Abstract
Cronobacter sakazakii is an opportunistic pathogen that can cause severe infections. Serotyping provides a basis for the categorization of bacterial strains and is an important tool for epidemiological and surveillance purposes. In this study, of the 135 Cronobacter strains tested initially, 119 were identified as C. sakazakii and used. A serotyping scheme for C. sakazakii that classifies strains based on their different O antigens was developed. Seven antisera that exhibited high agglutinin titers (>640) were produced. O2 and O6 antisera were specific for their homologous strains, O4 and O7 antisera gave heterologous titers with O1 and O6 antigens, respectively, and O1, O3, and O5 antisera cross-reacted with each other and require preabsorption with the other two antigens. All of these 119 C. sakazakii strains were clearly assigned to these seven serotypes. O1 and O2 are the dominant serotypes, comprising 69.7% of the isolates. We also characterized the O-antigen gene clusters using restriction fragment length polymorphism (RFLP). The grouping of C. sakazakii strains based on their RFLP banding patterns correlated well with the grouping of strains based on our serotyping scheme. The serotype scheme presented here could prove to be a useful tool for serotyping C. sakazakii isolates.
Cronobacter sakazakii is a Gram-negative bacterium that can cause severe infection in neonates and may lead to necrotizing enterocolitis, sepsis, and meningitis (11, 13, 28). In addition to neonatal infection, recent reports have highlighted the risk posed to immunocompromised adults with bacteremia and osteomyelitis (8). The primary reservoir of Cronobacter spp. has not been determined, but it is postulated that plant material may be an important source (13). Cronobacter spp. have been isolated from a wide variety of foods, including milk, cheese, dried foods, meats, water, vegetables, rice, bread, tea, herbs, spices, and powdered infant formula (PIF) (3, 5, 13). Surveillance studies have detected Cronobacter spp. in households, livestock facilities, food factories, and PIF production facilities (20, 22).
Cronobacter (formerly Enterobacter sakazakii) was originally defined as a species and included 16 different biogroups (10, 19). Recently, Cronobacter has been reclassified as a genus and the strains reorganized into six distinct genomospecies, i.e., C. sakazakii, C. malonaticus, C. muytjensii, C. dublinensis, C. turicensis, and C. genomospecies 1. The original classified biogroups 1 to 4, 7, 8, 11, and 13 are proposed as C. sakazakii (18).
O antigen is a heat-stable antigenic component of the lipopolysaccharide (LPS) found in the outer membranes of Gram-negative bacteria. It is a major target of both the host immune system and bacteriophages and therefore is one of the most variable constituents of the cell (32). The diversity of O-antigen structures provides the primary basis for serotyping schemes that are used to classify Gram-negative bacteria. Since it was established by Kauffman in the 1940s, O-antigen serotyping has become the most widely used method of identifying strains for epidemiological and surveillance purposes. Although molecular typing approaches, such as pulsed-field gel electrophoresis (PFGE), multilocus sequence typing (MLST), and plasmid fingerprinting (PF), have been developed, serotyping remains valuable to characterize isolates for monitoring outbreaks and general bacterial surveillance and is still widely used. For example, the World Health Organization (WHO), jointly with the U.S. Centers for Disease Control and Prevention (CDC), has conducted routine Salmonella serotyping for public health purposes (14). Molecular methods for serotyping, including restriction fragment length polymorphism (RFLP), gene-specific PCR, and microarrays, now have been utilized to identify genes that define serotype (7, 12, 37).
Information related to Cronobacter O antigen is lacking. Only two serotypes, O1 and O2, have been designated so far, and their O-antigen gene clusters were sequenced and analyzed (27). Antisera against these two serotypes have not been produced or tested. Four C. sakazakii O-antigen chemical structures, including those of the O1 and O2 serotypes, have been characterized, and this indicated the existence of multiple O serotypes in C. sakazakii (1, 2, 9, 25). The genome sequence of C. sakazakii type strain ATCC BAA-894, which was previously assigned to the O1 serotype, was published recently (23, 27). So far there is no comprehensive scheme for O-antigen classification of C. sakazakii.
In this study, a total of 135 strains collected from the American Type Culture Collection (ATCC), the Czech Collection of Microorganisms (CCM), and various foods were characterized using a variety of phenotypic tests and sequence analysis of the 16S rRNA gene (>1,300 bp). Two methods, antiserum reactivity and O-antigen gene cluster RFLP (using long-range PCR to amplify the O-antigen gene clusters, followed by enzymatic restriction), were used to classify these strains. A serotyping scheme for the classification of C. sakazakii that includes seven different serotypes was developed.
MATERIALS AND METHODS
Bacterial strains.
A total of 135 strains were included in this study, including 4 strains (ATCC 29004, ATCC 29544, ATCC 12868, and ATCC 51329) from the ATCC, 3 strains (CCM 3460, CCM 3461, and CCM 3479) from the CCM, and 128 strains collected at 11 different Entry-Exit Inspection and Quarantine Bureaus in China from 2005 to 2010 (Table 1). The 128 strains collected at the various Entry-Exit Inspection and Quarantine Bureaus in China were originally identified as strains of the Cronobacter genus by using the API 20E, Vitek, or Biolog system.
TABLE 1.
Biogroups and sources of strains used in this study
| Species and biogroup | No. and sources of strains | Total no. of strains |
|---|---|---|
| C. sakazakii | 3,a 2,b 39,c 3,d 4,e 2,f 2,g 3,h 4,i 4,j 5,k 48m | 119 |
| 1 | 3,a 2,b 21,c 2,d 2,e 1,f 2,g 1,h 2,i 2,j 1,k 27m | 66 |
| 2 | 12,c 1,d 2,e 2,h 2i, 1,j 3,k 17m | 40 |
| 3 | 1,c 1,j 1m | 3 |
| 4 | 3,c 1,f 1m | 5 |
| 7 | 1m | 1 |
| 8 | 2,c 1m | 3 |
| 13 | 1k | 1 |
| C. malonaticus | 2e | 2 |
| 5 | 1e | 1 |
| 9 | 1e | 1 |
| C. muytjensii | 1a | 1 |
| 15 | 1a | 1 |
| C. dublinensis | 1,b 1,e 3,i 2m | 7 |
| 6 | 1,b 1,e 1,i 1m | 4 |
| 10 | 2,i 1m | 3 |
| C. turicensis | 2,e 3,i 1l | 6 |
| 16 | 2,e 3,i 1l | 6 |
| Total | 135 |
American Type Culture Collection (ATCC).
Czech Collection of Microorganisms (CCM).
Environmental isolates from food obtained at the Tianjin Entry-Exit Inspection and Quarantine Bureau, China.
Environmental isolates from food obtained at the Liaoning Entry-Exit Inspection and Quarantine Bureau, China.
Chinese Academy of Inspection and Quarantine, Beijing, China.
Environmental isolates from food obtained at the Jilin Entry-Exit Inspection and Quarantine Bureau, China.
Environmental isolates from food obtained at the Neimenggu Entry-Exit Inspection and Quarantine Bureau, China.
Environmental isolates from food obtained at the Shenyang Entry-Exit Inspection and Quarantine Bureau, China.
Environmental isolates from food obtained at the Xinjiang Entry-Exit Inspection and Quarantine Bureau, China.
Environmental isolates from food obtained at the Guangdong Entry-Exit Inspection and Quarantine Bureau, China.
Environmental isolates from food obtained at the Hunan Entry-Exit Inspection and Quarantine Bureau, China.
Environmental isolates from food obtained at the Hubei Entry-Exit Inspection and Quarantine Bureau, China.
Environmental isolates from food obtained at the Beijing Entry-Exit Inspection and Quarantine Bureau, China.
Bacterial identification.
Strains were initially assessed based on the color of their colonies (a blue-green color indicates α-glucosidase activity) on a chromogenic agar (DFI) (Beijing Land Bridge Technology Co., Ltd., China). A blue-green color indicated that a strain was likely a Cronobacter sp. 16S rRNA gene sequencing was then carried out as described previously (17, 18, 23). Amplification was performed using Taq DNA polymerase (TaKaRa Biotechnology Dalian Co. Ltd., China). PCR-generated DNA was purified using an agarose gel DNA fragment recovery kit (TaKaRa Biotechnology, Dalian Co. Ltd., China), and the purified PCR products were sequenced using an ABI 3730 automated DNA sequencer (Applied Biosystems). Ten phenotypic tests (Voges-Proskauer, methyl red, nitrate reduction, ornithine utilization, motility at 37°C, acid production from inositol, acid production from dulcitol, indole production, malonate utilization, and gas production from glucose) were also conducted using Microbiochemical tubes (Hangzhou Tianhe Microorganism Reagent Co. Ltd., China) (17, 19).
DNA preparation.
DNA was purified from Cronobacter cultures using a bacterial genomic DNA purification kit (Tiangen Biotech Co., Ltd. Beijing, China) according to the manufacturer's protocol. DNA samples were stored at −20°C.
Preparation of antigens for immunization.
All strains used for immunization were grown for 18 to 24 h in 400 ml of 2YT medium at 37°C. Cultures were harvested by centrifugation at 5,000 × g for 20 min, washed in 20 ml 0.85% NaCl, and finally suspended in 20 ml 0.85% NaCl (ca. 4 × 109 cells/ml). The cellular suspension was subsequently heated at 121°C for 30 min and then cooled. Formaldehyde (0.5%, vol/vol) was added, and the autoagglutination of suspensions was tested. Only strains that did not exhibit autoagglutination were used for immunization. The resultant antigens were stored at 4°C.
Production of antisera.
Adult New Zealand White rabbits (12 weeks of age) were injected intravenously with the prepared antigens. Two rabbits were used for each strain. At 3-day intervals, injections were given at doses of 0.8 ml, 1.5 ml, 2.5 ml, and 4.5 ml. One week after the final injection, a test bleed was taken from the marginal ear vein. If the titer was greater than 320, the rabbit was exsanguinated the next day. If antibody levels were inadequate, the rabbit was not used further. The separated antisera were stored at −20°C.
Tube agglutination.
Both titer determination and serotyping were carried out via tube agglutination. O antigens were prepared by autoclaving cultures taken from agar plates after growth for 18 to 24 h. Tube agglutination was performed in 96-well U-bottom microtiter plates with 25 μl of antigen and 25 μl of 2-fold-diluted serum in phosphate-buffered saline (PBS). Microtiter plates were incubated at 37°C for 4 h and then at 4°C overnight. The titer was taken to be the most diluted concentration of a serum to give a positive reaction compared to the PBS (50 mM phosphate buffer, 150 mM NaCl, pH 7.2) negative control.
Serum absorption.
Cell suspensions for absorption of agglutinins were prepared by inoculating moist, thickly poured (about 1 ml medium per plate) 90-mm infusion agar plates. Plates were incubated in an upright position for 18 to 24 h. The culture from the plate was then suspended in PBS and autoclaved at 121°C for 30 min before centrifugation at 5,000 × g for 20 min. The pellet was washed gently with PBS three times and resuspended in 2 ml of antiserum. The mixture was incubated at 37°C for 2 h. The mixture was centrifuged at 5,000 × g for 15 min and the supernatant collected. This absorbed antiserum was then tested against all type strains which reacted with the unabsorbed antiserum. This process was repeated until cross-reactions no longer occurred.
O-antigen gene cluster RFLP.
Long-range PCR analysis of the O-antigen gene cluster was performed with the Expand long-template PCR system (Roche Applied Science) using primers wl-10324 (5′-GCA CTG GTA GCT ATT GAG CCA GGG GCG GTA GCA T-3′) and wl-2211 (5′-ACT GCC ATA CCG ACG ACG CCG ATC TGT TGC TTG G-3′). These primer sequences are derived from the JUMPStart site and gnd gene, which flank the O-antigen gene cluster (15, 16). Thirty-two cycles of PCR were performed as follows: denaturation at 94°C for 30 s, annealing at 61°C for 30 s, and extension at 68°C for 15 min. About 800 ng PCR product was digested with 2.5 U MboII (Fermentas Canada Inc., Burlington, Ontario, Canada) in the supplied buffer. The mixture was incubated at 37°C for 2 h, followed by a final denaturation step at 65°C for 20 min. Five microliters of the digest was visualized on a 1.5% agarose gel. DNA fingerprints were stored as tagged image format files (TIFFs) and were analyzed with BioNumerics software (Applied Maths, Belgium). Cluster analysis was performed by the unweighted-pair group method with an arithmetic averages with BioNumerics software, based on the Dice coefficient with a 1% tolerance parameter.
Nucleotide sequence accession numbers.
A total of 135 full-length 16S rRNA gene sequences of Cronobacter spp. were deposited in GenBank under the accession numbers HQ880287 to HQ880421.
RESULTS
Identification of bacteria.
Chromogenic agar, 16S rRNA sequencing, and phenotypic characterization were used to identify each strain as to biogroup and species. Of the 135 strains used initially, 119 are of biogroups 1 to 4, 7, 8, and 13 and identified as C. sakazakii, 7 are of biogroups 6 and 10 and identified as C. dublinesis, 6 are of biogroup 16 and identified as C. turicensis, 2 are of biogroups 5 and 9 and identified as C. malonaticus, and 1 is of biogroup 15 and identified as C. muytjensii (Table 1). All of these 119 C. sakazakii strains were used in the subsequent study.
Establishment of C. sakazakii serotypes.
Seven antisera were ultimately selected for the current C. sakazakii serotype scheme, and all 119 tested C. sakazakii strains were clearly assigned to one of these seven serotypes.
Strains ATCC 29544 and ATCC 12868 have previously been assigned to the O1 and O2 serotypes, respectively (27). In this study, strains ATCC 29544 and ATCC 12868 were initially used as standard antigen strains to produce O1 and O2 antisera, respectively. The O1 and O2 antisera were then used to test all 119 C. sakazakii strains by tube agglutination. When the heterologous titer of a tested strain was similar (within one-quarter) to the homologous titer of the standard antigen strain, the tested strain was used to absorb the corresponding antiserum. If the homologous titer of absorbed antiserum significantly decreased, the tested strain was assigned to this particular serotype. In total, 49 strains were assigned to serotype O1 and 34 strains to serotype O2. Of the other 36 strains, 15 were selected randomly to prepare new antisera. These 15 experimental antisera together with the O1 and O2 antisera were evaluated by cross-agglutination and cross-absorption tests in which the antisera were titrated with homologous and heterologous antigens. When two or more antisera exhibited strong cross-reactions and cross-absorptions, only one was selected for inclusion in the provisional serotype scheme. Five of 15 experimental antisera were eventually selected for the present C. sakazakii serotype scheme. All strains that previously showed no reaction were tested once again for reactivity to the new antisera, and all were clearly assigned into these five serotypes.
Of the 119 C. sakazakii strains, 49 (41.2%) were assigned to serotype O1, 34 (28.6%) were assigned to O2, 8 (6.7%) were assigned to O3, 8 (6.7%) were assigned to O4, 4 (3.4%) were assigned to O5, 8 (6.7%) were assigned to O6, and 8 (6.7%) were assigned to O7.
Titer determination.
The homologous and heterologous titers of these seven antisera are shown in Table 2. In general, homologous titers were high, varying from 640 to 2,560. The heterologous reactions were fewer, and the titers were low (below 320). The O2 and O6 antisera were specific for their homologous strains. The O4 and O7 antisera gave heterologous titers with O1 and O6 antigens, respectively. A single absorption was required to render the O4 and O7 antisera specific. Serotypes O1, O3, and O5 appeared to represent an antigen pool, as these antisera agglutinated all the members of the pool. Their specific antisera need to be absorbed with the other two strains (Table 3).
TABLE 2.
Homologous and heterologous agglutinin titers of C. sakazakii antisera
| Antiserum type | Agglutinin titer to strains of serotypea: |
||||||
|---|---|---|---|---|---|---|---|
| O1 (ATCC 29544) | O2 (ATCC 12868) | O3 (G2726) | O4 (G2594) | O5 (G2706) | O6 (G2704) | O7 (G2592) | |
| O1 | 2,560 | 160 | 320 | ||||
| O2 | 640 | ||||||
| O3 | 40 | 640 | 40 | ||||
| O4 | 80 | 640 | |||||
| O5 | 160 | 160 | 1,280 | ||||
| O6 | 640 | ||||||
| O7 | 40 | 640 | |||||
Strain names in parentheses indicate the reference strains used for preparation of the antisera. Strains with G numbers are isolates collected at 11 different Entry-Exit Inspection and Quarantine Bureaus in China.
TABLE 3.
Agglutination of absorbed C. sakazakii antisera
| Antiserum type | Absorbing antigen(s) | Agglutinin titer to strains of serotype: |
||||
|---|---|---|---|---|---|---|
| O1 | O3 | O4 | O5 | O7 | ||
| O1 | O3, O5 | 1,280 | ||||
| O3 | O1, O5 | 640 | ||||
| O4 | O1 | 640 | ||||
| O5 | O1, O3 | 1,280 | ||||
| O7 | O6 | 640 | ||||
RFLP analysis of the O-antigen gene cluster.
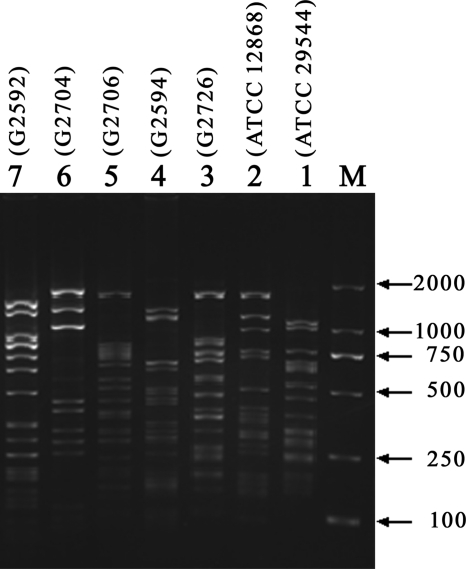
Long PCR products, with sizes of approximately 9 to 12 kb, were amplified in all 119 C. sakazakii strains (data not shown). Clearly identifiable and reproducible patterns were obtained for all of these 119 C. sakazakii strains after MboII digestion of amplified products. A dendrogram for all 119 C. sakazakii strains was generated using fragment analysis software, and these strains were grouped into seven clusters, with clusters 1 to 7 having 49, 34, 8, 8, 4, 8, and 8 strains, respectively (Fig. 1). The grouping of C. sakazakii strains based on their RFLP patterns correlated well with the grouping of strains based on our serotyping scheme (Fig. 1). The RFLP profiles for all seven O serotypes are shown in Fig. 2. The number of bands composing each pattern varied from 9 to 16 (Fig. 2).
FIG. 1.
Dendrogram for all 119 C. sakazakii strains, generated using the BioNumerics software program. Cluster analysis was performed by the unweighted-pair group method using arithmetic averages. Dotted lines indicate weak bands. Seven clusters, corresponding to the seven serotypes, were observed. Numbers in parentheses indicate the number of strains.
FIG. 2.
Gel electrophoresis showing RFLP binding patterns for all seven C. sakazakii O serotypes. Lane M, DL2000 DNA marker; lanes 1 to 7, serotypes O1 to O7, respectively. Strain names are given in parentheses.
DISCUSSION
In this study, 119 C. sakazakii strains were identified and used to develop this O-antigen serotyping scheme. These strains were collected from a wide range of sources, including 3 ATCC strains, 2 CCM strains, and 114 strains obtained from 11 different Entry-Exit Inspection and Quarantine Bureaus in China. The 114 C. sakazakii strains obtained within China were isolated from various foods, including milk powder, skim milk powder, whey powder, shrimp strips, soy bean, onion rings, chocolate, tea, cookies, instant noodles, ice cream, butter, cheese, and flavorings. Additionally, these foods were produced in many countries, including the United States, Ireland, France, Argentina, the Netherlands, India, Switzerland, Singapore, New Zealand, Denmark, Australia, and China (Fig. 1). These strains represent seven of the eight currently known biogroups of C. sakazakii, i.e., all except the rarely isolated biogroup 11 (19). Therefore, more strains, especially strains of biogroup 11, should be collected and analyzed in the future.
Differences in serotype distribution occur between regions, and serotype distribution may also change within the same region over time. In this study, the O1 and O2 serotypes comprised approximately 67% of all the isolates, making them the dominant serotypes. This kind of situation often happens and is not surprising. For example, in Kolkata, India, Vibrio parahaemolyticus O3:K6 was the most emergent serotype during 1996 to 1999, while O1:K25 changed to be the most emergent serotype during 2000 to 2004 (29). Of 114 environmental isolates, 56 can be traced to their country origins. Analysis revealed that the serotype distribution doses not have a clear association with country origins with one exception: six of nine C. sakazakii strains from India are assigned to serotype O3, two are O2 strains, and the last one is an O7 strain, which implies that O3 may be the prevalent serotype in India.
All 119 C. sakazakii strains can be clearly assigned to one of the seven antisera produced. Compared with 174 serotypes of Escherichia coli (36), 34 serotypes of Shigella (24), 46 serotypes of Salmonella (31), and more than 200 serotypes of Vibrio cholerae (35), the C. sakazakii O antigen is relatively unvaried. This may be due to adaptation to a simple niche, resulting in reduced selective pressure (33). Considering that E. coli, Shigella, and Salmonella colonize primarily animal intestine and Cronobacter spp. are usually associated with plant material (30), we presume that the reservoir of Cronobacter spp. is unlikely to be a complex environment, such as animal intestine. Recently, surveillance of Cronobacter spp. in farming and domestic environments, food production animals, and retail foods also demonstrated that Cronobacter spp. were not carried by food production animals (26).
A previous study showed that genes involved in O-antigen synthesis in C. sakazakii are located between the galF and gnd genes (27). In this study, the grouping of C. sakazakii strains based on their O-antigen gene cluster RFLP patterns correlated well with the grouping of strains based on our serotyping scheme, further proving that the region between the galF and gnd genes is indeed involved in C. sakazakii O-antigen synthesis and that the criteria we adopted for establishing new serotypes are correct. However, these two grouping results could differ due to the difference of O-antigen structures caused by genes located outside the O-antigen gene cluster. For instance, Shigella flexneri has the same O-antigen gene clusters, but the O-antigen structures actually presented on the bacterial surface differ in glucose side branches and O-acetyl residues. These different residues are attached by enzymes encoded by prophage genes (6, 24). In that case, the RFLP and antiserum analyses would not have agreed. In this study, no such inconsistencies were observed.
O antigen is on the cell surface and becomes one of the most variable cell constituents. The genome of C. sakazakii ATCC BAA-894 has been sequenced, and comparative genomic hybridization (CGH) has been undertaken. The results revealed that the genes responsible for O-antigen synthesis are highly divergent (23). The variety of O-antigen gene clusters can arise in many ways, such as homologous recombination, mutation, insertion or deletion mediated by insertion sequence (IS) elements or plasmids, and modification induced by phages (4, 7, 21, 24, 34, 38). Sequencing O-antigen gene clusters and determining the O-antigen chemical structures could help further define C. sakazakii serotypes.
In conclusion, an O-antigen serotyping scheme for C. sakazakii based on O-antigen antisera and O-antigen gene cluster RFLP was developed, and all the tested strains were adequately differentiated into seven clearly defined serotypes. Though more serotypes may be found with additional testing of more C. sakazakii strains, we believe that this simple and reliable method should be adopted as a routine procedure in typing C. sakazakii strains.
Acknowledgments
This study was supported by grants from the National Science Foundation of China (31000044), the Chinese National Science Fund for Distinguished Young Scholars (30788001), the National 863 Program of China (2006AA020703 and 2009AA06Z403), the National Key Program for Infectious Diseases of China (2008ZX10004-002), and the Fundamental Research Funds for the Central Universities (65010721).
Footnotes
Published ahead of print on 4 February 2011.
REFERENCES
- 1.Arbatsky, N. P., et al. 2010. Structure of the O-polysaccharide of Cronobacter sakazakii O2 with a randomly O-acetylated l-rhamnose residue. Carbohydr. Res. 345:2090-2094. [DOI] [PubMed] [Google Scholar]
- 2.Arbatsky, N. P., et al. 2010. Structure of the O-polysaccharide of Cronobacter sakazakii O1 containing 3-(N-acetyl-l-alanyl)amino-3,6-dideoxy-d-glucose. Carbohydr. Res. 345:2095-2098. [DOI] [PubMed] [Google Scholar]
- 3.Baumgartner, A., M. Grand, M. Liniger, and C. Iversen. 2009. Detection and frequency of Cronobacter spp. (Enterobacter sakazakii) in different categories of ready-to-eat foods other than infant formula. Int. J. Food Microbiol. 136:189-192. [DOI] [PubMed] [Google Scholar]
- 4.Bennett, P. M. 2004. Genome plasticity: insertion sequence elements, transposons and integrons, and DNA rearrangement. Methods Mol. Biol. 266:71-113. [DOI] [PubMed] [Google Scholar]
- 5.Chap, J., et al. 2009. International survey of Cronobacter sakazakii and other Cronobacter spp. in follow up formulas and infant foods. Int. J. Food Microbiol. 136:185-188. [DOI] [PubMed] [Google Scholar]
- 6.Coimbra, R. S., F. Grimont, and P. A. Grimont. 1999. Identification of Shigella serotypes by restriction of amplified O-antigen gene cluster. Res. Microbiol. 150:543-553. [DOI] [PubMed] [Google Scholar]
- 7.Coimbra, R. S., et al. 2000. Identification of Escherichia coli O-serogroups by restriction of the amplified O-antigen gene cluster (rfb-RFLP). Res. Microbiol. 151:639-654. [DOI] [PubMed] [Google Scholar]
- 8.Corti, G., I. Panunzi, M. Losco, and R. Buzzi. 2007. Postsurgical osteomyelitis caused by Enterobacter sakazakii in a healthy young man. J. Chemother. 19:94-96. [DOI] [PubMed] [Google Scholar]
- 9.Czerwicka, M., et al. 2010. Structure of the O-polysaccharide isolated from Cronobacter sakazakii 767. Carbohydr. Res. 345:908-913. [DOI] [PubMed] [Google Scholar]
- 10.Farmer, J. J., III, M. A. Asbury, F. W. Hickman, and D. J. Brenner. 1980. Enterobacter sakazakii: a new species of “Enterobacteriaceae” isolated from clinical specimens. Int. J. Syst. Bacteriol. 30:569-584. [Google Scholar]
- 11.Giovannini, M., et al. 2008. Enterobacter sakazakii: an emerging problem in paediatric nutrition. J. Int. Med. Res. 36:394-399. [DOI] [PubMed] [Google Scholar]
- 12.Han, W., et al. 2007. DNA microarray-based identification of serogroups and virulence gene patterns of Escherichia coli isolates associated with porcine postweaning diarrhea and edema disease. Appl. Environ. Microbiol. 73:4082-4088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Healy, B., et al. 2010. Cronobacter (Enterobacter sakazakii): an opportunistic foodborne pathogen. Foodborne Pathog. Dis. 7:339-350. [DOI] [PubMed] [Google Scholar]
- 14.Herikstad, H., Y. Motarjemi, and R. V. Tauxe. 2002. Salmonella surveillance: a global survey of public health serotyping. Epidemiol. Infect. 129:1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hobbs, M., and P. R. Reeves. 1994. The JUMPstart sequence: a 39 bp element common to several polysaccharide gene clusters. Mol. Microbiol. 12:855-856. [DOI] [PubMed] [Google Scholar]
- 16.Iguchi, A., et al. 2008. Genomic comparison of the O-antigen biosynthesis gene clusters of Escherichia coli O55 strains belonging to three distinct lineages. Microbiology 154:559-570. [DOI] [PubMed] [Google Scholar]
- 17.Iversen, C., et al. 2007. The taxonomy of Enterobacter sakazakii: proposal of a new genus Cronobacter gen. nov. and descriptions of Cronobacter sakazakii comb. nov., Cronobacter sakazakii subsp. sakazakii comb. nov., Cronobacter sakazakii subsp. malonaticus subsp. nov., Cronobacter turicensis sp. nov., Cronobacter muytjensii sp. nov., Cronobacter dublinensis sp. nov. and Cronobacter genomospecies 1. BMC Evol. Biol. 7:64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iversen, C., et al. 2008. Cronobacter gen. nov., a new genus to accommodate the biogroups of Enterobacter sakazakii, and proposal of Cronobacter sakazakii gen. nov., comb. nov., Cronobacter malonaticus sp. nov., Cronobacter turicensis sp. nov., Cronobacter muytjensii sp. nov., Cronobacter dublinensis sp. nov., Cronobacter genomospecies 1, and of three subspecies, Cronobacter dublinensis subsp. dublinensis subsp. nov., Cronobacter dublinensis subsp. lausannensis subsp. nov. and Cronobacter dublinensis subsp. lactaridi subsp. nov. Int. J. Syst. Evol. Microbiol. 58:1442-1447. [DOI] [PubMed] [Google Scholar]
- 19.Iversen, C., M. Waddington, J. J. Farmer III, and S. J. Forsythe. 2006. The biochemical differentiation of Enterobacter sakazakii genotypes. BMC Microbiol. 6:94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kandhai, M. C., M. W. Reij, L. G. Gorris, O. Guillaume-Gentil, and M. van Schothorst. 2004. Occurrence of Enterobacter sakazakii in food production environments and households. Lancet 363:39-40. [DOI] [PubMed] [Google Scholar]
- 21.Kido, N., and H. Kobayashi. 2000. A single amino acid substitution in a mannosyltransferase, WbdA, converts the Escherichia coli O9 polysaccharide into O9a: generation of a new O-serotype group. J. Bacteriol. 182:2567-2573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kilonzo-Nthenge, A., F. C. Chen, and S. L. Godwin. 2008. Occurrence of Listeria and Enterobacteriaceae in domestic refrigerators. J. Food Prot. 71:608-612. [DOI] [PubMed] [Google Scholar]
- 23.Kucerova, E., et al. 2010. Genome sequence of Cronobacter sakazakii BAA-894 and comparative genomic hybridization analysis with other Cronobacter species. PLoS One 5:e9556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu, B., et al. 2008. Structure and genetics of Shigella O antigens. FEMS Microbiol. Rev. 32:627-653. [DOI] [PubMed] [Google Scholar]
- 25.Maclean, L. L., E. Vinogradov, F. Pagotto, J. M. Farber, and M. B. Perry. 2010. The structure of the O-antigen of Cronobacter sakazakii HPB 2855 isolate involved in a neonatal infection. Carbohydr. Res. 345:1932-1937. [DOI] [PubMed] [Google Scholar]
- 26.Molloy, C., et al. 2009. Surveillance and characterisation by pulsed-field gel electrophoresis of Cronobacter spp. in farming and domestic environments, food production animals and retail foods. Int. J. Food Microbiol. 136:198-203. [DOI] [PubMed] [Google Scholar]
- 27.Mullane, N., et al. 2008. Molecular analysis of the Enterobacter sakazakii O-antigen gene locus. Appl. Environ. Microbiol. 74:3783-3794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mullane, N. R., et al. 2007. Enterobacter sakazakii an emerging bacterial pathogen with implications for infant health. Minerva Pediatr. 59:137-148. [PubMed] [Google Scholar]
- 29.Nair, G. B., et al. 2007. Global dissemination of Vibrio parahaemolyticus serotype O3:K6 and its serovariants. Clin. Microbiol. Rev. 20:39-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Osaili, T., and S. Forsythe. 2009. Desiccation resistance and persistence of Cronobacter species in infant formula. Int. J. Food Microbiol. 136:214-220. [DOI] [PubMed] [Google Scholar]
- 31.Popoff, M. Y., J. Bockemuhl, and L. L. Gheesling. 2003. Supplement 2001 (no. 45) to the Kauffmann-White scheme. Res. Microbiol. 154:173-174. [DOI] [PubMed] [Google Scholar]
- 32.Reeves, P. P., and L. Wang. 2002. Genomic organization of LPS-specific loci. Curr. Top. Microbiol. Immunol. 264:109-135. [PubMed] [Google Scholar]
- 33.Reeves, P. R. 1992. Variation in O-antigens, niche-specific selection and bacterial populations. FEMS Microbiol. Lett. 79:509-516. [DOI] [PubMed] [Google Scholar]
- 34.Selander, R. K., P. Beltran, and N. H. Smith. 1991. Evolutionary genetics of Salmonella, p. 25-27. In R. K. Selander, A. G. Clark, and T. S. Whittam (ed.), Evolution at the molecular level. Sinauer Associates, Sunderland, MA.
- 35.Shimada, T., et al. 1994. Extended serotyping scheme for Vibrio cholerae. Curr. Microbiol. 28:175-178. [Google Scholar]
- 36.Stenutz, R., A. Weintraub, and G. Widmalm. 2006. The structures of Escherichia coli O-polysaccharide antigens. FEMS Microbiol. Rev. 30:382-403. [DOI] [PubMed] [Google Scholar]
- 37.Wang, L., and P. R. Reeves. 1998. Organization of Escherichia coli O157 O antigen gene cluster and identification of its specific genes. Infect. Immun. 66:3545-3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang, Q., et al. 2009. Genetic and structural analyses of Escherichia coli O107 and O117 O-antigens. FEMS Immunol. Med. Microbiol. 55:47-54. [DOI] [PubMed] [Google Scholar]