Abstract
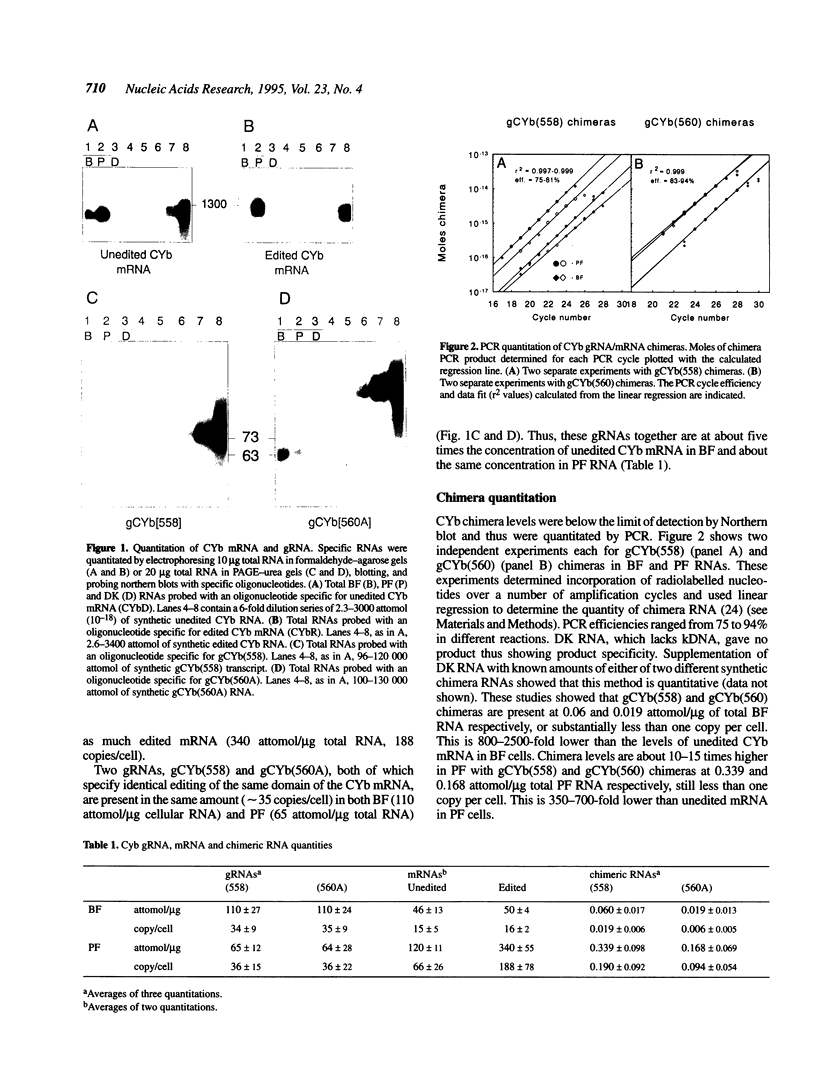
Kinetoplast mitochondrial RNA editing is the developmentally regulated post-transcriptional process of uridine insertion and deletion in mRNAs directed by short guide RNAs (gRNAs), which creates functional mRNAs. Two mechanisms are proposed: transesterification which predicts gRNA/mRNA chimeric intermediates, and enzymatic steps which allow but do not require chimeric intermediates. We quantitated the copy number of apocytochrome b (CYb) gRNAs, edited/unedited mRNAs and gRNA/mRNA chimeras in bloodstream and procyclic form cells of Trypanosoma brucei. Both forms have 35 copies/cell of two gRNAs. Bloodstream forms contain 15 unedited and edited CYb mRNA molecules/cell while procyclic forms have four times as much unedited and over 10 times as much edited mRNA. Chimera levels are very low, 350-5000-fold lower than unedited mRNA or gRNAs, but are over 10 times more abundant in procyclic than bloodstream forms. These results are consistent with chimeras being editing intermediates if their resolution is rapid in respect to their formation, although they could be non-productive byproducts of the editing reaction. Bloodstream chimera sequences differ from procyclic chimeras. These results indicate that developmental regulation is not by gRNA abundance and suggest that it occurs at the level of gRNA utilization possibly by changing abundance of unedited CYb mRNA.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Benne R., Van den Burg J., Brakenhoff J. P., Sloof P., Van Boom J. H., Tromp M. C. Major transcript of the frameshifted coxII gene from trypanosome mitochondria contains four nucleotides that are not encoded in the DNA. Cell. 1986 Sep 12;46(6):819–826. doi: 10.1016/0092-8674(86)90063-2. [DOI] [PubMed] [Google Scholar]
- Bhat G. J., Koslowsky D. J., Feagin J. E., Smiley B. L., Stuart K. An extensively edited mitochondrial transcript in kinetoplastids encodes a protein homologous to ATPase subunit 6. Cell. 1990 Jun 1;61(5):885–894. doi: 10.1016/0092-8674(90)90199-o. [DOI] [PubMed] [Google Scholar]
- Blum B., Bakalara N., Simpson L. A model for RNA editing in kinetoplastid mitochondria: "guide" RNA molecules transcribed from maxicircle DNA provide the edited information. Cell. 1990 Jan 26;60(2):189–198. doi: 10.1016/0092-8674(90)90735-w. [DOI] [PubMed] [Google Scholar]
- Blum B., Simpson L. Formation of guide RNA/messenger RNA chimeric molecules in vitro, the initial step of RNA editing, is dependent on an anchor sequence. Proc Natl Acad Sci U S A. 1992 Dec 15;89(24):11944–11948. doi: 10.1073/pnas.89.24.11944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum B., Simpson L. Guide RNAs in kinetoplastid mitochondria have a nonencoded 3' oligo(U) tail involved in recognition of the preedited region. Cell. 1990 Jul 27;62(2):391–397. doi: 10.1016/0092-8674(90)90375-o. [DOI] [PubMed] [Google Scholar]
- Blum B., Sturm N. R., Simpson A. M., Simpson L. Chimeric gRNA-mRNA molecules with oligo(U) tails covalently linked at sites of RNA editing suggest that U addition occurs by transesterification. Cell. 1991 May 17;65(4):543–550. doi: 10.1016/0092-8674(91)90087-f. [DOI] [PubMed] [Google Scholar]
- Cech T. R. RNA editing: world's smallest introns? Cell. 1991 Feb 22;64(4):667–669. doi: 10.1016/0092-8674(91)90494-j. [DOI] [PubMed] [Google Scholar]
- Chomczynski P., Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987 Apr;162(1):156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Corell R. A., Myler P., Stuart K. Trypanosoma brucei mitochondrial CR4 gene encodes an extensively edited mRNA with completely edited sequence only in bloodstream forms. Mol Biochem Parasitol. 1994 Mar;64(1):65–74. doi: 10.1016/0166-6851(94)90135-x. [DOI] [PubMed] [Google Scholar]
- Decker C. J., Sollner-Webb B. RNA editing involves indiscriminate U changes throughout precisely defined editing domains. Cell. 1990 Jun 15;61(6):1001–1011. doi: 10.1016/0092-8674(90)90065-m. [DOI] [PubMed] [Google Scholar]
- Feagin J. E., Jasmer D. P., Stuart K. Developmentally regulated addition of nucleotides within apocytochrome b transcripts in Trypanosoma brucei. Cell. 1987 May 8;49(3):337–345. doi: 10.1016/0092-8674(87)90286-8. [DOI] [PubMed] [Google Scholar]
- Feagin J. E., Shaw J. M., Simpson L., Stuart K. Creation of AUG initiation codons by addition of uridines within cytochrome b transcripts of kinetoplastids. Proc Natl Acad Sci U S A. 1988 Jan;85(2):539–543. doi: 10.1073/pnas.85.2.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feagin J. E., Stuart K. Developmental aspects of uridine addition within mitochondrial transcripts of Trypanosoma brucei. Mol Cell Biol. 1988 Mar;8(3):1259–1265. doi: 10.1128/mcb.8.3.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koslowsky D. J., Göringer H. U., Morales T. H., Stuart K. In vitro guide RNA/mRNA chimaera formation in Trypanosoma brucei RNA editing. Nature. 1992 Apr 30;356(6372):807–809. doi: 10.1038/356807a0. [DOI] [PubMed] [Google Scholar]
- Koslowsky D. J., Riley G. R., Feagin J. E., Stuart K. Guide RNAs for transcripts with developmentally regulated RNA editing are present in both life cycle stages of Trypanosoma brucei. Mol Cell Biol. 1992 May;12(5):2043–2049. doi: 10.1128/mcb.12.5.2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard V. W., Harris M. E., Hajduk S. L. Native mRNA editing complexes from Trypanosoma brucei mitochondria. EMBO J. 1992 Dec;11(12):4429–4438. doi: 10.1002/j.1460-2075.1992.tb05543.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard V. W., Rohrer S. P., Michelotti E. F., Hancock K., Hajduk S. L. Organization of minicircle genes for guide RNAs in Trypanosoma brucei. Cell. 1990 Nov 16;63(4):783–790. doi: 10.1016/0092-8674(90)90144-4. [DOI] [PubMed] [Google Scholar]
- Read L. K., Corell R. A., Stuart K. Chimeric and truncated RNAs in Trypanosoma brucei suggest transesterifications at non-consecutive sites during RNA editing. Nucleic Acids Res. 1992 May 11;20(9):2341–2347. doi: 10.1093/nar/20.9.2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley G. R., Corell R. A., Stuart K. Multiple guide RNAs for identical editing of Trypanosoma brucei apocytochrome b mRNA have an unusual minicircle location and are developmentally regulated. J Biol Chem. 1994 Feb 25;269(8):6101–6108. [PubMed] [Google Scholar]
- Souza A. E., Myler P. J., Stuart K. Maxicircle CR1 transcripts of Trypanosoma brucei are edited and developmentally regulated and encode a putative iron-sulfur protein homologous to an NADH dehydrogenase subunit. Mol Cell Biol. 1992 May;12(5):2100–2107. doi: 10.1128/mcb.12.5.2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart K., Gobright E., Jenni L., Milhausen M., Thomashow L., Agabian N. The IsTaR 1 serodeme of Trypanosoma brucei: development of a new serodeme. J Parasitol. 1984 Oct;70(5):747–754. [PubMed] [Google Scholar]
- Stuart K. Mitochondrial DNA of an African trypanosome. J Cell Biochem. 1983;23(1-4):13–26. doi: 10.1002/jcb.240230103. [DOI] [PubMed] [Google Scholar]
- Wiesner R. J. Direct quantification of picomolar concentrations of mRNAs by mathematical analysis of a reverse transcription/exponential polymerase chain reaction assay. Nucleic Acids Res. 1992 Nov 11;20(21):5863–5864. doi: 10.1093/nar/20.21.5863. [DOI] [PMC free article] [PubMed] [Google Scholar]