Abstract
Nine thermophilic cellulolytic clostridial isolates and four other noncellulolytic bacterial isolates were isolated from self-heated biocompost via preliminary enrichment culture on microcrystalline cellulose. All cellulolytic isolates grew vigorously on cellulose, with the formation of either ethanol and acetate or acetate and formate as principal fermentation products as well as lactate and glycerol as minor products. In addition, two out of nine cellulolytic strains were able to utilize xylan and pretreated wood with roughly the same efficiency as for cellulose. The major products of xylan fermentation were acetate and formate, with minor contributions of lactate and ethanol. Phylogenetic analyses of 16S rRNA and glycosyl hydrolase family 48 (GH48) gene sequences revealed that two xylan-utilizing isolates were related to a Clostridium clariflavum strain and represent a distinct novel branch within the GH48 family. Both isolates possessed high cellulase and xylanase activity induced independently by either cellulose or xylan. Enzymatic activity decayed after growth cessation, with more-rapid disappearance of cellulase activity than of xylanase activity. A mixture of xylan and cellulose was utilized simultaneously, with a significant synergistic effect observed as a reduction of lag phase in cellulose degradation.
Plant biomass represents an abundant and valuable renewable natural resource that may be used for a wide range of purposes, including the production of fuels and chemicals in addition to food and feed (41). Microbial conversion of cellulosic biomass is a promising strategy for low-cost biomass processing (21). After cellulose, xylan is the most abundant polymer in plants (40). Because of this, microbial conversion of hemicellulose and hemicellulose sugars is a subject of active research (29) and xylanases are of interest for application in pulp and paper (18) as well as products of other industries (4).
Several anaerobic thermophiles have been shown to utilize cellulose, including Clostridium thermocellum, Clostridium straminisolvens, Clostridium stercorarium, Caldicellulosiruptor saccharolyticus, and Caldicellulosiruptor obsidiansis (8, 12, 15, 23, 27, 44). C. thermocellum exhibits a high growth rate on crystalline cellulose (22) but does not utilize xylan, does not grow on xylose and other pentoses, and grows poorly on glucose (20). Extremely thermophilic cellulolytic Caldicellulosiruptor saccharolyticus can coutilize glucose and xylose (42), and its close relative Caldicellulosiruptor bescii DSM 6725 has been found to degrade xylan and xylose by Yang et al. (45), although the original report (36) about this organism described it to be unable to grow on xylose. C. bescii was recently shown to utilize cellulose and hemicellulose originating from lignocellulose (45), although with a relatively low yield of fermentation products, indicating a degree of substrate conversion below 20%. Several mesophilic Clostridium strains have been reported to utilize both cellulose and xylan, including strains of Clostridium phytofermentans and C. cellulovorans (17, 34, 43). However, C. stercorarium is the only thermophilic cellulolytic Clostridium species reported to utilize xylan, and the rate of cellulose degradation by C. stercorarium is modest compared to the rate for C. thermocellum (1, 46).
In our preceding study (13), we obtained three cellulolytic consortia from self-heated biocompost. They contained cumulatively 30 and 11 operational taxonomic units (OTUs), based on a survey of the 16S rRNA and GH48 (glycosyl hydrolase family 48) gene clone libraries, respectively. Here, we report on the next step of our biocompost study that resulted in isolation, purification, and characterization of nine thermophilic, cellulolytic bacterial isolates. Most of the isolates turned out to be different from C. thermocellum and display similarly high cellulolytic activity. Two isolates are able to degrade cellulose, xylan, and their mixture with the same efficiency and rates.
MATERIALS AND METHODS
Field site.
Samples were collected from the Middlebury College compost facility in May 2008 at locations 40 to 50 cm below the surface and temperatures ranging from 52 to 72°C across sampling sites. The original compost material consisted of three roughly equal parts: wood chips, horse manure, and food waste.
Sampling and enrichment procedure.
Strictly anaerobic conditions were maintained by flushing with pure N2 throughout primary sampling and subsequent inoculation. Compost samples were obtained with a T-shaped steel corer with a 2-cm diameter at three distinct spots, designated CO-4, CO-5, and CO-6. Between 8 and 15 g of extracted material was transferred into serum bottles containing 100 ml of mineral medium, pH 7, and 1 gram of microcrystalline cellulose (Avicel PH105; FMC Corp., Philadelphia, PA) or filter paper (no. 1; Whatman). After inoculation, each bottle was immediately flushed with N2. In addition, “cellulosic traps” consisting of stainless steel tea strainers containing about 1 g of filter paper strips were placed in the same compost. Traps were placed at a depth of about 40 cm and left in situ to obtain growth of cellulolytic microorganisms. After 1 week of incubation, the traps were retrieved and delivered to the laboratory, and partly decayed filter paper was used for inoculation of cellulose-mineral media.
Nutrient medium.
The enrichment medium did not contain sulfates and nitrates to preclude development of sulfate-reducing or denitrifying bacteria competing with fermenting cellulose-degrading microorganisms. PE medium for primary enrichments contained the following concentrations (g/liter): KH2PO4, 2.08; K2HPO4, 2.22; MgCl2·6H2O, 0.1; NH4Cl, 0.4; and CaCl2·2H2O, 0.05. Upon arriving at the laboratory, primary enrichments were incubated at 55°C and transferred to fresh medium every 4 to 6 days. For subsequent transfers, chemically defined minimal medium M was used. This medium contained the following concentrations (g/liter): Avicel, 3; KH2PO4, 1.04; K2HPO4, 1.11; NaHCO3, 2.5; MgCl2·6H2O, 0.2; NH4Cl, 0.4; CaCl2·2H2O, 0.05; FeCl2·4H2O, 0.05; l-cysteine-HCl, 0.5; and resazurin, 0.0025. SL-10 trace element (1 ml/liter) (3) and vitamin (4 ml/liter) solutions were added as concentrated solutions. The vitamin solution contained the following concentrations (g/liter): pyridoxamine dihydrochloride, 0.2; p-aminobenzoic acid (PABA), 0.1; d-biotin, 0.05; vitamin B12, 0.05; thiamine-HCl, 0.0125; folic acid, 0.5; Ca-pantothenate, 0.125; nicotinic acid, 0.125; pyridoxine-HCl, 0.025; thioctic acid, 0.125; and riboflavin, 0.0125 (all purchased from Sigma-Aldrich).
Vitamins were sterilized by filtration. Phosphates were autoclaved separately to avoid precipitation. Other minerals and l-cysteine were prepared as a 100×-concentrated stock solution, flushed with N2 and autoclaved. All sterile and reduced ingredients were combined in serum bottles inside an anaerobic glove box, sealed and crimped.
Isolation of pure cultures and total cell counts.
Isolation of pure cultures of cellulose-utilizing bacteria was performed on agar-Avicel and agar-cellobiose media after 10 consecutive transfers on M medium with the following changes: 15 g/liter of agar was added, vitamins were replaced with 2.0 g/liter of yeast extract, and the concentration of Avicel was increased to 20 g/liter. Cellulolytic consortia were serially diluted into agar-Avicel medium that had preliminarily been melted and cooled to 55°C and then plated under strictly anoxic conditions in anaerobic glove box. After solidifying, plates were incubated under anaerobic conditions at 55°C.
Cellulose-utilizing bacteria formed colonies with clearing zones. CFU were counted after 4 to 7 days of incubation at 55°C. Single colonies were picked with a syringe needle and inoculated into cellulose- and cellobiose-mineral liquid M medium. Isolates first grown on cellobiose medium were transferred onto Avicel medium to evaluate the ability to utilize cellulose.
Cultivation and growth measurement.
Batch fermentation of various substrates was carried out in serum bottles with a total volume of 240 ml filled with 100 ml of defined M medium and 1.5 to 5.0 g/liter of Avicel, xylan (from Birchwood, Sigma, or Oats Spelts, Tokyo Chemical Industry Co., Ltd., Japan), xylose, pretreated wood (obtained from Mascoma Corp. as a mixed steam-pretreated hardwood and then washed, grounded, and dried before addition to media), or a mixture of Avicel and xylan. One volume percent of actively growing Avicel batch cultures was used as an inoculum. Two or three bottles of each culture were incubated at 55°C with agitation at 180 rpm. Two replicate 2-ml subsamples were taken periodically from each bottle and immediately centrifuged at 10,000 × g. Supernatants were used to measure pH (pH-meter UB-10 [Denver Instrument] and Orion 410 [Orion Research, Inc.]) and fermentation products by high-performance liquid chromatography (HPLC) (19). Pellets were washed and analyzed for total pellet C (TPC) and total pellet N (TPN) content with a Shimadzu TOC-V combustion analyzer coupled with a TNM-1 total nitrogen module (26). A glycine solution (1 g/liter) was used as a standard. The TPC and TPN data allowed calculation of cell mass and residual insoluble substrate as described earlier (26): the TPN was interpreted as microbial N, assuming that contribution of extracellular enzymes to the pellet N was negligible and that cellulose, xylan, and pretreated wood were N free. In preliminary tests, it was found that the majority of N compounds interfering with cell N determination (NH4+, amino acids, peptides, and extracellular proteins) were completely removed by pellet washing. The bacterial N content in cells of Clostridium thermocellum and isolate 4-2a grown on cellobiose was measured, and this content was observed to differ in a relatively narrow range (0.27 ± 0.02 g N/g C). The yield of tested strains was calculated within a period of intensive batch growth as a cell mass increment (based on TPN with the above-indicated N/C ratio) per unit of consumed insoluble substrate (with respective declines in residual TPC of cellulose or xylan corrected for cell mass).
Enzyme assay.
Cellulase activity was analyzed with a MarkerGene fluorescent-cellulase assay kit (MarkerGene Technologies, Inc.) with the fluorescent substrate resorufin cellobioside (6). Fluorescence was recorded using a Gen5 microtiter plate reader (BioTeck Instruments, Inc.) with a 530/25-nm excitation filter and a 595/35-nm emission filter. Fluorescence reading was taken at 2-min intervals for 60 min.
Xylanase activity analysis was performed with an EnzChek Ultra xylanase assay kit (Molecular Probes, Inc.) using the fluorescent substrate 6,8-difluoro-4-methylumbelliferyl β-d-xylobioside (DiFMUX2) (9). Fluorescence was recorded with the same instrument, using a 360/40-nm excitation filter and a 460/40-nm emission filter. Fluorescence reading was taken at 2-min intervals for 90 min. The activities of both enzymes (cellulase and xylanase) in the original cultural liquid (sum of free and bound enzymes) and in the supernatant after centrifugation of the suspension at 13,000 rpm (number of free enzymes) were recorded. Enzymatic activity was calculated from the slope of the linear segment of the product accumulation curve or as the initial rate of fluorescent-product release when the hydrolysis rate was progressively declining during the measurement period. Control for self-decay of fluorescent substrate was set up according to the manufacturer's instructions. Three to six replicates of cultural liquid and spent medium were used to determine enzymatic activity at each time point to calculate the mean and standard deviation.
DNA extraction, PCR amplification, and phylogenetic analysis.
Genomic DNA was extracted from microbial biomass with a GenElute genomic DNA kit (Sigma) according to the manufacturer's instructions. PCR amplification of the 16S rRNA gene and sequencing were done as described before (33). Amplification of GH48 genes was performed with the GH48F and GH48R degenerate primers as previously described (13). For C. thermocellum and C. straminisolvens-type isolates, coamplification of celS and celY genes was resolved by constructing clone libraries with amplicons by use of the pGEM-T-easy vector (Promega, Madison, WI) as previously described (13). C. thermocellum ATCC 27405 was used as a positive control. Amplified PCR products were sequenced at Agencourt Bioscience Corporation (Beverly, MA). Nucleotide sequences were aligned with sequences from GenBank using BioEdit version 7.0.5 (11) or CLUSTAL_X (39). Phylogenetic trees were reconstructed using the ME algorithm (28) via the MEGA4 program package (37). Screening for sequence similarity was carried out with BLAST (2).
Microscopy.
Microscopic observation was performed after smear staining with DTAF [5-(4,6-dichlorotriazin-2-yl)amino-fluorescein] using a Leica DMLB microscope equipped with a mercury short-arc photooptic lamp (33). A K3 filter (illumination path, 470 to 490 nm; observation path, 515 nm) was used to visualize DTAF-stained microbial cells.
Statistical calculations and data analysis.
Descriptive statistics of primary data, including mean, confidence interval, and standard deviation were performed with MS Excel. We used 2 to 5 replicates for all analytical measurements (HPLC, TPC, and TPN), and the relative error did not exceed 5 to 10%. The batch growth experiments were done at least twice, with two or three replicate bottles.
Phylogenetic trees were assembled using a bootstrap test with 1,000 replicates to evaluate robustness.
Accession numbers.
The sequences generated within this study have been deposited in GenBank under accession numbers FJ808599 to FJ808612, GQ265352 to GQ265362, and HM171683 to HM171687. Strain 4-2a was deposited in the ATCC under deposition number PTA10114.
RESULTS
Isolation of pure cultures.
Enrichment cultures after 5 to 10 consecutive transfers exhibited reproducible cellulose fermentation comparable to that of pure cultures (e.g., of Clostridium thermocellum), as indicated by the fermentation products, extent, and time required for cellulose utilization. These mixed cultures are referred to below as “consortia,” which are distinct from “primary enrichment,” having nonstabilized variable composition. In total, we obtained four anaerobic thermophilic cellulolytic consortia: three (CO-4, CO-5, and CO-6) were derived directly from three respective biocompost samples as described earlier (13), and one, named CT, was obtained from a cellulose trap. Plating of the serially diluted consortium cultures on cellulose agar revealed bacterial colonies with extensive clearing zones, indicating cellulose solubilization by extracellular enzymes (see Fig. S1 in the supplemental material).
Cell numbers for consortia were estimated by plate count and by direct microscopy. Both methods produced converging results. For example, at the end of active growth in liquid batch culture, when cellulose was used nearly completely, the consortia CO-4, CO-5, and CT contained about 2.5 × 108 CFU/ml, while CO-6 had 1 order of magnitude higher density, 2.5 × 109 CFU/ml. Direct microscopic counts in the same liquid cultures had 1 × 109 and 3 × 109 cell/ml, respectively. In total, nine cellulolytic and four noncellulolytic bacterial strains were isolated from four tested consortia. Strains CT1 and CT2 were isolated from the CT consortium, strains 4-1 and 4-2a came from CO-4, strain 5-8 was isolated from CO-5, and the rest of the strains (6-17a, 6-24, 6-26, and 6-29) were isolated from CO-6. Noncellulolytic strains 6-12, 6-16, 6-30, and 6-31 were obtained from CO-6.
Phylogenetic analysis of isolates.
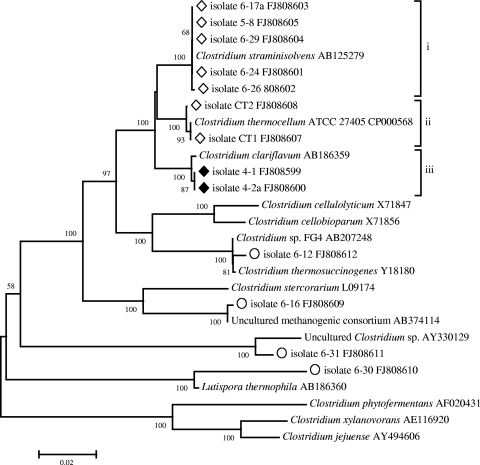
Phylogenetic analysis of cellulolytic and noncellulolytic isolates based on 16S rRNA gene sequences (Fig. 1) revealed cellulolytic bacteria belonging to three particular groups, all of them belonging to the genus Clostridium: (i) bacteria closely related to C. straminisolvens (15), (ii) bacteria related to C. thermocellum, and (iii) bacteria related to C. clariflavum recently isolated from methanogenic sludge (30, 32).
FIG. 1.
Phylogenetic tree of anaerobic thermophilic cellulolytic (⋄), cellulolytic/xylanolytic (♦), and noncellulolytic (○) isolates from biocompost based on 16S rRNA gene sequence comparisons.
The 16S rRNA gene sequence similarities for strains 4-1 and 4-2a were 100% with respect to each other and 99.7% with respect to C. clariflavum. In spite of the 16S rRNA gene sequence identity, these two strains showed phenotypic differences in cell morphology and fermentation pattern (see below); therefore, we consider them distinct although closely related organisms diverged significantly from other members of the Clostridium genus. The sequence similarity was only 96% with respect to C. thermocellum and C. straminisolvens. Cellulolytic strains 6-17a, 5-8, 6-24, 6-26, and 6-29 formed a tight cluster (99 to 100% similarity) with C. straminisolvens, while isolates CT1 and CT2 clustered with C. thermocellum (99 to 100% similarity). Noncellulolytic isolate 6-12 was related to Clostridium thermosuccinogenes and Clostridium sp. strain FG4 (16), strain 6-30 was similar to a recently described Lutispora thermophila strain (31), and strains 6-16 and 6-31 turned out to be related to previously uncultured microorganisms from glucose- or cellulose-degrading consortia of methanogenic reactors (5, 38).
GH48 phylogeny.
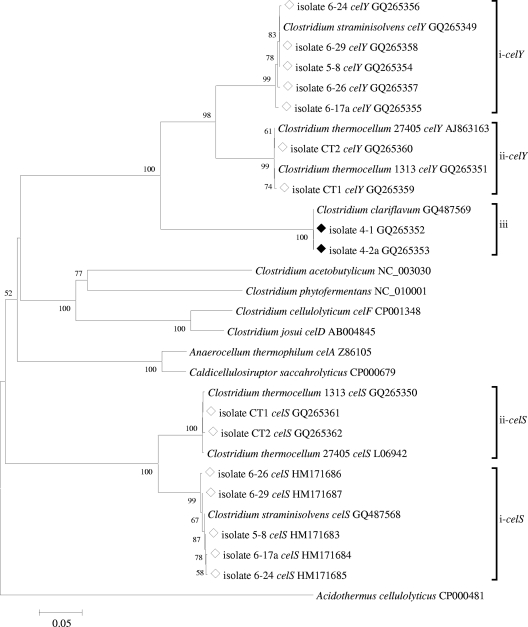
Phylogenetic analysis was also carried out with respect to exocellulases of glycosyl hydrolase family 48 (GH48), a major enzyme of interest within cellulolytic microorganisms. PCR amplification of this region yielded an amplicon of variable length, between 1,143 and 1,167 nucleotides. Consistent with observations from 16S rRNA gene analyses, cellulolytic isolates formed three groups (Fig. 2): (i) isolates 6-17a, 5-8, 6-24, 6-26, and 6-29 were most closely related to the glycosyl hydrolase family 48 gene sequences in C. straminisolvens celS- and celY-like genes; (ii) isolates CT1 and CT2, related to C. thermocellum, formed two separate clusters of sequences closely related to the celS and celY genes of C. thermocellum type strain ATCC 27405 and strain DSM 1313; and (iii) strains 4-1 and 4-2a, related to C. clariflavum, formed a distinct cluster of identical nucleotide sequences, with no known sequences closely related to them. The matches closest to Clostridium clariflavum-like sequences were C. thermocellum celY (74.1% nucleotide similarity and 87% deduced amino acid sequence similarity) and C. straminisolvens (73.4% nucleotide similarity and 87% deduced amino acid sequence similarity). No GH48 genes were detected in noncellulolytic isolates.
FIG. 2.
Phylogenetic tree of thermophilic anaerobic cellulolytic (⋄) and cellulolytic and xylanolytic (♦) isolates from biocompost based on GH48 gene sequence comparisons.
Novel isolates related to C. thermocellum and C. straminisolvens were determined to have both free (celY-like) and cellulosome-bound (celS-like) exoglucanases from glycosyl hydrolase family 48, as seen in reference strains (13). GH48 genes in C. straminisolvens- and C. clariflavum-related isolates displayed very similar grouping, as observed in 16S rRNA gene analyses, suggesting a very strict conservation of this particular family of glycosyl hydrolases within cellulolytic clostridia. GH48 genes retrieved from isolates 4-1 and 4-2a represent novel exoglucanases.
Cell morphology.
The bacteria were found to be straight and curved rods of different lengths, long filamentous cells, and cells with terminal spores. All strains related to C. straminisolvens were represented by curved rods (5 to 15 by 0.1 to 0.3 μm) or filamentous cells (10 to 40 by 0.1 to 0.3 μm) attached to cellulose particles. We observed that isolates having almost identical 16S rRNA sequences displayed significant morphological diversity, even under uniform growth conditions. C. thermocellum-related strains were represented by slightly curved rods (5 to 15 by 0.2 to 0.5 μm). The strains 4-2a and 4-1 were different from each other: strain 4-2a was represented by long curved rods (2 to 10 by 0.1 to 0.2 μm), while 4-1 strain formed short and mostly straight rods (2 to 4 by 0.1 to 0.3 μm). Both strains changed their morphology, depending on the carbon substrate used; the cellulose-grown cells were 2 to 3 times longer than the cells grown on xylan. The phylogenetic identity of strains 4-1 and 4-2a grown on different substrates was confirmed by 16S rRNA gene analyses. All cultures grown on Avicel produced a bright yellow or light cream pigment.
Fermentation of cellulose.
Table 1 shows the major fermentation products found in all tested strains on the second day of batch culture growth on 3 g/liter cellulose. All strains produced acetate, ethanol, and lactate. With the exception of strain CT2, all isolates produced formate. The ethanol concentration at the end of the fermentation ranged from 0.3 to 10.5 mM. The most active strains, 6-29 and 6-24 (C. straminisolvens group), produced 9.2 to 10.5 mM ethanol, which accounted 22 to 30% of the theoretical maximal yield (assuming 2:1 stoichiometry between ethanol and glucose), with the acetate-to-ethanol ratio being close to 1:1. In other strains, acetate was the predominant fermentation product. In the C. clariflavum group, the acetate/ethanol ratios reached values between 10:1 (strain 4-2a) and 22:1 (strain 4-1); these strains had formate (not ethanol) as a second major fermentation product. There was a very clear relationship between phylogenetic position and fermentation profile. Only members of the C. clariflavum group generated significant amounts of formate, while all isolates related to C. straminisolvens and C. thermocellum fermented Avicel mostly to acetate and ethanol. Lactate accumulated at the late stage of fermentation as the pH dropped.
TABLE 1.
Fermentation products formed by various isolates from cellulosea
| Isolate | Closest known relative | Avg concn (mM) ± SD |
Acetate/ethanol ratio | |||
|---|---|---|---|---|---|---|
| Lactate | Formate | Acetate | Ethanol | |||
| 4-1 | Clostridium clariflavum | 0.2 ± 0.04 | 2.74 ± 0.07 | 7.78 ± 0.55 | 0.35 ± 0.03 | 22.2 |
| 4-2a | Clostridium clariflavum | 0.97 ± 0.02 | 3.53 ± 0.09 | 9.2 ± 0.61 | 0. 9 ± 0.04 | 10.3 |
| 6-24 | Clostridium straminisolvens | 0.31 ± 0.03 | 0.37 ± 0.05 | 8.58 ± 1.65 | 9.21 ± 0.91 | 0.9 |
| 6-26 | Clostridium straminisolvens | 0.06 ± 0.001 | 0.13 ± 0.02 | 4.87 ± 0.99 | 3.82 ± 1.96 | 1.3 |
| 6-29 | Clostridium straminisolvens | 0.41 ± 0.03 | 0.26 ± 0.08 | 8.19 ± 1.59 | 10.49 ± 0.99 | 0.8 |
| 6-17a | Clostridium straminisolvens | 0.4 ± 0.006 | 0.31 ± 0.002 | 9.57 ± 1.86 | 6.79 ± 0.5 | 1.4 |
| 5-8 | Clostridium straminisolvens | 0.03 ± 0.01 | 0.12 ± 0.02 | 2.75 ± 0.31 | 1.22 ± 0.09 | 2.2 |
| CT1 | Clostridium thermocellum | 0.08 ± 0.01 | 0.38 ± 0.03 | 8.64 ± 1.0 | 4.17 ± 0.3 | 2.1 |
| CT2 | Clostridium thermocellum | 0.26 ± 0.04 | 0.0 | 3.77 ± 0.41 | 1.43 ± 0.07 | 2.6 |
Values shown are averages of results from two replicas, each using 3 g/liter of Avicel.
Fermentation and enzymatic activity of strain 4-2a grown on various carbon substrates.
The most intriguing members of our collection were strains 4-1 and 4-2a, related to C. clariflavum. Isolated from the CO-4 consortium developed on cellulose, both strains were able to utilize a wide spectrum of plant polymers, including xylan, xylose, and pretreated wood (Table 2). Xylan supported very intensive growth of these bacteria. Growth on pretreated wood remained quite vigorous although less intensive. Xylose supported anaerobic growth of both strains, but its fermentation was extremely slow and incomplete; after 10 days of incubation at 55°C, about 50% of xylose remained unutilized. The major fermentation products of xylan were acetate and formate; pretreated wood was transformed mainly into acetate. The ethanol concentrations ranged from 0.8 to 0.9 mM. The acetate/ethanol ratios ranged from 10:1 to 16:1, very similar to the ratios observed in cellulose fermentations. The major fermentation products of xylose were acetate and lactate, with no ethanol detected.
TABLE 2.
Fermentation products formed by isolates 4-1 and 4-2a from xylan, pretreated wood, and xylosea
| Strain and growth substrate | Avg concn (mM) ± SD |
Acetate/ethanol ratio | |||
|---|---|---|---|---|---|
| Lactate | Formate | Acetate | Ethanol | ||
| 4-1 | |||||
| Xylan | 0.32 ± 0.03 | 3.63 ± 0.09 | 12.83 ± 0.43 | 0.79 ± 0.13 | 16.3 |
| Xylose | 0.21 ± 0.19 | 0 | 2.63 ± 0.76 | 0 | |
| Pretreated wood | 0.88 ± 0.88 | 0.57 ± 0.17 | 10.28 ± 2.1 | 0.87 ± 0.3 | 11.8 |
| 4-2a | |||||
| Xylan | 0.43 ± 0.03 | 3.05 ± 0.13 | 12.18 ± 0.47 | 0.88 ± 0.15 | 13.7 |
| Xylose | 0.41 ± 0.06 | 0 | 2.85 ± 0.25 | 0 | |
| Pretreated wood | 0.44 ± 0.43 | 0.22 ± 0.06 | 9.4 ± 1.47 | 0.91 ± 0.26 | 10.3 |
Values shown are averages of results from two replicas, each using 3 g/liter of each growth substrate.
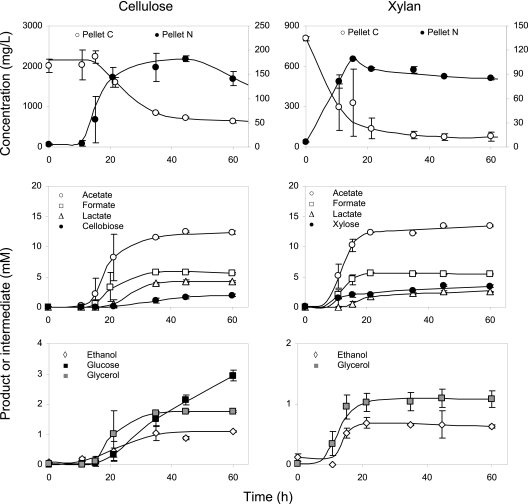
The dynamics of Avicel and xylan fermentation was observed for strain 4-2a in batch culture with 5 g/liter of separately added polymeric substrates (Fig. 3). Although inocula were obtained on medium with cellulose, degradation of xylan started immediately, while fermentation of Avicel was delayed by 11 to 15 h. Beyond the lag phase, both polymeric substrates were consumed within 20 h, indicating an exceptionally high degradation rate, comparable to the reported cellulolytic activity of C. thermocellum (22). Growth efficiencies turned out to be roughly the same on cellulose and xylan, with a cell yield of 0.14 g C biomass per g C for each substrate. After utilization of more than 50% of the added C substrate, the solubilization process slowed down, accompanied by acidification of cultural liquid (pH drop from 8 to 6 units; not shown). Metabolic acidity was the most probable reason for microbial growth cessation before complete utilization of xylan and cellulose. The major fermentation products (>5 mM) from Avicel were acetate, formate, and lactate; the minor products (<2 mM) were ethanol and glycerol. The two main intermediates of Avicel depolymerization were cellobiose and glucose. All listed products formed two distinct clusters: (i) acetate, formate, glycerol, and ethanol closely followed the dynamics of cell mass, and (ii) lactate, cellobiose, and glucose lagged behind the growth. With xylan, the product dynamics was very similar. Xylose, the main depolymerization intermediate of xylan, was accumulated up to 3.5 mM during stationary phase and eventually degraded after prolonged culture starvation (data not shown).
FIG. 3.
Fermentation of cellulose (left column) and xylan (right column) by isolate 4-2a. Top panels, total pellet carbon (TPC; C substrate plus C cell) and total pellet nitrogen (TPN; measure of cell mass concentration); bottom panels, fermentation products and depolymerization intermediates.
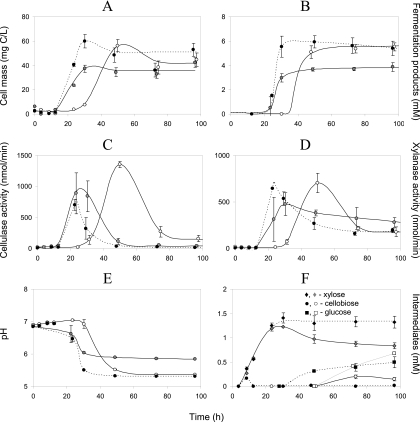
In the second experiment, strain 4-2a was grown on (i) 1.5 g/liter of Avicel, (ii) 1.5 g/liter of xylan, and (iii) mixture of 1.5 g/liter Avicel and 1.5 g/liter xylan to see a possible competition or synergy in the conversion of the two substrates. Apart from cell mass, fermentation products, and pH, we also monitored enzymatic activity of cellulase and xylanase (Fig. 4). There was a high degree of synchrony between cell growth (Fig. 4A), accumulation of fermentation products (Fig. 4B), and levels of cellulase (Fig. 4C) and xylanase (Fig. 4D) activity. It becomes evident from these data that synthesis of cellulases and xylanases was induced by both polymeric substrates. Cellulase activity correlated with cell mass, and although it was the highest in the culture with Avicel, its level remained high enough in culture grown on xylan and the xylan-cellulose mixture. In a similar way, xylanase activity was expressed to the same degree in the medium with xylan or cellulose alone, the difference being mainly explained (r2 = 0.86) by the variation in bacterial cell mass concentration. Again (as in the first experiment), xylan started to be degraded without lag phase, unlike the delay of about 1 day in the Avicel-containing medium. In the culture grown on a mixture of two substrates (dotted lines in all panels of Fig. 4), growth was even more intensive and instant than on xylan. It was accompanied by faster acidification (Fig. 4E), with pH drop to the tolerance limit of pH 5.4, with a concomitant abrupt decline in enzymatic activity and a slowing down of the fermentation. The indicated acidification prevented complete utilization of the cellulose-xylan mixture. Nevertheless, we have clear indication (Fig. 4F) that the presence of xylan stimulated degradation of cellulose: cellobiose (unique intermediate deriving from cellulose) was released as early as after 8 h of bacterial growth on the mixture of two substrates, while, with cellulose alone, it was detected after 50 h. In the xylan and xylan-cellulose media (first day of incubation) (Fig. 4F, black and gray diamonds), the initial rates of xylose release were the same; however, the positive effect of cellulose became clear on the 2nd day, being expressed as a stimulation of xylose uptake.
FIG. 4.
Growth dynamics of isolate 4-2a on medium with cellulose (open symbols, continuous lines), xylan (gray-filled symbols, continuous lines), or a mixture of cellulose and xylan (black-filled symbols, dotted lines). Top, cell biomass concentration (A) and sum of fermentation products (B) (see Fig. 3 for individual products); middle, cellulase (C) and xylanase (D) activity; bottom, pH (E) and intermediate degradation products (F).
The majority of enzymatic activity in the strain 4-2a was found to be associated with particulate matter, with no more than 15% of enzymatic activity being detected in a supernatant. The only exception was the cellulase activity in xylan-grown culture, where up to 75% of the enzymes were detected in a supernatant. This indicates that strain 4-2a produces free extracellular enzymes which are easily bound to insoluble substrates. Xylan forms smaller sediment because of partial thermal degradation and solubilization during autoclaving; therefore, it binds fewer enzymes and mainly xylanase, leaving the most of cellulase free. The activities of both enzymes (cellulase and xylanase) quickly declined after growth cessation, indicating abrupt inactivation of extracellular enzymes.
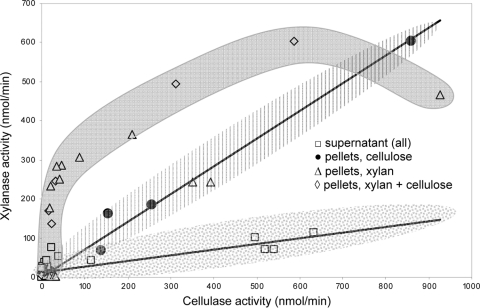
Close correlation between cellulase and xylanase activities could be explained by either their coexpression in the same operon(s) or that both polymers are degraded by a single enzyme with wide substrate specificity (10). To test the second option, we plotted the pooled data on xylanase and cellulase activities versus each other (Fig. 5). Three domains of data points were formed: (i) in cellulose-grown culture, the pellet-associated activities of xylanase and cellulase were closely correlated (r2 = 0.987); (ii) in xylan and xylan-cellulose media, the relationship between the activities of the two enzymes was nonlinear; and (iii) in supernatants of all cultures, the activities of free xylanases and cellulases displayed a weak correlation (r2 = 0.671), with the respective regression line having a much shallower slope than in the first group. Behind the wide variation in the xylanase/cellulase ratio, there seems to be a slower inactivation of xylanase activity than of cellulase activity in the stationary phase of batch culture (Fig. 4C and D). The revealed difference is sufficient to decline the hypothesis that the same hydrolytic enzymes catalyze a breakdown of both polymers.
FIG. 5.
Relationship between cellulase and xylanase activities. The pooled xylanase activity is plotted versus respective cellulase activity. The shaded areas indicate three domains with different relationships between the activities of two enzymes, allowing rejection of the hypothesis that degradation of xylan and cellulose is catalyzed by the same enzyme.
DISCUSSION
In this paper, we describe the isolation, characterization, and primary testing of anaerobic thermophilic bacteria from mixed cellulolytic culture.
The isolation of a single colony of anaerobic microorganisms is an intricate procedure, so we paid special attention to methodology in an attempt to find optimal plating conditions for cellulolytic thermophiles. Our experience supports the following major conclusions. (i) Serial dilution of bacterial suspension before plating should be done with full-strength reduced medium. The use of buffer or water for serial dilution resulted in lowered plating efficiency. (ii) Consortia or enriched cultures should be growing actively before plating. Substrate-depleted cultures resulted in lower cell counts and poor reproducibility of plating. (iii) Plating and single colony isolation should be done several times to maximize the probability of isolating pure cultures of cellulolytic organisms. There is a high likelihood that noncellulolytic colonies happen to be present within a clearing zone produced by a cellulose degrader. We found repetition of plating 2 to 3 times to be effective in eliminating noncellulolytic contaminants.
In natural microbial communities, plate counts typically result in 100- to 1,000-fold-lower microbial abundance than microscopic counts, and this discrepancy stems from the broad metabolic diversity of indigenous populations and the presence of “viable but nonculturable” cells (35). The lack of such discrepancy in our consortia indicates the narrow metabolic diversity and absence of stressed organisms, which have most probably been lost in the course of consecutive transfers. Low diversity was also detected in the 16S rRNA gene and glycosyl hydrolase family 48 sequences of these consortia (13). Limited diversity can be also explained by high cultivation temperature in combination with cellulose recalcitrance: only limited numbers of species are able to develop under such conditions. Similar observations of restricted diversity and a lack of significant difference between plated and counted microbial abundances were found in extreme natural communities, like polar sea ice (14).
Contrary to previous studies (25), we have found wider phylogenetic and metabolic diversity of clostridia, probably due to methodological improvements, such as the use of reduced media for serial dilution, avoiding starvation and oxygen stress and use of pour-plating techniques instead of rolling tubes.
The isolated strains formed three distinct groups. The first group of moderately thermophilic bacteria contained four isolates that were not able to utilize cellulose. These isolates were related to different taxonomic groups, including noncellulolytic clostridia (C. thermosuccinogenes), the recently described anaerobic bacterium Lutispora thermophila (31), and previously uncultured microorganisms from cellulose-degrading methanogenic reactors (5, 38). All of these isolates were able to grow as pure cultures on cellobiose medium with l-cysteine as a reducing agent, and they sustained ∼10 transfers within cellulolytic consortia on defined cellulose-mineral medium. There are two tentative explanations for their stable coexistence with cellulolytic microorganisms in consortia: (i) they have higher affinity for depolymerization products (cellobiose and glucose) than cellulolytic bacteria and therefore successfully compete with them for common carbon substrates, or (ii) they are able to utilize noncellulosic organic compounds in nutrient medium (l-cysteine and vitamins) as a carbon and energy source and therefore do not compete or stimulate cellulolytic bacteria. The concentration of vitamins was too low to sustain stable growth, and therefore, l-cysteine was the most probable candidate for the noncellulosic carbon source. It is known that C. thermosuccinogenes is able to use glucose, xylose, and cellobiose (7). On the other hand, L. thermophila did not utilize any of the detected Avicel fermentation products or intermediates but was able to degrade cysteine (31). Thus, it is likely that strain 6-12, related to C. thermosuccinogenes, survived in consortia by utilizing cellobiose and glucose and that strain 6-30, related to L. thermophila, had been enriched on l-cysteine.
The second, and most abundant, group of isolates were related to C. straminisolvens and C. thermocellum. These isolates are moderately thermophilic bacteria specializing on utilization of cellulose only. Finally, two out of nine obtained cellulolytic isolates identified as members of C. clariflavum group were able to degrade cellulose, xylan, and xylose, representing the most versatile group. Cultivation of strain 4-2a on individual polymers and their mixtures led us eventually to a tentative explanation of this versatility: cellulose, even without xylan, provokes synthesis of xylanase while xylan induces biosynthesis of cellulase. Taking an ecological viewpoint, such a tight coupling in expression of hydrolytic activity does not look wasteful, because under natural conditions, cellulose is never supplied separately from hemicellulose or lignin. Therefore, bacteria related to C. clariflavum represent a specialized group of microorganisms adapted to degrade the entire plant litter under hot and anoxic conditions. Xylan, as the most soluble component of lignocellulose, probably acts as a signaling factor initiating cell growth on lignocellulose. Such assumption agrees with the fact that xylan stimulates degradation of cellulose by reducing lag phase to a minimum. Under natural conditions, these bacteria probably stay dormant most of the time and can be triggered to “wake up” by the combined effect of chemical signal from xylan and the rise in temperature above 50°C. A warming-up should be at least a short-term event occurring locally at decomposition hotspots, due to biogenic heat production by other soil microorganisms. In a biocompost pile, the process of self-heating is scaled up both in time and in space, giving a chance for thermophilic bacteria to approach high cell density.
C. thermocellum, one of the most successful cellulose degraders known, combines traits of cellulase and xylanase activities (24, 46). However, contrary to strain 4-2a and C. stercorarium (1, 46), it does not utilize xylan and xylose (24). Synthesis of xylan-degrading enzymes without the ability to assimilate all of the released xylose is another subject worthy of discussion and further studies.
Supplementary Material
Acknowledgments
This research was supported by a grant from the BioEnergy Science Center (BESC), Oak Ridge National Laboratory, a U.S. Department of Energy (DOE) Bioenergy Research Center supported by the Office of Biological and Environmental Research in the DOE Office of Science, and by Mascoma Corporation.
We thank Anna Guseva for technical assistance as well as Melissa Beckwith and Norm Cushman for providing us with access to their composting facility at Middlebury College, VT.
Footnotes
Published ahead of print on 11 February 2011.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Adelsberger, H., C. Hertel, E. Glawischnig, V. V. Zverlov, and W. H. Schwarz. 2004. Enzyme system of Clostridium stercorarium for hydrolysis of arabinoxylan: reconstitution of the in vivo system from recombinant enzymes. Microbiology 150:2257-2266. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., et al. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Atlas, R. M. 1996. Handbook of microbiological media, 2nd ed. CRC Press LLC, Boca Raton, FL.
- 4.Beg, Q. K., M. Kapoor, L. Mahajan, and G. S. Hoondal. 2001. Microbial xylanases and their industrial applications: a review. Appl. Microbiol. Biotechnol. 56:326-338. [DOI] [PubMed] [Google Scholar]
- 5.Burrell, P. C., C. O'Sullivan, H. Song, W. P. Clarke, and L. L. Blackall. 2004. Identification, detection, and spatial resolution of Clostridium populations responsible for cellulose degradation in a methanogenic landfill leachate bioreactor. Appl. Environ. Microbiol. 70:2414-2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coleman, D. J., M. J. Studler, and J. J. Naleway. 2007. A long-wavelength fluorescent substrate for continuous fluorometric determination of cellulase activity: resorufin-beta-D-cellobioside. Anal. Biochem. 371:146-153. [DOI] [PubMed] [Google Scholar]
- 7.Drent, W. J., G. A. Lahpor, W. M. Wiegant, and J. C. Gottschal. 1991. Fermentation of inulin by Clostridium thermosuccinogenes sp. nov., a thermophilic anaerobic bacterium isolated from various habitats. Appl. Environ. Microbiol. 57:455-462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Freier, D., C. P. Mothershed, and J. Wiegel. 1988. Characterization of Clostridium thermocellum JW20. Appl. Environ. Microbiol. 54:204-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ge, Y., E. G. Antoulinakis, K. R. Gee, and I. Johnson. 2007. An ultrasensitive, continuous assay for xylanase using the fluorogenic substrate 6,8-difluoro-4-methylumbelliferyl [beta]-d-xylobioside. Anal. Biochem. 362:63-68. [DOI] [PubMed] [Google Scholar]
- 10.Gilbert, H. J., and G. P. Hazelwood. 1993. Bacterial cellulases and xylanases. J. Gen. Microbiol. 139:187-194. [Google Scholar]
- 11.Hall, T. A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41:95-98. [Google Scholar]
- 12.Hamilton-Brehm, S. D., et al. 2010. Caldicellulosiruptor obsidiansis sp. nov., an anaerobic, extremely thermophilic, cellulolytic bacterium isolated from Obsidian Pool, Yellowstone National Park. Appl. Environ. Microbiol. 76:1014-1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Izquierdo, J. A., M. V. Sizova, and L. R. Lynd. 2010. Diversity of bacteria and glycosyl hydrolase family 48 genes in cellulolytic consortia enriched from thermophilic biocompost. Appl. Environ. Microbiol. 76:3545-3553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Junge, K., F. Imhoff, T. Staley, and J. W. Deming. 2002. Phylogenetic diversity of numerically important arctic sea-ice bacteria cultured at subzero temperature. Microb. Ecol. 43:315-328. [DOI] [PubMed] [Google Scholar]
- 15.Kato, S., et al. 2004. Clostridium straminisolvens sp. nov., a moderately thermophilic, aerotolerant and cellulolytic bacterium isolated from a cellulose-degrading bacterial community. Int. J. Syst. Evol. Microbiol. 54:2043-2047. [DOI] [PubMed] [Google Scholar]
- 16.Kato, S., S. Haruta, Z. J. Cui, M. Ishii, and Y. Igarashi. 2005. Stable coexistence of five bacterial strains as a cellulose-degrading community. Appl. Environ. Microbiol. 71:7099-7106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kosugi, A., K. Murashima, and R. H. Doi. 2001. Characterization of xylanolytic enzymes in Clostridium cellulovorans: expression of xylanase activity dependent on growth substrates. J. Bacteriol. 183:7037-7043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kulkarni, N., A. Shendye, and M. Rao. 1999. Molecular and biotechnological aspects of xylanases. FEMS Microbiol. Rev. 23:411-456. [DOI] [PubMed] [Google Scholar]
- 19.Lu, Y., Y. H. Zhang, and L. R. Lynd. 2006. Enzyme-microbe synergy during cellulose hydrolysis by Clostridium thermocellum. Proc. Natl. Acad. Sci. U. S. A. 103:16165-16169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lynd, L. R., D. Currie, N. Ciazza, C. Herring, and N. Orem. 2008. Consolidated bioprocessing of cellulosic biomass to ethanol using thermophilic bacteria, p. 55-74. In J. D. Wall et al. (ed.), Bioenergy. ASM Press, Washington, DC.
- 21.Lynd, L. R., et al. 2008. How biotech can transform biofuels. Nat. Biotechnol. 26:169-172. [DOI] [PubMed] [Google Scholar]
- 22.Lynd, L. R., P. J. Weimer, W. H. van Zyl, and I. S. Pretorius. 2002. Microbial cellulose utilization: fundamentals and biotechnology. Microbiol. Mol. Biol. Rev. 66:506-577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Madden, R. H. 1983. Isolation and characterization of Clostridium stercorarium sp. nov., cellulolytic thermophile. Int. J. Syst. Bacteriol. 33:837-840. [Google Scholar]
- 24.Morag, E., E. A. Bayer, and R. Lamed. 1990. Relationship of cellulosomal and noncellulosomal xylanases of Clostridium thermocellum to cellulose-degrading enzymes. J. Bacteriol. 172:6098-6105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ozkan, M., et al. 2001. Characterization of 13 newly isolated strains of anaerobic, cellulolytic, thermophilic bacteria. J. Ind. Microbiol. Biotechnol. 27:275-280. [DOI] [PubMed] [Google Scholar]
- 26.Panikov, N. S., and L. R. Lynd. 2010. Physiological and methodological aspects of cellulolytic microbial cultures, p 644-656. In R. H. Baltz et al. (ed.), Manual of industrial microbiology and biotechnology, 3rd ed. ASM Press, Washington, DC.
- 27.Rainey, F. A., et al. 1994. Description of Caldicellulosiruptor saccharolyticus gen. nov., sp. nov: an obligately anaerobic, extremely thermophilic, cellulolytic bacterium. FEMS Microbiol. Lett. 120:263-266. [DOI] [PubMed] [Google Scholar]
- 28.Rzhetsky, A., and M. Nei. 1992. Statistical properties of the ordinary least-squares, generalized least-squares, and minimum-evolution methods of phylogenetic inference. J. Mol. Evol. 35:367-375. [DOI] [PubMed] [Google Scholar]
- 29.Saha, B. C. 2003. Hemicellulose bioconversion. J. Ind. Microbiol. Biotechnol. 30:279-291. [DOI] [PubMed] [Google Scholar]
- 30.Shiratori, H., et al. 2006. Isolation and characterization of a new Clostridium sp. that performs effective cellulosic waste digestion in a thermophilic methanogenic bioreactor. Appl. Environ. Microbiol. 72:3702-3709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shiratori, H., et al. 2008. Lutispora thermophila gen. nov., sp. nov., a thermophilic, spore-forming bacterium isolated from a thermophilic methanogenic bioreactor digesting municipal solid wastes. Int. J. Syst. Evol. Microbiol. 58:964-969. [DOI] [PubMed] [Google Scholar]
- 32.Shiratori, H., et al. 2009. Clostridium clariflavum sp. nov. and Clostridium caenicola sp. nov., moderately thermophilic, cellulose-/cellobiose-digesting bacteria isolated from methanogenic sludge. Int. J. Syst. Evol. Microbiol. 59:1764-1770. [DOI] [PubMed] [Google Scholar]
- 33.Sizova, M. V., N. S. Panikov, T. P. Tourova, and P. W. Flanagan. 2003. Isolation and characterization of oligotrophic acido-tolerant methanogenic consortia from a Sphagnum peat bog. FEMS Microbiol. Ecol. 45:301-315. [DOI] [PubMed] [Google Scholar]
- 34.Sleat, R., R. A. Mah, and R. Robinson. 1984. Isolation and characterization of an anaerobic, cellulolytic bacterium, Clostridium cellulovorans sp. nov. Appl. Environ. Microbiol. 48:88-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Staley, J. T., and J. J. Gosink. 1999. Poles apart: biodiversity and biogeography of sea ice bacteria. Annu. Rev. Microbiol. 53:189-215. [DOI] [PubMed] [Google Scholar]
- 36.Svetlichny, V. A., T. P. Svetlichnaya, N. Chernykh, and G. A. Zavarzin. 1990. Anaerocellum thermophilum gen. nov. sp. nov, an extreme thermophilic celluloselytic eubacterium isolated from hot springs in the Valley of Geysers. Microbiology (Moscow) 59:871-879. [Google Scholar]
- 37.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596-1599. [DOI] [PubMed] [Google Scholar]
- 38.Tang, Y. Q., T. Matsui, S. Morimura, X. L. Wu, and K. Kida. 2008. Effect of temperature on microbial community of a glucose-degrading methanogenic consortium under hyperthermophilic chemostat cultivation. J. Biosci. Bioeng. 106:180-187. [DOI] [PubMed] [Google Scholar]
- 39.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thomson, J. A. 1993. Molecular biology of xylan degradation. FEMS Microbiol. Rev. 10:65-82. [DOI] [PubMed] [Google Scholar]
- 41.U.S. Environmental Protection Agency. 2007. Biomass conversion: emerging technologies, feedstocks, and products. EPA/600/R-07/144. U.S. Environmental Protection Agency, Washington, DC.
- 42.van de Werken, H. J. G., et al. 2008. Hydrogenomics of the extremely thermophilic bacterium Caldicellulosiruptor saccharolyticus. Appl. Environ. Microbiol. 74:6720-6729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Warnick, T. A., B. A. Methe, and S. B. Leschine. 2002. Clostridium phytofermentans sp. nov., a cellulolytic mesophile from forest soil. Int. J. Syst. Evol. Microbiol. 52:1155-1160. [DOI] [PubMed] [Google Scholar]
- 44.Wiegel, J., and M. Dykstra. 1984. Clostridium thermocellum: adhesion and sporulation while adhered to cellulose and hemicellulose. Appl. Microbiol. Biotechnol. 20:59-65. [Google Scholar]
- 45.Yang, S.-J., et al. 2009. Efficient degradation of lignocellulosic plant biomass, without pretreatment, by the thermophilic anaerobe “Anaerocellum thermophilum” DSM 6725. Appl. Environ. Microbiol. 75:4762-4769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zverlov, V. V., and W. H. Schwarz. 2008. Bacterial cellulose hydrolysis in anaerobic environmental subsystems—Clostridium thermocellum and Clostridium stercorarium, thermophilic plant-fiber degraders. Ann. N. Y. Acad. Sci. 1125:298-307. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.