Abstract
The present study uses a mathematical-empirical approach to estimate the cardinal growth temperature parameters (Tmin, the temperature below which growth is no longer observed; Topt, the temperature at which the μmax equals its optimal value; μopt, the optimal value of μmax; and Tmax, the temperature above which no growth occurs) of 27 yeast strains belonging to different Saccharomyces and non-Saccharomyces species. S. cerevisiae was the yeast best adapted to grow at high temperatures within the Saccharomyces genus, with the highest optimum (32.3°C) and maximum (45.4°C) growth temperatures. On the other hand, S. kudriavzevii and S. bayanus var. uvarum showed the lowest optimum (23.6 and 26.2°C) and maximum (36.8 and 38.4°C) growth temperatures, respectively, confirming that both species are more psychrophilic than S. cerevisiae. The remaining Saccharomyces species (S. paradoxus, S. mikatae, S. arboricolus, and S. cariocanus) showed intermediate responses. With respect to the minimum temperature which supported growth, this parameter ranged from 1.3 (S. cariocanus) to 4.3°C (S. kudriavzevii). We also tested whether these physiological traits were correlated with the phylogeny, which was accomplished by means of a statistical orthogram method. The analysis suggested that the most important shift in the adaptation to grow at higher temperatures occurred in the Saccharomyces genus after the divergence of the S. arboricolus, S. mikatae, S. cariocanus, S. paradoxus, and S. cerevisiae lineages from the S. kudriavzevii and S. bayanus var. uvarum lineages. Finally, our mathematical models suggest that temperature may also play an important role in the imposition of S. cerevisiae versus non-Saccharomyces species during wine fermentation.
The estimation of the temperature range in which microorganisms are able to grow is very important for the food industry, to guarantee food safety or optimize fermentative conditions, for example, but also in ecological and taxonomic studies to classify and identify the different species of microorganisms. In this way, several works have shown the marked importance of temperature for the growth of industrial yeasts (1, 4, 22, 24), as well as the influence of this environmental factor in determining the natural distribution of wild species (12, 21, 23, 25). Specifically, there is an increasing interest in determining the influence of temperature in the adaptation of Saccharomyces species to both wild and fermentative environments (8, 21, 25).
The Saccharomyces genus includes several species associated only with natural habitats (S. cariocanus, S. kudriavzevii, S. mikatae, S. paradoxus, and S. arboricolus) and others that are present in both fermentative and wild habitats (S. cerevisiae and S. bayanus). The ability of the latter to ferment a broad range of beverages (cider, beer, and wines) and foods (bread, vegetables, etc.) (19) has unconsciously favored their selection by humans for thousands of years. It is thought that temperature could play an important role in the imposition and presence of S. cerevisiae in human activities (8). Several studies have shown that S. cerevisiae is well adapted to grow at higher temperatures, while other species, such as S. bayanus and S. kudriavzevii, are better adapted to grow at lower temperatures (2, 3, 11, 15, 22). However, there is a lack of quantitative information on this respect, and many of these studies do not include the whole biological temperature range in which Saccharomyces yeasts are able to grow. Thus, a more detailed study of the influence of temperature on Saccharomyces growth is necessary, including a larger number of strains isolated from different origins.
In this endeavor, predictive microbiology could be a very useful tool. This discipline uses mathematical models to quantitatively describe the behavior of microorganisms as a function of environmental variables (14). In the specific case of temperature, a primary model (usually a sigmoidal function) is first required to estimate the yeast growth parameters under diverse isothermal conditions. Then, a secondary model is necessary to appropriately describe the effects in the whole biotemperature range assayed (dynamic conditions). Fortunately, temperature has been a factor widely studied, and diverse secondary mathematical models are available in the references (14, 18, 29). Specifically, we have used in the present study the cardinal temperature model with inflection (CTMI) developed by Rosso et al. (20), because of the simplicity of its use and the easy and direct biological interpretation of results.
The study of the evolution of a determined phenotype (in our case, yeast response versus temperature changes) within a taxon (Saccharomyces genus) is essential to understand the evolutionary history of the group. A meaningful approach for a comprehensive understanding is to first generate a well supported molecular phylogenetic tree of the group and then interpret the evolution of morphological traits in the light of this phylogeny. The evolution of the trait may be plastic and stochastic or, on the contrary, may evolve according to a trend tightly linked to the phylogeny. Methods have been developed recently to detect phylogenetic dependence in comparative data (6, 17). These tests have the advantage that they only use the topological structure of the tree. Specifically, the orthogram method developed by Ollier et al. (17) represents a relevant approach because it detects and characterizes phylogenetic dependence of quantitative data and, at the same time, highlights different patterns of evolution along a phylogenetic tree.
The main goal of the present study was to determine the whole biological temperature range in which the different Saccharomyces species are able to grow. For this purpose, primary (reparameterized Gompertz equation) and secondary (CTMI) models are used to fit experimental data collected at different isothermal conditions. We also evaluate the phylogenetic dependence of the CTMI parameters by means of a canonical procedure which allows variance decomposition along the phylogenetic tree (orthogram method). Finally, CTMI models are also used to describe the effect of temperature on a hypothetical sympatric association between S. cerevisiae and the rest of the Saccharomyces species.
MATERIALS AND METHODS
Yeast strains and inoculum preparation.
A total of 27 yeast strains belonging to different Saccharomyces and non-Saccharomyces species were used in the present study. Yeasts were selected to obtain representative isolates from natural (10 strains) and fermentative (17 strains) habitats where possible. Their origins and designations are listed in Table 1.
TABLE 1.
Origin and designation of the 27 yeast strains used in this study
| Species | Straina | Origin (country) |
|---|---|---|
| S. cerevisiae | CECT 10131 | Centaurea alba flower (Spain) |
| T73L | Wine fermentation (Spain) | |
| PE35 M | Masato fermentation (Peru) | |
| CPE7 | Sugarcane fermentation (Brazil) | |
| KYOKAI/CBS 6412 | Sake fermentation (Japan) | |
| TEMOHAYA-MI26 | Agave fermentation (Mexico) | |
| Qa23L | Wine fermentation (Portugal) | |
| TTAM | Wine fermentation (France) | |
| PDMM | Wine fermentation (France) | |
| RVAM | Wine fermentation (Spain) | |
| S. paradoxus | CECT 1939NT/CBS 432NT | Tree exudate (Russia) |
| 120 M | Pulque fermentation (Mexico) | |
| K54 | Wine fermentation (Croatia) | |
| S. bayanus var. uvarum | NCAIM 789 | Carpinus betulus exudate (Hungary) |
| BM58L | Wine fermentation (Spain) | |
| S. kudriavzevii | CA111 | Quercus ilex bark (Spain) |
| CR85 | Quercus ilex bark (Spain) | |
| CR89 | Quercus faginea bark (Spain) | |
| CR90 | Quercus faginea bark (Spain) | |
| S. mikatae | NBRC 1815T/CBS 8839T | Soil (Japan) |
| S. arboricolus | CBS 10644T | Quercus fabri bark (China) |
| S. cariocanus | CBS 8841T | Fruit fly (Drosophila sp) (Brazil) |
| Non-Saccharomyces | Hanseniaspora uvarum CECT 10389 | Wine fermentation (Spain) |
| Candida stellata CECT 11108 | Wine fermentation (Spain) | |
| Torulaspora delbrueckii CECT 11199 | Wine fermentation (Spain) | |
| Kluyveromyces marxianus CECT 10585 | Wine fermentation (Spain) | |
| Pichia fermentans CECT 10064 | Wine fermentation (Spain) |
Superscript annotations: T, type strain; NT, neotype strain; L, commercial strain from Lallemand, Inc.; M, commercial strain from Mauri Yeast Australia.
Inocula were prepared by introducing one single colony from a pure culture of each strain into 5 ml of YM broth medium (Difco; Becton Dickinson Company, Sparks, MD). After 48 h of incubation at room temperature (25 ± 2°C [mean ± standard deviation]), 1 ml from each tube was centrifuged at 9,000 × g for 10 min and the pellet washed with sterile saline solution (9 g/liter), centrifuged, and resuspended again in 0.5 ml of sterile saline solution to obtain a concentration of about 7.3 log10 CFU ml−1, which was confirmed by surface spread on a YM agar plate. These yeast suspensions were used to inoculate the different experiments as described below.
Yeast growth conditions.
The basal culture medium selected for all experiments was yeast nitrogen base broth (YNB, Difco), supplemented with 20 g/liter of glucose as carbon source. The medium was sterilized by filtration (0.2 μm) and stored at 4°C until use. The final pH of the medium was 5.4 ± 0.1.
Growth was monitored at 600 nm in a SPECTROstar Omega instrument (BMG Labtech, Offenburg, Germany) at different temperatures (4, 8, 10, 14, 22, 29, 33, 37, 40, 42, and 46°C). Measurements were taken every hour for 3 days after a preshaking of 20 s for the temperatures included between 22 and 46°C. However, for lower temperatures (from 4 to 14°C), microtiter plates (96 wells) had to be incubated outside the SPECTROstar spectrophotometer and then transferred into it to take the measurements every 8 h for 14 days. The wells of the microplate were filled with 0.01 ml of inoculum and 0.25 ml of YNB medium, always reaching an initial optical density (OD) of approximately 0.2 (inoculum level of ∼6.0 log10 CFU ml−1). The inocula were always above the detection limit of the apparatus, which was determined by comparison with a previously established calibration curve. OD measurements could be used to estimate growth parameters because there was always a linear relationship between OD and yeast plate counts within the range studied (data not shown). Uninoculated wells for each experimental series were also included in the microtiter plates to determine and, consequently, subtract the noise signal. All experiments were carried out in triplicate. Therefore, a total of 891 growth curves (11 levels of temperature times 27 strains times 3 replicates) were obtained and analyzed.
Primary modeling.
Growth parameters were obtained from each treatment by directly fitting OD measurements versus time to the reparameterized Gompertz equation proposed by Zwietering et al. (28), which was originally introduced in predictive microbiology by Gibson et al. (7). It has the following expression:
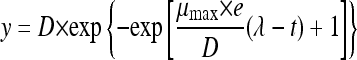 |
(1) |
where y = ln(ODt/OD0), with OD0 being the initial OD and ODt the OD at time t; D = ln(ODmax/OD0), with ODmax being the maximum optical density reached; μmax is the maximum specific growth rate (h−1); and λ the lag phase period (h). Growth data from each temperature and strain were fitted by a nonlinear regression procedure, minimizing the sum of squares of the difference between experimental data and the fitted model, i.e., loss function (observed − predicted)2. This task was accomplished using the nonlinear module of the Statistica 7.0 software package (StatSoft, Inc., Tulsa, OK) and its Quasi-Newton option.
Secondary modeling.
The cardinal temperature model with inflection (CTMI) (18, 20) was used to describe the μmax changes of yeasts as a function of temperature T (°C). CTMI is a descriptive model purely based on empirical observations and includes the three cardinal temperature values often used in microbiology. It has the following expression:
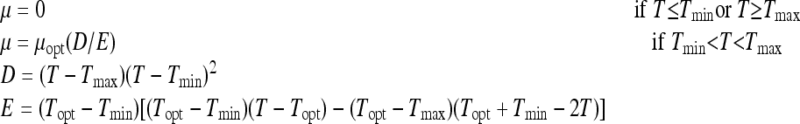 |
(2) |
where Tmax is the temperature above which no growth occurs, Tmin is the temperature below which growth is no longer observed, and Topt is the temperature at which the μmax equals its optimal value (μopt). As in the case of the primary modeling, the CTMI parameters were estimated by a nonlinear regression procedure using the Statistica 7.0 software package. The adequacy of the fit was checked by the proportion of variance explained by the model (R2) with respect to the experimental data.
ANOVAs.
First, strains were checked for significant differences among them by means of analysis of variance (ANOVA) using the one-way ANOVA module of Statistica 7.0 software. The dependent variables introduced for the analysis were the CTMI parameters obtained from secondary modeling. The post hoc comparison was carried out using the Scheffé test, which is considered to be one of the most conservative post hoc tests (27). An alternative advantage of the Scheffé test is that it can also be used with unequal sample sizes.
A second ANOVA was then performed by grouping the different Saccharomyces strains as a function of their respective species. In this way, the CTMI average parameters for the species S. cerevisiae, S. paradoxus, S. bayanus var. uvarum, and S. kudriavzevii were estimated from data for 10, 3, 2, and 4 strains, respectively, isolated from diverse origins. In this way, the biological temperature range obtained for each species represents a general behavior rather than a single-strain trait.
Identification of phylogenetic dependence.
We use the orthogram method developed by Ollier et al. (17) to determine the existence of dependence among the CTMI model parameters (Topt, Tmax, and μopt) and Saccharomyces phylogeny. This method, based on the variance decomposition along the phylogenetic tree, considers the null hypothesis to be the complete absence of phylogenetic dependence and uses the following statistics to corroborate this hypothesis (17): R2Max, which tests whether a significant change in the quantitative trait appeared at one node of the phylogenetic tree while being conserved in the deriving branches; SkR2k, which assesses to what extent the variance distribution across the phylogenetic tree is skewed to the root or to the tips; Dmax, which corresponds to the Kolmogorov-Smirnov statistic; and SCE, which measures the average local variation of the orthogram values. The method consists essentially of three steps. First, phylogenetic relations between the units of the tree are described by means of dummy variables taking values equal to 1 (for descendant tips within a node) or 0 (for the remaining tips). In a second step, these dummy variables are ordered in decreasing phylogenetic dissimilarity. Finally, since these dummy variables are not linearly independent, an orthonormal decomposition is necessary to obtain a matrix of linearly independent vectors (orthonormal vectors) which are used as regressors against CTMI model parameters. The method provides two graphical tools, called orthogram and cumulative orthogram, obtained by plotting the squared coefficients and the cumulative squared coefficients against the orthonormal vectors, respectively, that are very useful to interpret and identify the possible phylogenetic dependence. A complete description of the procedure and interpretation of statistics can be found in Ollier et al. (17) and Covain et al. (6). Orthograms and associated tests were conducted using the ade4 package (5) in R 2.4.0 software (10).
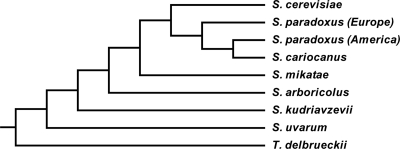
The topology of the phylogenetic tree of the genus Saccharomyces (Fig. 1) was obtained from Wang and Bai (26), who used sequences of the ITS-5.8S ribosomal DNA (rDNA) region and 26S rDNA D1/D2 domains to determine the phylogenetic relationships among Saccharomyces species. The phylogenetic positions of S. paradoxus populations from America (in our case, strain S. paradoxus 120 M) and Europe (strains S. paradoxus 1939 and S. paradoxus K54) were obtained from Liti et al. (13). Torulaspora delbrueckii was included as the outgroup.
FIG. 1.
Phylogenetic tree of the Saccharomyces genus. The topology was obtained from Liti et al. (13) and Wang and Bai (26). Torulaspora delbrueckii was used as the outgroup. S. uvarum, S. bayanus var. uvarum.
RESULTS
Estimation of the temperature range where yeasts are able to grow.
In this work, we have studied and compared the response of 27 yeast strains in a wide range of temperature values (from 4 to 46°C). For this purpose, yeasts were monitored by means of OD measurements, and their respective biological growth parameters (μmax and λ) estimated at each value. A total of 891 growth curves were fitted using a nonlinear regression procedure with the reparameterized Gompertz equation proposed by Zwietering et al. (28), which represents an empirical approach for the estimation of the growth parameters. In all cases the fit was good, with an R2 ranging from 0.95 to 0.99. Although the lag phase duration was also calculated for all experiments, it was not possible to appropriately model this parameter as a function of this environmental variable. Even at low temperatures, the lag phase of the different yeast species was very short, always below 15 h (data not shown). However, changes in μmax as a function of temperature could be fitted well by means of the CTMI secondary model.
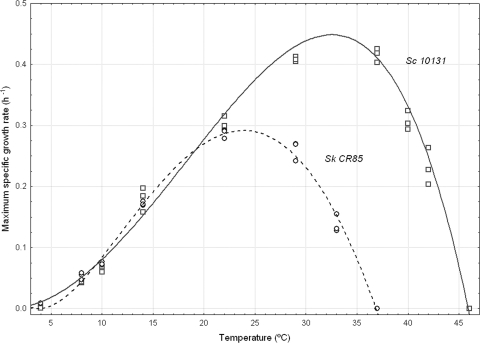
As an example, Fig. 2 shows the fit of the CTMI model to μmax experimental data obtained for the wild strains S. cerevisiae 10131 and S. kudriavzevii CR85. Clearly, a considerable difference was noticed between the two microorganisms, which was especially evident with respect to the optimum and maximum growth temperatures. In this way, S. cerevisiae 10131 was able to grow at up to 45°C and had its optimum growth around 33°C, with an estimated μmax at this temperature of 0.45 h−1. Conversely, S. kudriavzevii CR85 was not able to grow at 37°C and showed an optimum growth at around 24°C, with a μmax of 0.29 h−1. Below 20°C, we can see that both fits are practically overlapped. The minimum temperature to support growth was around 4°C in both cases (Fig. 2).
FIG. 2.
Fit of the cardinal temperature model to experimental data obtained for strains S. cerevisiae CECT10131 (squares) and S. kudriavzevii CR85 (circles). The quantitative parameters of both fits are shown in Table 2.
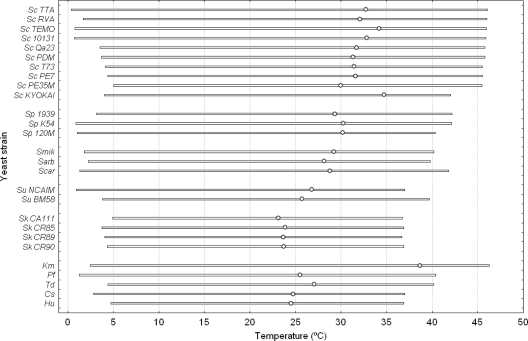
An advantage of the use of the CTMI model is that all its parameters have biological meanings, so the interpretation of this type of model is very easy and direct. Table 2 shows the temperature-dependent parameters obtained with the CTMI model for the 27 yeast strains assayed in this work. For each strain, average values were obtained from 3 independent experiments. The Scheffé post hoc comparison test was used to distinguish significant differences among strains for each parameter. Totals of 13, 11, and 5 different groups were obtained for parameters μopt, Topt, and Tmax, respectively. Conversely, no significant differences were found among strains in the case of Tmin. These parameters ranged from 0.096 (Candida stellata) to 0.449 h−1 (S. cerevisiae 10131) for μopt, from 23.1 (S. kudriavzevii CA111) to 38.7°C (Kluyveromyces marxianus) for Topt, from 36.7 (S. kudriavzevii CR89) to 46.3°C (K. marxianus) for Tmax, and finally, from 0.4 (S. cerevisiae TTA) to 5.0°C (S. cerevisiae PE35M) for Tmin. Figure 3 provides a graphical representation of the biological temperature range in which yeast strains were able to grow. In general, the S. cerevisiae strains exhibited the highest maximum and optimum growth temperatures within the Saccharomyces genus, with the exception of strain S. cerevisiae PE35M, which had a lower Topt (∼30°C). However, S. kudriavzevii strains clearly showed the lowest maximum and optimum growth temperatures, although these were very similar to the values obtained for the S. bayanus var. uvarum strains. With the exception of the species K. marxianus, whose results showed it to be the most thermotolerant microorganism assayed in this work (see Table 2), the rest of the non-Saccharomyces yeasts exhibited optimum growth temperatures ranging from 24 to 27°C and maximum growth temperatures from 36 to 40°C.
TABLE 2.
Estimated parameters (μopt, Tmax, Tmin, and Topt) of the cardinal temperature model for the 27 yeast strains assayed in this work
| Species | Strain | Avg (SD)a |
|||
|---|---|---|---|---|---|
| μopt(h−1) | Topt(°C) | Tmax(°C) | Tmin(°C) | ||
| S. cerevisiae | CECT 10131 | 0.449 (0.009) M | 32.86 (0.61) IJK | 45.90 (0.10) DE | 0.74 (0.10) A |
| T73L | 0.425 (0.043) LM | 31.42 (0.59) GHIJK | 45.51 (0.11) D | 4.11 (1.65) A | |
| PE35 M | 0.375 (0.008) JKLM | 29.99 (0.22) EFGHI | 45.48 (0.12) D | 5.04 (0.23) A | |
| CPE7 | 0.298 (0.010) CDEFGHI | 31.62 (0.39) HIJK | 45.50 (0.14) D | 4.38 (0.61) A | |
| KYOKAI/CBS 6412 | 0.284 (0.014) CDEFG | 34.75 (0.76) K | 42.00 (0.01) C | 4.02 (2.45) A | |
| TEMOHAYA-MI26 | 0.377 (0.006) JKLM | 34.19 (0.35) JK | 45.96 (0.01) DE | 0.76 (0.44) A | |
| Qa23L | 0.358 (0.020) HIJKL | 31.72 (0.95) HIJK | 45.79 (0.09) DE | 3.55 (0.85) A | |
| TTAM | 0.384 (0.024) KLM | 32.71 (0.75) IJK | 46.06 (0.12) DE | 0.43 (0.33) A | |
| PDMM | 0.347 (0.003) FGHIJK | 31.33 (0.43) GHIJ | 45.77 (0.09) DE | 3.72 (0.69) A | |
| RVAM | 0.382 (0.020) JKLM | 32.09 (0.69) HIJK | 45.99 (0.16) DE | 1.69 (1.23) A | |
| S. paradoxus | CECT 1939NT/CBS 432NT | 0.313 (0.008) EFGHIJK | 29.32 (0.50) EFGH | 42.20 (0.13) C | 3.15 (0.14) A |
| 120 M | 0.357 (0.009) GHIJKL | 30.18 (0.82) FGHI | 40.36 (0.51) B | 1.05 (0.14) A | |
| K54 | 0.288 (0.007) CDEFGHI | 30.27 (0.65) FGHI | 42.10 (0.14) C | 0.89 (1.21) A | |
| S. bayanus var. uvarum | NCAIM 789 | 0.307 (0.012) DEFGHIJ | 26.78 (0.23) BCDE | 37.02 (0.10) A | 0.93 (0.39) A |
| BM58L | 0.283 (0.007) CDEF | 25.70 (0.51) ABCD | 39.71 (0.05) B | 3.81 (0.44) A | |
| S. kudriavzevii | CA111 | 0.198 (0.002) B | 23.14 (0.11) A | 36.75 (0.03) A | 4.94 (0.15) A |
| CR85 | 0.291 (0.004) CDEFGHI | 23.88 (0.09) ABC | 36.87 (0.08) A | 3.77 (0.40) A | |
| CR89 | 0.258 (0.015) BCDE | 23.68 (0.34) AB | 36.69 (0.06) A | 4.07 (0.73) A | |
| CR90 | 0.284 (0.004) CDEFGH | 23.74 (0.28) AB | 36.91 (0.15) A | 4.37 (1.12) A | |
| S. mikatae | NBRC 1815T/CBS 8839T | 0.318 (0.002) EFGHIJK | 29.20 (1.46) EFGH | 40.20 (0.10) B | 1.79 (0.95) A |
| S. arboricolus | CBS 10644T | 0.328 (0.009) EFGHIJK | 28.14 (0.61) DEFG | 39.80 (0.04) B | 2.26 (0.99) A |
| S. cariocanus | CBS 8841T | 0.329 (0.006) EFGHIJK | 28.81 (0.73) DEFGH | 41.79 (0.06) C | 1.31 (1.10) A |
| Non-Saccharomyces | Hanseniaspora uvarum CECT 10389 | 0.198 (0.013) B | 24.51 (0.30) ABC | 36.87 (0.07) A | 4.71 (2.81) A |
| Candida stellata CECT 11108 | 0.096 (0.004) A | 24.77 (0.54) ABC | 37.00 (0.01) A | 2.81 (1.02) A | |
| Torulaspora delbrueckii CECT 11199 | 0.235 (0.006) BCD | 27.08 (0.48) CDEF | 40.19 (0.07) B | 4.42 (1.63) A | |
| Kluyveromyces marxianus CECT 10585 | 0.362 (0.004) IJKL | 38.68 (0.36) L | 46.28 (0.03) E | 2.43 (1.26) A | |
| Pichia fermentans CECT 10064 | 0.232 (0.008) BC | 25.51 (0.82) ABCD | 40.38 (0.28) B | 1.25 (0.81) A | |
Standard deviations for each parameter (in parentheses) were obtained from 3 independent nonlinear fits. Values within the same column followed by different letters are significantly different according to a Scheffé post hoc comparison test.
FIG. 3.
Temperature range in which the 27 yeast strains assayed in this work were able to grow. Optimum growth temperatures are marked by the circles on the bars. Sc, S. cerevisiae; Sp, S. paradoxus; Smik, S. mikatae; Sarb, S. arboricolus; Scar, S. cariocanus; Su, S. bayanus var. uvarum; Sk, S. kudriavzevii; Km, Kluyveromyces marxianus; Pf, Pichia fermentans; Td, Torulaspora delbrueckii; Cs, Candida stellata; Hu, Hanseniaspora uvarum.
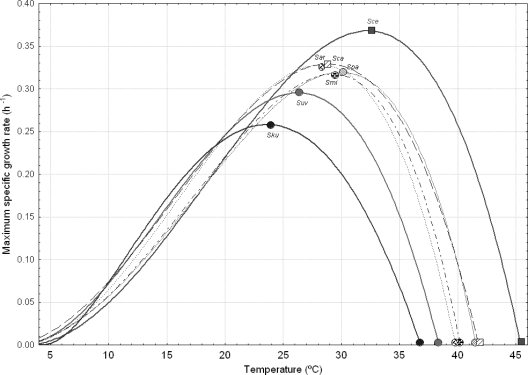
The second ANOVA, grouping strains according to their respective Saccharomyces species, showed that S. cerevisiae was the species with the statistically highest Topt and Tmax values (average of 32.3 and 45.4°C, respectively) (Table 3). Conversely, S. kudriavzevii exhibited the lowest Topt and Tmax values (significantly different with respect to the rest of the Saccharomyces species in the case of Topt), with 23.6 and 36.8°C, respectively. Again, no significant differences were found among yeasts for their Tmin values. A clear and more intuitive interpretation of the overall response of the Saccharomyces species as a function of temperature can be obtained by the graphical representations of the models, which are shown in Fig. 4. S. cerevisiae clearly appears as the most thermotolerant Saccharomyces species, exhibiting the highest μmax, Topt, and Tmax values. The species S. paradoxus, S. mikatae, S. cariocanus, and S. arboricolus formed a group with very similar responses (no significant differences among them). Conversely, S. kudriavzevii and S. bayanus var. uvarum showed the lowest μopt, Topt, and Tmax values. However, both species had the highest μmax when temperature was included in the interval from 10 to 20°C. The minimum temperatures to support growth were very similar among all Saccharomyces species, although S. kudriavzevii showed a slightly but not significantly higher Tmin value. We also studied a possible relationship among the CTMI model parameters obtained for the diverse Saccharomyces species. In this way, we found a linear correlation (R2 always above 0.90) between Topt and Tmax, μopt and Tmax, and μopt and Topt. However, there was no correlation between Topt and Tmin or μopt and Tmin. This means that the species which supported the highest growth temperatures also had the highest Topt and μopt values, but species with the lowest Topt and μopt values did not exhibit the minimum growth temperatures. This fact can also be graphically observed in Fig. 4.
TABLE 3.
Results of ANOVA for the parameters of the cardinal temperature model (μopt, Tmax, Tmin, and Topt), grouping strains according to their respective Saccharomyces species
| Species | No. of strains/no. of cases included in ANOVA | Avg (SD)a |
|||
|---|---|---|---|---|---|
| μopt(h−1) | Topt(°C) | Tmax(°C) | Tmin(°C) | ||
| S. cerevisiae | 10/30 | 0.368 (0.051) B | 32.27 (1.45) D | 45.39 (1.17) D | 2.84 (1.91) A |
| S. bayanus var. uvarum | 2/6 | 0.295 (0.016) A | 26.24 (0.69) B | 38.36 (1.47) BC | 2.37 (1.61) A |
| S. kudriavzevii | 4/12 | 0.258 (0.039) A | 23.61 (0.35) C | 36.80 (0.12) C | 4.29 (0.74) A |
| S. paradoxus | 3/9 | 0.319 (0.031) AB | 29.92 (0.73) A | 41.55 (0.93) A | 1.69 (1.25) A |
| S. mikatae | 1/3 | 0.318 (0.002) AB | 29.20 (1.46) A | 40.20 (0.10) AB | 1.79 (0.95) A |
| S. arboricolus | 1/3 | 0.328 (0.009) AB | 28.14 (0.61) AB | 39.80 (0.04) AB | 2.26 (0.99) A |
| S. cariocanus | 1/3 | 0.329 (0.006) AB | 28.81 (0.73) AB | 41.79 (0.06) A | 1.31 (1.10) A |
Values within the same column followed by different superscript letters are significantly different according to a Scheffé post hoc comparison test.
FIG. 4.
Changes of the maximum specific growth rates of the yeast species S. cerevisiae (Sce), S. paradoxus (Spa), S. mikatae (Smi), S. arboricolus (Sar), S. cariocanus (Sca), S. bayanus var. uvarum (Suv), and S. kudriavzevii (Sku) as a function of temperature. The graph was built using the average Saccharomyces parameters of the cardinal temperature model shown in Table 3.
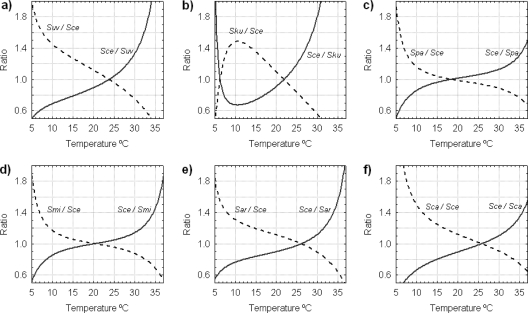
Figure 5 shows the theoretical evolution of the μmax ratios of the Saccharomyces species as a function of temperature. This parameter was obtained by dividing the μmax of S. cerevisiae by the μmax of each of the rest of the Saccharomyces species and vice versa. A ratio of 1 is indicative of both yeasts growing with similar μmax at a specific temperature value. On the other hand, a ratio of 2 means that one yeast grows 2-fold faster than the other. Thus, this parameter provides valuable information on the effects of temperature on a hypothetical sympatric association between S. cerevisiae and the rest of the Saccharomyces species. In all cases, the results showed S. cerevisiae to be the most competitive yeast at high temperatures. The models show that S. cerevisiae grows faster than S. bayanus var. uvarum above 24°C, and at 35°C, its μmax almost doubles compared to that of S. bayanus var. uvarum (Fig. 5a). In the rest of the comparisons, S. cerevisiae grows faster than S. kudriavzevii, S. paradoxus, S. mikatae, S. arboricolus, and S. cariocanus above 22, 19, 20, 26, and 25°C, respectively (Fig. 5b, c, d, e, and f). Below these values, S. cerevisiae progressively grows more slowly than the other species and is less competitive. In fact, at 5°C, the μmax of S. bayanus var. uvarum is double the μmax of S. cerevisiae (Fig. 5a).
FIG. 5.
Model predictions for the ratios of the maximum specific growth rates of S. cerevisiae (Sce) versus those of S. bayanus var. uvarum (Suv) (a), S. kudriavzevii (Sku) (b), S. paradoxus (Spa) (c), S. mikatae (Smi) (d), S. arboricolus (Sar) (e), and S. cariocanus (Sca) (f) as a function of temperature.
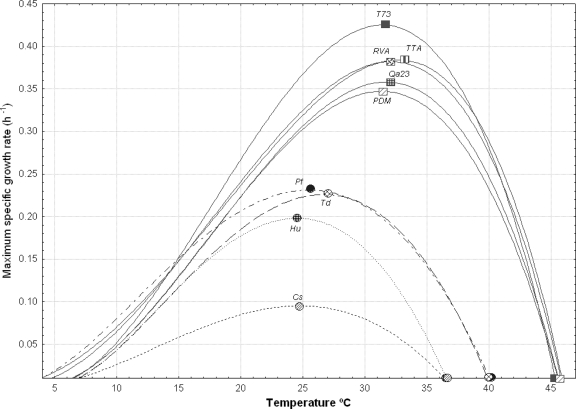
We also determined the influence of temperature on the growth of S. cerevisiae and non-Saccharomyces strains isolated from wine fermentations. Figure 6 shows the μmax evolution of 5 commercial S. cerevisiae strains and 4 non-Saccharomyces wine yeasts as a function of temperature. The S. cerevisiae strains were able to grow at up to 45°C, and their Topt was around 32°C, with a μopt ranging from 0.35 to 0.42 h−1 (Table 2). However, the strains of T. delbrueckii and Pichia fermentans were unable to grow above 40°C, while this value was even lower (around 36°C) for the species Hanseniaspora uvarum and Candida stellata. The optimum growth temperatures of these 4 strains were included in the interval from 24 to 27°C, with a μopt that was always lower than for S. cerevisiae strains (between 0.096 and 0.235 h−1). As can be deduced from the results in Fig. 6, at temperatures above 20°C, the difference between the μmax of S. cerevisiae and the μmax of non-Saccharomyces strains increased progressively.
FIG. 6.
Changes of the maximum specific growth rates of the non-Saccharomyces species H. uvarum (Hu), C. stellata (Cs), T. delbrueckii (Td), and P. fermentans (Pf) and different S. cerevisiae wine strains (T73, Qa23, TTA, RVA, and PDM) as a function of temperature.
Analysis of phylogenetic dependence.
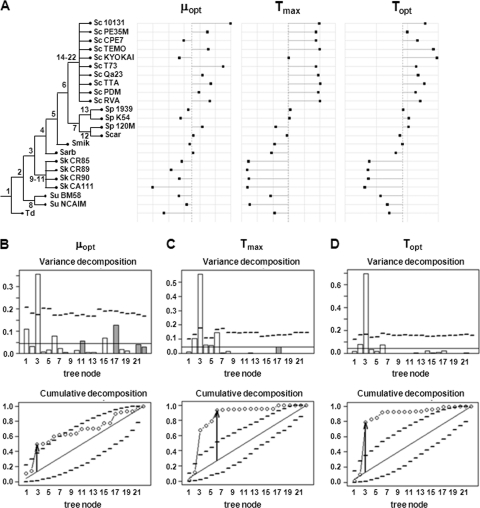
The CTMI model parameters μopt, Topt, and Tmax, which are quantitative variables, were tested using the orthogram approach (17) to determine a possible relationship with Saccharomyces phylogeny. We did not analyze Tmin because no significant differences were observed among strains. The topology of the phylogenetic tree, with node identifications (left side), and the corresponding values for each tested variable and strain (right side) are shown in Fig. 7 A. The orthogram analysis showed that the orthonormal vector representing node 3 (which differentiates S. arboricolus, S. mikatae, S. cariocanus, S. paradoxus, and S. cerevisiae from S. kudriavzevii and S. bayanus var. uvarum) explained the greatest part of the variance for the three parameters analyzed (Fig. 7B, C, and D). This vector peaked in all cases outside the confidence limits (represented by dashed lines in Fig. 7B, C, and D, upper panels) and showed a strong departure from the value expected under the hypothesis of absence of phylogenetic dependence (represented by a solid straight line). Cumulative variance representation confirmed the preponderance of the orthonormal vector 3 in the variance distribution, and again, a significant departure from the null hypothesis was registered for parameters μopt, Tmax, and Topt (solid line in Fig. 7B, C, and D, lower panels). The maximum deviations from the expected values were given for the sum of the first three and six orthonormal vectors for the Topt and Tmax parameters, respectively (vertical arrows in Fig. 7C and D, lower panels). Phylogenetic dependence was also confirmed by the results of the statistical tests R2Max, SkR2k, Dmax, and SCE. For the Topt and Tmax parameters, all test results were significant, showing P values lower than 0.0001. In the case of μopt, 3 of the 4 tests displayed significant values (0.042, 0.030, and 0.017 for R2Max, SkR2k, and Dmax, respectively). Only the SCE test was not able to indicate a significant departure from the null hypothesis, with a P value of 0.061. For the three CTMI parameters, the results of the R2Max test indicated that a significant part of the variance was explained by a single vector's contribution, indicative of a single punctual modification event having occurred in the evolutionary history of the genus. Relating this result to the orthogram plots, we determined that the significant change in the variables occurred after the divergence of the S. arboricolus, S. cariocanus, S. mikatae, S. paradoxus, and S. cerevisiae lineages from the S. kudriavzevii and S. bayanus var. uvarum lineages (Fig. 7A, node 3). Moreover, the decomposed variance plot and the SkR2k statistics also indicated that a significant variance explanation occurred in nodes toward the root. However, according to the orthogram plot and the cumulative orthogram plot of parameters Tmax and Topt (Fig. 7C and D), a secondary modification of parameters could also have occurred at node 6, after the divergence of the S. paradoxus and S. cerevisiae lineages, although the statistics tests did not support the presence of this second event. In any case, these results suggest that these three parameters have been modeled during the evolution of the Saccharomyces genus.
FIG. 7.
Phylogenetic dependence analysis for the cardinal temperature model parameters (μopt, Tmax, and Topt) of the 22 Saccharomyces strains. Torulaspora delbrueckii was used as an outgroup. (A) The topology of the phylogenetic relationships of the different strains is depicted on the left, and the dot plot of temperature model parameters for each strain on the right. Sc, S. cerevisiae; Sp, S. paradoxus; Scar, S. cariocanus; Smik, S. mikatae; Sarb, S. arboricolus; Sk, S. kudriavzevii; Su, S. bayanus var. uvarum; Td, Torulaspora delbrueckii. (B, C, and D) Variance decomposition using orthograms (top) and cumulative orthograms (bottom) for μopt, Tmax, and Topt, respectively. In the orthogram plots, abscissas correspond to the vector numbers associated with the nodes indicated in the phylogenetic topology of panel A, while ordinates show the contribution of each vector to the variance of the parameter by the squared regression coefficients (positive in white and negative in gray). Dashed lines correspond to upper confidence limits at 95%, deduced from Monte Carlo permutations, and solid lines represent the mean value. In the cumulative orthogram plots, ordinates show the cumulated contribution of successive vectors to the variance. Diagonal solid lines represent the expected values in the absence of phylogenetic dependence. Dashed lines correspond to the bilateral 95% confidence intervals. Circles show the observed values of cumulated squared regression coefficients. Vertical arrows mark the maximum deviations from the expected values (diagonal lines).
DISCUSSION
The influence of temperature on microorganism growth has been widely studied by microbiologists, and different mathematical models have been developed to quantify and predict its effects. Rosso et al. (20) and, more recently, Oscar (18) compared several temperature secondary models on the basis of different criteria (simplicity and biological significance of parameters, applicability, quality of fit, and ease of determination of parameters), concluding that the CTMI model was better than its competitors to fit a total of 48 data sets belonging to different species of microorganisms. In the present study, the CTMI model was also very useful to fit the experimental growth data of 27 yeast strains in the whole biological temperature range.
S. bayanus var. uvarum and S. kudriavzevii are considered the most psychrotrophic species of the Saccharomyces genus (3, 11, 22). Our results corroborate these observations, and S. bayanus var. uvarum and S. kudriavzevii were the yeasts with the lowest Topt and Tmax values. Unfortunately, little information is available in the literature to carry out a quantitative comparison with data obtained in this study. Serra et al. (22) mentioned that the optimum growth temperature for the wine strain S. bayanus var. uvarum P3 was attained around 28°C, close to the value obtained from this work for the average of the two S. bayanus var. uvarum strains assayed (26.3°C). In the case of S. kudriavzevii, Arroyo-López et al. (2) estimated, using response surface methodology, that the optimum growth temperature of the type strain IFO 1802T was attained at 24°C, while in this study, the average obtained for four S. kudriavzevii strains was 23.6°C. Belloch et al. (3) mentioned that both species were able to grow at 30°C but not at 37°C. Sampaio and Gonçalves (21) reported that the maximum growth temperatures for wild S. kudriavzevii and S. bayanus var. uvarum strains were 35°C and 36°C, respectively. In this work, the Tmax values estimated were slightly higher, 36.8°C for S. kudriavzevii and 38.3°C for S. bayanus var. uvarum.
Conversely, S. cerevisiae was the most thermotolerant species within the genus Saccharomyces, with the highest optimum (32.3°C) and maximum (45.4°C) growth temperatures. Several authors (2, 22) have reported that the optimum growth temperature of S. cerevisiae wine strains was around 34°C. The association of S. cerevisiae with fermentations established by humans is well known, and preliminary data suggest that temperature could play an important role in the predominance of this species. Heard and Fleet (9) observed that S. cerevisiae dominated traditional grape juice fermentations when they were carried out at higher temperatures. Recently, Goddard (8) also showed that temperature was an important factor in the imposition of S. cerevisiae versus non-Saccharomyces yeasts during wine fermentations. An increase of temperature from 16 to 23°C as a consequence of the highly vigorous respirofermentative consumption of sugars (Crabtree effect) favored the rapid growth of S. cerevisiae cells and final imposition of the species, although the initial frequency of this species was very low (<1%) (8). Such data clearly correlated with our results. We found that at temperatures above 20°C, the μmax of the S. cerevisiae wine strains increased faster than the μmax of the non-Saccharomyces wine species. These differences were especially evident at 32°C. Even under the optimum growth temperature conditions of the non-Saccharomyces species (∼25°C), the μmax values of the S. cerevisiae wine strains were always significantly higher.
Unfortunately, our knowledge of the ecology and distribution of the Saccharomyces species in wild environments is still very limited, but the present study could shed some light on the influence of temperature on the ecological interactions among Saccharomyces species. Diverse studies have shown that S. cerevisiae and its sibling species S. paradoxus occupy the same ecological niches (oak exudates, oak bark, and oak-associated soils) in widely separated woodland sites (16, 23). Sweeney et al. (25) reported that the growth temperature profiles of diverse S. paradoxus and S. cerevisiae wild strains, isolated from a single natural site, were different. S. paradoxus wild isolates exhibited Topt values of around 30°C (similar to those presented in this work), while for S. cerevisiae, Topt was above 37°C. Sampaio and Gonçalves (21) also carried out an interesting study on the influence of temperature on the sympatric association of four Saccharomyces species (S. cerevisiae, S. paradoxus, S. bayanus var. uvarum, and S. kudriavzevii) isolated from oak bark samples. Their study showed that temperature played a fundamental role in the interactions among the Saccharomyces species. They suggested that circadian temperature changes could provide a range of temperatures, allowing the sympatric association involving a species more adapted to grow at high temperatures (S. cerevisiae or S. paradoxus) and another species more adapted to grow at low temperatures (S. bayanus var. uvarum or S. kudriavzevii). Our results also support the hypothesis that adaptation to grow at different temperatures could be a very important factor in the ecology of Saccharomyces species. Mathematical models developed in the present study reveal that S. cerevisiae grows faster than the rest of the Saccharomyces species at high temperatures but exhibits a loss of competitiveness at low temperatures. Our models estimate the limits of the temperatures at which S. cerevisiae grows faster than the other species. These temperature values are 24°C for S. cerevisiae with respect to S. bayanus var. uvarum, 22°C with respect to S. kudriavzevii, 19°C with respect to S. paradoxus, 20°C with respect to S. mikatae, 26°C with respect to S. arboricolus, and 26°C with respect to S. cariocanus. In this way, in a hypothetical sympatric association between the species S. cerevisiae and S. kudriavzevii, if temperature were the only limiting factor, the growth of S. cerevisiae would be selectively favored over that of S. kudriavzevii at temperatures above 22°C. However, circadian temperature changes around this value would favor the growth of both species even in the same ecological niche. It is worth noting that this is a hypothetical case based on theoretical models. However, these predictions confirm previous studies of relative fitness tests carried out by Sampaio and Gonçalves (21) with the species S. cerevisiae and S. kudriavzevii at 10 and 30°C. If the culture was incubated at 10°C, only the species S. kudriavzevii was found at the end of the fermentation, while S. cerevisiae dominated the fermentations at 30°C. However, both species coexist in nature in the same microhabitats (oak barks), where a circadian cycle of temperatures is present. Another argument presented by Sampaio and Gonçalves (21) for the sympatric association of Saccharomyces species is the absence of overlap in the geographic distribution of S. kudriavzevii and S. bayanus var. uvarum, the two species more adapted to low temperatures.
According to the competitive exclusion principle, niche differentiation is necessary for sympatric coexistence of closely related species, and the different temperature growth profiles exhibited for the Saccharomyces species could explain this phenomenon. The analysis of the phylogenetic dependence of the CTMI parameters showed that a single event, which occurred after the divergence of the S. arboricolus, S. mikatae, S. cariocanus, S. paradoxus, and the S. cerevisiae lineages from the S. kudriavzevii and S. bayanus var. uvarum lineages, favored the adaptation of the former species to grow at higher temperatures. The analysis also suggests that a second event could now be occurring in the S. cerevisiae lineage after its divergence from S. paradoxus and S. cariocanus, which would explain the higher thermotolerance exhibited by this species. Another interesting point, although it was not revealed by the phylogenetic dependence analysis, is the progressive adaptation of S. kudriavzevii to grow at lower temperatures. This was evidenced because S. bayanus var. uvarum, the first species to diverge within the Saccharomyces genus, exhibited higher Topt and Tmax values than S. kudriavzevii. In light of these results, temperature has influenced the evolution of the Saccharomyces genus, favoring the adaptation of some species to grow at lower (S. kudriavzevii) and higher (especially S. cerevisiae) temperatures.
Acknowledgments
This work was supported by Generalitat Valenciana (project PROMETEO/2009/019) and the Spanish Government (projects AGL2009-12673-CO2-01, AGL2009-12673-CO2-02, and AGL2007-65498-C02-02 to A.Q., E.B., and J.M.G., respectively). F. N. Arroyo-López thanks the Spanish Government (MICINN) for his Juan de la Cierva postdoctoral research contract.
Footnotes
Published ahead of print on 11 February 2011.
REFERENCES
- 1.Arroyo-López, F. N., M. C. Durán Quintana, and A. Garrido Fernández. 2006. Use of the generalized z-value concept to study the effects of temperature, NaCl concentration and pH on Pichia anomala, a yeast related to table olive fermentation. Int. J. Food Microbiol. 106:45-51. [DOI] [PubMed] [Google Scholar]
- 2.Arroyo-López, F. N., S. Orlic, A. Querol, and E. Barrio. 2009. Effects of temperature, pH and sugar concentration on the growth parameters of Saccharomyces cerevisiae, S. kudriavzevii and their interspecific hybrid. Int. J. Food Microbiol. 131:120-127. [DOI] [PubMed] [Google Scholar]
- 3.Belloch, C., S. Orlic, E. Barrio, and A. Querol. 2008. Fermentative stress adaptation of hybrids within the Saccharomyces sensu stricto complex. Int. J. Food Microbiol. 122:188-195. [DOI] [PubMed] [Google Scholar]
- 4.Charoenchai, C., G. Fleet, and P. A. Henschke. 1998. Effects of temperature, pH and sugar concentration on the growth rates and cell biomass of wine yeasts. Am. J. Enol. Viticult. 49:283-288. [Google Scholar]
- 5.Chessel, D., A. Dufour, and J. Thioulouse. 2004. The ade4 Package-I-One-table methods. R News 4:10-15. [Google Scholar]
- 6.Covain, R., S. Dray, S. Fisch-Muller, and J. I. Montoya-Burgos. 2008. Assessing phylogenetic dependence of morphological traits using co-inertia prior to investigate character evolution in Loricariinae catfishes. Mol. Phylogenet. Evol. 46:986-1002. [DOI] [PubMed] [Google Scholar]
- 7.Gibson, A. M., N. Bratchell, and T. A. Roberts. 1987. The effect of sodium chloride and temperature on the rate and extent of growth of Clostridium botulinum type A in pasteurized pork slurry. J. Appl. Bacteriol. 62:479-490. [DOI] [PubMed] [Google Scholar]
- 8.Goddard, M. R. 2008. Quantifying the complexities of Saccharomyces cerevisiae's ecosystem engineering via fermentation. Ecology 89:2077-2082. [DOI] [PubMed] [Google Scholar]
- 9.Heard, G. M., and G. H. Fleet. 1998. The effects of temperature and pH on the growth of yeast species during fermentation of grape juice. J. Appl. Bacteriol. 65:23-28. [Google Scholar]
- 10.Ihaka, R., and R. Gentleman. 1996. R: a language for data analysis and graphics. J. Comput. Graph. Stat. 5:299-314. [Google Scholar]
- 11.Kishimoto, M., and S. Goto. 1995. Growth temperatures and electrophoretic karyotyping as tools for practical discrimination of Saccharomyces bayanus and Saccharomyces cerevisiae. J. Gen. Appl. Microbiol. 41:239-247. [Google Scholar]
- 12.Lachance, M. A., J. M. Bowles, and W. T. Starmer. 2003. Geography and niche occupancy as determinants of yeast biodiversity: the yeast-insect-morning glory ecosystem of Kipuka Puaulu, Hawai'i. FEMS Yeast Res. 4:105-111. [DOI] [PubMed] [Google Scholar]
- 13.Liti, G., D. B. H. Barton, and E. J. Louis. 2006. Sequence diversity, reproductive isolation and species concepts in Saccharomyces. Genetics 174:839-850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McMeekin, T. A., J. N. Olley, T. Ross, and D. A. Ratkowsky. 1993. Predictive microbioloy: theory and application. John Wiley & Sons, Inc., New York, NY.
- 15.Naumov, G. I. 1996. Genetic identification of biological species in the Saccharomyces sensu stricto complex. J. Ind. Microbiol. 17:295-302. [Google Scholar]
- 16.Naumov, G. I., E. S. Naumova, and P. Sniegowski. 1998. Saccharomyces paradoxus and Saccharomyces cerevisiae are associated with exudates of North American oaks. Can. J. Microbiol. 44:1045-1050. [PubMed] [Google Scholar]
- 17.Ollier, S., P. Couteron, and D. Chessel. 2006. Orthonormal transform to decompose the variance of a life-history trait across a phylogenetic tree. Biometrics 62:471-477. [DOI] [PubMed] [Google Scholar]
- 18.Oscar, T. P. 2002. Development and validation of a tertiary simulation model for predicting the potential growth of Salmonella typhimurium on cooked chicken. Int. J. Food Microbiol. 76:177-190. [DOI] [PubMed] [Google Scholar]
- 19.Querol, A., and G. Fleet. 2006. Yeasts in food and beverages. Springer-Verlag, Berlin, Germany.
- 20.Rosso, L., J. R. Lobry, and J. P. Flandrois. 1993. An unexpected correlation between cardinal temperatures of microbial growth highlighted by a new model. J. Theor. Biol. 162:447-463. [DOI] [PubMed] [Google Scholar]
- 21.Sampaio, J. P., and P. Gonçalves. 2008. Natural populations of Saccharomyces kudriavzevii in Portugal are associated with oak bark and sympatric with S. cerevisiae and S. paradoxus. Appl. Environ. Microbiol. 74:2144-2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Serra, A., P. Strehaiano, and P. Taillandier. 2005. Influence of temperature and pH on Saccharomyces bayanus var. uvarum growth; impact of a wine yeast interspecific hybridization on these parameters. Int. J. Food Microbiol. 104:257-265. [DOI] [PubMed] [Google Scholar]
- 23.Sniegowski, P. D., P. G. Dombrowski, and E. Fingerman. 2002. Saccharomyces cerevisiae and Saccharomyces paradoxus coexist in a natural woodland site in North America and display different levels of reproductive isolation from European conspecifics. FEMS Yeast Res. 1:299-306. [DOI] [PubMed] [Google Scholar]
- 24.Sørensen, B. J., and M. Jakobsen. 1997. The combined effects of temperature, pH and NaCl on growth of Debaryomyces hansenii analyzed by flow cytometry and predictive microbiology. Int. J. Food Microbiol. 34:209-220. [DOI] [PubMed] [Google Scholar]
- 25.Sweeney, J. Y., H. A. Kuehne, and P. D. Sniwgowski. 2004. Sympatric natural Saccharomyces cerevisiae and S. paradoxus populations have different thermal growth profiles. FEMS Yeast Res. 4:521-525. [DOI] [PubMed] [Google Scholar]
- 26.Wang, S. A., and F. Y. Bai. 2008. Saccharomyces arboricolus sp. nov., a yeast species from tree bark. Int. J. Syst. Evol. Microbiol. 58:510-514. [DOI] [PubMed] [Google Scholar]
- 27.Winer, B. J. 1962. Statistical principles in experimental design. McGraw-Hill, New York, NY.
- 28.Zwietering, M. H., I. Jongerburger, F. M. Rombouts, and K. Van't Riet. 1990. Modeling of the bacterial growth curve. Appl. Environ. Microbiol. 56:1875-1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zwietering, M. H., J. T. Koos, B. E. Hasenack, J. C. Wit, and K. Van't Riet. 1991. Modeling of bacterial growth as a function of temperature. Appl. Environ. Microbiol. 57:1094-1101. [DOI] [PMC free article] [PubMed] [Google Scholar]