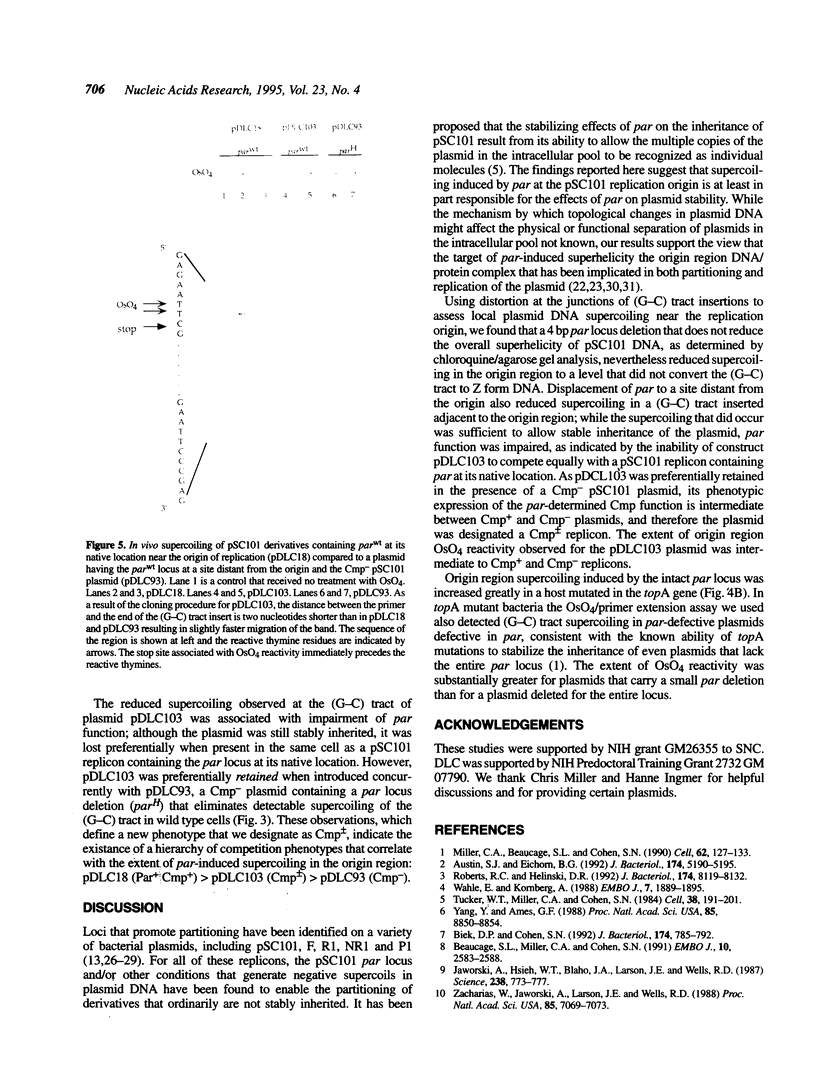
Previous work has shown that deletion of the partition (par) locus of plasmid pSC101 results in decreased overall superhelical density of plasmid DNA and concommitant inability of the plasmid to be stably inherited in populations of dividing cells. We report here that the biological effects of par correlate specifically with its ability to generate supercoils in vivo near the origin of pSC101 DNA replication. Using OsO4 reactivity of nucleotides adjoining 20 bp (G-C) tracts introduced into pSC101 DNA to measure local DNA supercoiling, we found that the wild type par locus generates supercoiling near the plasmid's replication origin adequate to convert a (G-C) tract in the region to Z form DNA. A 4 bp deletion that decreases par function, but produces no change in the overall superhelicity of pSC101 DNA as determined by chloroquine/agarose gel analysis, nevertheless reduced (G-C) tract supercoiling sufficiently to eliminate OsO4 reactivity. Mutation of the bacterial topA gene, which results in stabilized inheritance of par-deleted plasmids, restored supercoiling of (G-C) tracts in these plasmids and increased OsO4 reactivity in par+ replicons. Removal of par to a site more distant from the origin decreased supercoiling in a (G-C) tract adjacent to the origin and diminished par function. Collectively, these findings indicate that par activity is dependent on its ability to produce supercoiling at the replication origin rather than on the overall superhelical density of the plasmid DNA.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Austin S. J., Eichorn B. G. Random diffusion can account for topA-dependent suppression of partition defects in low-copy-number plasmids. J Bacteriol. 1992 Aug;174(16):5190–5195. doi: 10.1128/jb.174.16.5190-5195.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin S., Abeles A. Partition of unit-copy miniplasmids to daughter cells. II. The partition region of miniplasmid P1 encodes an essential protein and a centromere-like site at which it acts. J Mol Biol. 1983 Sep 15;169(2):373–387. doi: 10.1016/s0022-2836(83)80056-4. [DOI] [PubMed] [Google Scholar]
- Beaucage S. L., Miller C. A., Cohen S. N. Gyrase-dependent stabilization of pSC101 plasmid inheritance by transcriptionally active promoters. EMBO J. 1991 Sep;10(9):2583–2588. doi: 10.1002/j.1460-2075.1991.tb07799.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardi A., Bernardi F. Complete sequence of pSC101. Nucleic Acids Res. 1984 Dec 21;12(24):9415–9426. doi: 10.1093/nar/12.24.9415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biek D. P., Cohen S. N. Identification and characterization of recD, a gene affecting plasmid maintenance and recombination in Escherichia coli. J Bacteriol. 1986 Aug;167(2):594–603. doi: 10.1128/jb.167.2.594-603.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biek D. P., Cohen S. N. Involvement of integration host factor (IHF) in maintenance of plasmid pSC101 in Escherichia coli: characterization of pSC101 mutants that replicate in the absence of IHF. J Bacteriol. 1989 Apr;171(4):2056–2065. doi: 10.1128/jb.171.4.2056-2065.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biek D. P., Cohen S. N. Involvement of integration host factor (IHF) in maintenance of plasmid pSC101 in Escherichia coli: mutations in the topA gene allow pSC101 replication in the absence of IHF. J Bacteriol. 1989 Apr;171(4):2066–2074. doi: 10.1128/jb.171.4.2066-2074.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biek D. P., Cohen S. N. Propagation of pSC101 plasmids defective in binding of integration host factor. J Bacteriol. 1992 Feb;174(3):785–792. doi: 10.1128/jb.174.3.785-792.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabello F., Timmis K., Cohen S. N. Replication control in a composite plasmid constructed by in vitro linkage of two distinct replicons. Nature. 1976 Jan 29;259(5541):285–290. doi: 10.1038/259285a0. [DOI] [PubMed] [Google Scholar]
- Conley D. L., Cohen S. N. Isolation and characterization of plasmid mutations that enable partitioning of pSC101 replicons lacking the partition (par) locus. J Bacteriol. 1995 Feb;177(4):1086–1089. doi: 10.1128/jb.177.4.1086-1089.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983 Jun 5;166(4):557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- Ingmer H., Cohen S. N. Excess intracellular concentration of the pSC101 RepA protein interferes with both plasmid DNA replication and partitioning. J Bacteriol. 1993 Dec;175(24):7834–7841. doi: 10.1128/jb.175.24.7834-7841.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingmer H., Cohen S. N. The pSC101 par locus alters protein-DNA interactions in vivo at the plasmid replication origin. J Bacteriol. 1993 Sep;175(18):6046–6048. doi: 10.1128/jb.175.18.6046-6048.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaworski A., Hsieh W. T., Blaho J. A., Larson J. E., Wells R. D. Left-handed DNA in vivo. Science. 1987 Nov 6;238(4828):773–777. doi: 10.1126/science.3313728. [DOI] [PubMed] [Google Scholar]
- Linder P., Churchward G., Caro L. Plasmid pSC101 replication mutants generated by insertion of the transposon Tn1000. J Mol Biol. 1983 Oct 25;170(2):287–303. doi: 10.1016/s0022-2836(83)80149-1. [DOI] [PubMed] [Google Scholar]
- Meacock P. A., Cohen S. N. Partitioning of bacterial plasmids during cell division: a cis-acting locus that accomplishes stable plasmid inheritance. Cell. 1980 Jun;20(2):529–542. doi: 10.1016/0092-8674(80)90639-x. [DOI] [PubMed] [Google Scholar]
- Miki T., Easton A. M., Rownd R. H. Cloning of replication, incompatibility, and stability functions of R plasmid NR1. J Bacteriol. 1980 Jan;141(1):87–99. doi: 10.1128/jb.141.1.87-99.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller C. A., Beaucage S. L., Cohen S. N. Role of DNA superhelicity in partitioning of the pSC101 plasmid. Cell. 1990 Jul 13;62(1):127–133. doi: 10.1016/0092-8674(90)90246-b. [DOI] [PubMed] [Google Scholar]
- Miller C. A., Cohen S. N. The partition (par) locus of pSC101 is an enhancer of plasmid incompatibility. Mol Microbiol. 1993 Aug;9(4):695–702. doi: 10.1111/j.1365-2958.1993.tb01730.x. [DOI] [PubMed] [Google Scholar]
- Nordström K., Molin S., Aagaard-Hansen H. Partitioning of plasmid R1 in Escherichia coli. I. Kinetics of loss of plasmid derivatives deleted of the par region. Plasmid. 1980 Sep;4(2):215–227. doi: 10.1016/0147-619x(80)90011-6. [DOI] [PubMed] [Google Scholar]
- Ogura T., Hiraga S. Mini-F plasmid genes that couple host cell division to plasmid proliferation. Proc Natl Acad Sci U S A. 1983 Aug;80(15):4784–4788. doi: 10.1073/pnas.80.15.4784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahmouni A. R., Wells R. D. Stabilization of Z DNA in vivo by localized supercoiling. Science. 1989 Oct 20;246(4928):358–363. doi: 10.1126/science.2678475. [DOI] [PubMed] [Google Scholar]
- Roberts R. C., Helinski D. R. Definition of a minimal plasmid stabilization system from the broad-host-range plasmid RK2. J Bacteriol. 1992 Dec;174(24):8119–8132. doi: 10.1128/jb.174.24.8119-8132.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasse-Dwight S., Gralla J. D. Footprinting protein-DNA complexes in vivo. Methods Enzymol. 1991;208:146–168. doi: 10.1016/0076-6879(91)08012-7. [DOI] [PubMed] [Google Scholar]
- Tautz D., Renz M. An optimized freeze-squeeze method for the recovery of DNA fragments from agarose gels. Anal Biochem. 1983 Jul 1;132(1):14–19. doi: 10.1016/0003-2697(83)90419-0. [DOI] [PubMed] [Google Scholar]
- Tucker W. T., Miller C. A., Cohen S. N. Structural and functional analysis of the par region of the pSC 10 1 plasmid. Cell. 1984 Aug;38(1):191–201. doi: 10.1016/0092-8674(84)90540-3. [DOI] [PubMed] [Google Scholar]
- Vocke C., Bastia D. Primary structure of the essential replicon of the plasmid pSC101. Proc Natl Acad Sci U S A. 1983 Nov;80(21):6557–6561. doi: 10.1073/pnas.80.21.6557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vocke C., Bastia D. The replication initiator protein of plasmid pSC101 is a transcriptional repressor of its own cistron. Proc Natl Acad Sci U S A. 1985 Apr;82(8):2252–2256. doi: 10.1073/pnas.82.8.2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahle E., Kornberg A. The partition locus of plasmid pSC101 is a specific binding site for DNA gyrase. EMBO J. 1988 Jun;7(6):1889–1895. doi: 10.1002/j.1460-2075.1988.tb03022.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J. C. Interaction between DNA and an Escherichia coli protein omega. J Mol Biol. 1971 Feb 14;55(3):523–533. doi: 10.1016/0022-2836(71)90334-2. [DOI] [PubMed] [Google Scholar]
- Yamaguchi K., Masamune Y. Autogenous regulation of synthesis of the replication protein in plasmid pSC101. Mol Gen Genet. 1985;200(3):362–367. doi: 10.1007/BF00425718. [DOI] [PubMed] [Google Scholar]
- Yang Y., Ames G. F. DNA gyrase binds to the family of prokaryotic repetitive extragenic palindromic sequences. Proc Natl Acad Sci U S A. 1988 Dec;85(23):8850–8854. doi: 10.1073/pnas.85.23.8850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zacharias W., Jaworski A., Larson J. E., Wells R. D. The B- to Z-DNA equilibrium in vivo is perturbed by biological processes. Proc Natl Acad Sci U S A. 1988 Oct;85(19):7069–7073. doi: 10.1073/pnas.85.19.7069. [DOI] [PMC free article] [PubMed] [Google Scholar]