Abstract
The filamentous fungus Aspergillus niger is widely used in biotechnological applications. Strain CBS513.88 is known to harbor 21 copies of the nonautonomous transposon Vader. Upon selection of chlorate-resistant A. niger colonies, one Vader copy was found integrated in the nirA gene. This copy was used for vector construction and development of a transposon-tagging method. Vader showed an excision frequency of about 1 in 2.2 × 105 conidiospores. A total of 95 of 97 colonies analyzed exhibited an excision event at the DNA level, and Vader footprints were found. By employing thermal asymmetric interlaced (TAIL)-PCR, the reintegration sites of 21 independent excision events were determined. All reintegration events occurred within or very close to genes. Therefore, this method can be used for transposon mutagenesis in A. niger.
Transposons are ubiquitous mobile genetic elements and are present in all prokaryotic and eukaryotic genomes. While transposons were detected early on in bacteria, plants, animals, and yeasts, it was only in the 1990s that they were described for filamentous fungi (7, 15, 21). It became evident that fungi harbor the same types of transposable elements as other eukaryotes, i.e., class I and class II transposable elements. Of all the fungal transposons described so far, only one has been applied for transposon mutagenesis (8).
The genome sequencing project of Aspergillus niger CBS513.88 was completed a few years ago (20). As this species is of industrial significance, an unbiased random mutagenesis tool based on endogenous transposons and used to identify new genes and understand their function and to create new industrially important mutants would be of great value. In a recent survey, we characterized all transposable elements of A. niger CBS513.88 and analyzed them for activity during strain improvement cycles. In that study, only Vader transposons were found to be active (3). The A. niger CBS513.88 strain is known to harbor 21 copies of Vader, which is a nonautonomous transposon. It was originally detected in A. niger var. awamori (1, 19). The members of this transposon family share a very high degree of similarity, with lengths about 439 bp and terminal inverted repeats of 44 bp. Vader elements are believed to be transactivated by the Tan1-encoded transposase, which is also present in the A. niger CBS513.88 genome. With the present study, we show that Vader elements can be activated, as observed in the strain lineage (3), and we present a new method for transposon mutagenesis in A. niger based on the transactivation of a labeled Vader copy.
MATERIALS AND METHODS
Strains and culture conditions.
Aspergillus niger strain CBS513.88 was used in this study. For selection experiments, A. niger was grown on Aspergillus minimal medium (AMM; 6.9 mM KCl, 11.2 mM KH2PO4, 4 M KOH, 2.1 mM MgSO4·7H2O, 50.5 mM glucose, 37 mM NH4Cl, 0.1% (vol/vol) trace element solution I [76.5 mM ZnSO4·7H2O, 178 mM H3BO3, 18 mM FeSO4·7H2O, 7 mM CoCI2·6H2O, 6.4 mM CuSO4·5H2O, 25 mM MnCI2·4H2O], 0.1% (vol/vol) trace element solution II [6.2 mM Na2MoO4·2H2O], and 1% (wt/vol) agarose). For excision selection, hygromycin B was added at a concentration of 125 μg/ml.
Transformation of A. niger was done according to published procedures (23), with the following modifications: 5 × 107 A. niger conidiospores were used to grow mycelia for protoplast preparation, mycelia were filtered through sterile Miracloth (Calbiochem, La Jolla, CA), and protoplast formation was performed using the lytic enzyme mixture Glucanex (Sigma, Steinheim, Germany).
Oligonucleotides and PCR amplification.
Oligonucleotides used are given in Table S1 in the supplemental material and were synthesized by MWG-Biotech (Ebersberg, Germany). PCR was done as previously described (2-4).
Selection for excision of Vader.
Mycelia of transformant AnT-6(3) carrying vector pIB635 were grown on solid medium, and conidiospores were obtained. A total of 107 conidiospores were placed on agarose plates containing minimal medium with 125 μg/ml hygromycin B. Conidiospores were then taken from hygromycin B-resistant colonies and streaked again on minimal medium with 125 μg/ml hygromycin B for 4 to 7 days in order to select against the formation of heterokaryons.
TAIL-PCR.
Thermal asymmetric interlaced (TAIL)-PCR was based on published methods (17). Each 20-μl primary TAIL-PCR mixture contained 2 μl of 10× PCR buffer, 200 mM deoxynucleoside triphosphate (dNTP) mix, 250 nM oligonucleotide IB1345, 5 μM oligonucleotide IB1347, 0.02 to 0.025 U Taq DNA polymerase, and 20 to 60 ng DNA. For the secondary reaction, 1 μl of 40-fold-diluted primary TAIL-PCR product was used as the template, with 200 nM oligonucleotide EH1434 and 3 μM oligonucleotide IB1347. For the tertiary TAIL-PCR, again 1 μl of 40-fold-diluted secondary TAIL-PCR product was used as the template. A total of 100 nM oligonucleotide IB1336 and 3 μM oligonucleotide IB1347 were employed. Details are listed in Table 1. Specific products were gel purified and directly sequenced to confirm the identities of the integration sites.
TABLE 1.
Thermal cycling conditions used for primary, secondary, and tertiary TAIL-PCR
| TAIL-PCR | File no. | No. of cycles | Thermal setting(s) (time) |
|---|---|---|---|
| Primary | 1 | 1 | 92°C (2 min), 95°C (1 min) |
| 2 | 4 high-stringency cycles | 94°C (15 s), 55°C (1 min), 72°C (2 min) | |
| 3 | 1 low-stringency cycle | 94°C (15 s), 30°C (3 min), 30°C (+ 0.2°C/s) | |
| 18°C (+ 0.3°C/s), 72°C (2 min) | |||
| 4 | 9 reduced-stringency cycles | 94°C (5 s), 37°C (1 min), 72°C (2 min) | |
| 5 | 11 super cycles | 94°C (5 s), 55°C (1 min), 72°C (2 min) | |
| 94°C (5 s), 55°C (1 min), 72°C (2 min) | |||
| 94°C (5 s), 37°C (1 min), 72°C (2 min) | |||
| 6 | 1 | 72°C (5 min) | |
| Secondary | 7 | 1 | 94°C (2 min) |
| 8 | 1 | 94°C (5 s), 55°C (1 min), 72°C (2 min) | |
| 9 | 9 super cycles | 94°C (5 s), 55°C (1 min), 72°C (2 min) | |
| 94°C (5 s), 55°C (1 min), 72°C (2 min) | |||
| 94°C (5 s), 37°C (1 min), 72°C (2 min) | |||
| 6 | 1 | 72°C (5 min) | |
| Tertiary | 10 | 1 | 94°C (2 min) |
| 11 | 19 normal cycles | 94°C (30 s), 37°C (1 min), 72°C (2 min) | |
| 6 | 1 | 72°C (5 min) |
Transposon trap experiments.
The basic medium used for transposon trap experiments contained 4.2 mM MgSO4·6H2O, 6.7 mM KCl, 0.65 mM FeSO4·7H2O, 58 mM sucrose, and 0.143% (vol/vol) trace element solution (55.6 mM ZnSO4·7H2O, 2.5 mM CuSO4·5H2O, 6.5 mM FeSO4·7H2O, 0.7 mM MnSO4·H2O, 1.6 mM boric acid, 0.4 mM MoNa2O4·2H2O). Three additional media were prepared by dissolving 35.3 mM NaNO3 (solution 1), 7.3 mM hypoxanthine (solution 2), or 35.3 mM NaNO3 and 225.5 mM NaClO3 (solution 3) in 300 ml 75 mM K2HPO4/KH2PO4, pH 5.8. Solutions 1, 2, or 3 and 700 ml basic medium were autoclaved separately and mixed, and 2% agar was added. In the case of solution 3, 1% (vol/vol) of a 10% l-arginine solution was added after autoclaving. Primary selection for transposon trap experiments was done on solution 3-basic medium. Colonies obtained were then plated on solution 1-basic medium and solution 2-basic medium to exclude mutations in cofactor genes.
Vector construction.
The 5′ and 3′ flanking regions of the A. niger niaD gene (An08g05610) were amplified from genomic DNA using primer pair IB0932 and IB0933 and primer pair IB0934 and IB0935, respectively. The Aspergillus nidulans gpdA promoter (Z32524) (22) was obtained using primer pair IB0910 and IB0911, and the A. nidulans trpC terminator (ENU24705) (18) was obtained using primer pair IB1041 and IB1042. Primer pair IB1039 and IB1040 served to amplify the Escherichia coli hph gene (V01499) (11). The sequence of the transposon Vader was not obtained via PCR but was synthesized by Sloning BioTechnology (Puchheim, Germany). This sequence contained flanking target site duplications and, within the transposon sequence, an AsiSI recognition site and an artificial anchor sequence for binding of a specific oligonucleotide in subsequent PCR analyses. The resulting vector pIB635 is shown here (see Fig. 2a).
RESULTS AND DISCUSSION
Transposon mutagenesis in general can be a powerful tool for generating mutation libraries (reviewed in references 6, 12, and 13). For this purpose, a heterologous transposon may be introduced in a new host (9), or endogenous transposons may be employed (14). To identify a Vader copy which can be transactivated, we used chlorate selection as a transposon trap. Chlorate-resistant A. niger colonies may be the result of mutations in either the niaD or nirA gene or in uptake or cofactor genes. After performing several physiological tests and molecular analyses on obtained isolates (data not shown), a Vader insertion event was observed in the nirA gene of one mutant (Fig. 1). Sequence analysis revealed the presence of the expected target site duplications.
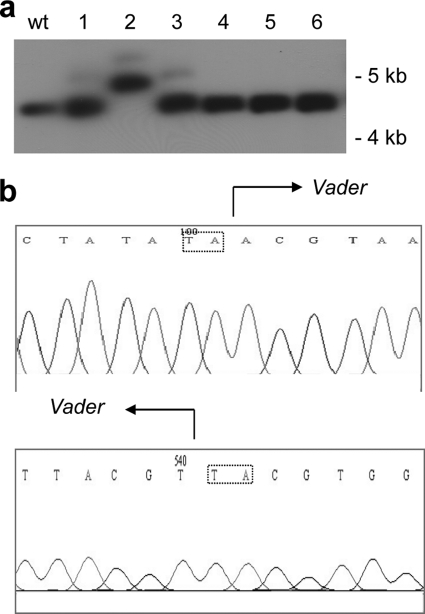
FIG. 1.
Transposition of a Vader copy in the A. niger nirA gene. (a) Southern blot with genomic DNA from wild-type (wt) A. niger and six chlorate-resistant mutants (1 to 6). The DNA was digested with the restriction enzyme XhoI. 32P[α-dCTP] was used to label nirA DNA. (b) Sequence analysis of the Vader insertion site in the nirA gene. The insertion site of Vader is indicated for both of the flanking sequences. The “TA” target site duplications are shown in boxes.
Based on the sequence of the activated Vader element inserted in the nirA gene, a synthetic element was synthesized which contained a unique 20-bp anchor, 5′-GAGTCCCGTCGTACTTCTGC-3′, allowing for specific amplification of this synthetic Vader element. The synthetic Vader element was then cloned between the gpdA promoter and the open reading frame of the hygromycin B resistance gene (Fig. 2 a). In this position, the element efficiently blocks the transcription of the resistance gene. The construct was targeted to the A. niger niaD locus using the 5′ and 3′ flanking sequences for homologous recombination. Several transformants which carried the synthetic Vader copy at the niaD locus were obtained (Fig. 2b). One transformant, AnT-6(3), was selected for all further experiments.
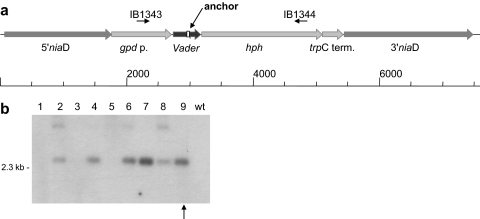
FIG. 2.
Vector construction and A. niger transformation. (a) Vector pIB635 for transformation of A. niger strain CBS513.88. This vector contains a synthetic Vader element surrounded by its target site duplication. Vader was cloned between the A. nidulans gpdA promoter and the E. coli hph gene. The 5′ and 3′ flanking sequences from the niaD gene allow homologous recombination into the A. niger niaD sequence. (b) Southern blot hybridization of genomic DNA (lanes 1 to 9) from A. niger transformants selected on chlorate-containing plates and the wild-type (wt) strain CBS513.88 digested with the restriction enzyme XbaI. 32P[α-dCTP] was used to label the probe. Six transformants were obtained. The arrow indicates transformant AnT-6(3), which was used for all further experiments.
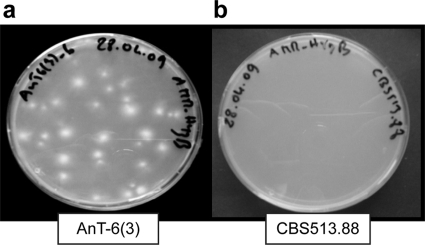
To select for mycelia in which a transposition event had occurred, 107 conidiospores each were plated on petri dishes containing minimal medium with 125 μg/ml hygromycin B. Under these conditions, transformant AnT-6(3) yielded 45 hygromycin B-resistant colonies, on average (Fig. 3 a), indicating an excision frequency of 1 in 2.2 × 105 conidiospores. As a control, the same amount of conidiospores from the untransformed CBS513.88 strain was plated, but hygromycin B-resistant colonies were never observed (Fig. 3b).
FIG. 3.
Selection assay for Vader excision. Example of selection assay on hygromycin B containing minimal medium. (a) Transformant AnT-6(3) yields numerous hygromycin B-resistant colonies; (b) untransformed CBS513.88 strain never yields colonies on hygromycin B.
The excision frequency of Vader is similar to that of the impala element from Fusarium gramineum (1 in 105 to 106 conidiospores) (10). The impala element has also been used in heterologous hosts with similar excision frequencies (8, 16, 25). The rather high excision frequency of Vader will facilitate to obtain large transposon mutant libraries of A. niger CBS513.88.
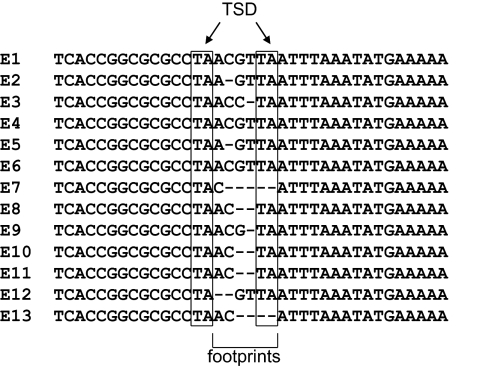
DNA was isolated from several hygromycin B-resistant isolates, and using oligonucleotides IB1343 and IB1344, part of the sequence flanking the synthetic Vader insertion site at the niaD locus was PCR amplified. Of 97 colonies examined, 95 gave evidence for excision of the Vader element from its original location (not shown). Sequence analysis was performed to confirm excision of transposon Vader from its donor site. Vader footprints are present in each excision event, as shown in Fig. 4. Each excision event is characterized by footprints of different types, such as ACGTTA, C, ACTA, ACCTA, AC, AGTTA, or ACGTA. One isolate exhibits the footprint GTTA, which was previously observed for Vader (3). While footprints like this are to be expected for most transposons, they are unusual for members of the Fot1/pogo subfamily, to which Vader belongs. Footprints of that subfamily typically contain a single copy of the target site duplication and the first or last nucleotide of the transposon sequence (7). Even more distantly related transposons appear to have a preference for this type of footprint (5, 24), including the mimp1 element (3, 9).
FIG. 4.
Molecular analysis of Vader excision. Sequence analysis of 13 excision events, each exhibiting an individual footprint (TSD = target site duplication).
From the 95 analyzed Vader excision events, none showed loss of the element (data not shown). To identify the genomic reintegration sites following the individual excision events, we established a protocol for thermal asymmetric interlaced PCR (TAIL-PCR) (17). Three examples of successful TAIL-PCRs are shown in Fig. 5. Specific TAIL-PCR amplicons from the third round were eluted from agarose gels and subjected to sequence analysis. Reintegration sites were determined by comparison of the obtained DNA sequences with the A. niger genome sequence found in the database. To reconfirm the location of the integration sites, oligonucleotides flanking the integration sites were employed for PCR amplification (see Fig. S1 in the supplemental material). Only three reintegration sites were not found in the database, while all others were identified. The very high reintegration frequency of Vader is very promising for development as a mutagenesis tool, as comparable experiments with the Restless transposon did not lead to reintegration after excision (26).
FIG. 5.
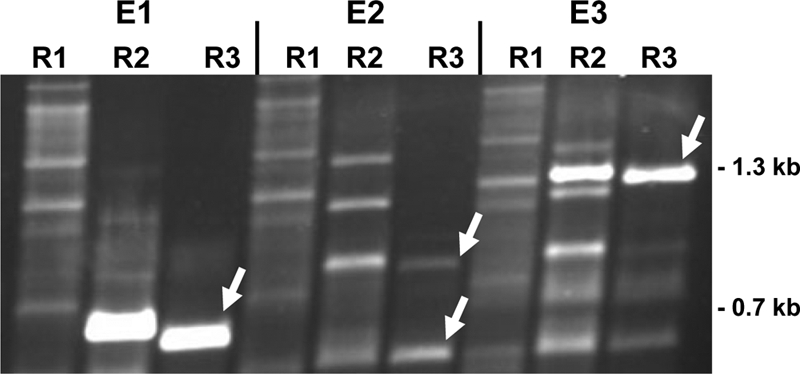
Vader reintegration. Gel electrophoresis of three different TAIL-PCR products from excision events E1, E2, and E3. Lane designations R1, R2, and R3 indicate products of the primary, secondary, and tertiary TAIL-PCRs, respectively. The random oligonucleotide IB1347 and nested oligonucleotides IB1345, EH1434, and IB1336 were employed. The white arrows indicate PCR fragments which were sequenced and found to contain A. niger genomic sequences flanking the transposon Vader at its reintegration site.
Using the TAIL-PCR method, 21 different integration sites were found on chromosomes 1, 2, 3, 4, 5, and 8, indicating that the reintegration site is not limited by the proximity of the donor site, which is localized on chromosome 8. Instead, transposition reintegration appears to be random with respect to genomic location. This finding highlights the usability of the method described here.
In the assembled A. niger genome of 33.9 Mb, there are 14,165 protein-coding genes, with an average length of 1.572 bp. The gene density (number of genes/kb) is 0.42. Of the 21 reintegration events analyzed, 1 occurred in an exon, 12 in annotated introns (in some concerning hypothetical genes), 3 within 400 bp downstream of an open reading frame, and 2 within 500 bp upstream of an open reading frame. Another three integration sites were more than 500 bp upstream of an open reading frame (Table 2). A few reintegration sites were found two or three times. These duplicates were found only in mycelia from the same experiment.
TABLE 2.
Reintegration sites of the synthetic Vader elementa
| GenBank accession no. | Locus tag | Reintegration site location | Chromosome |
|---|---|---|---|
| XM_001390097 | An03g02370 | Intronb | 5 |
| XM_001395060 | An12g00080 | 1,415 bp upstream | 3 |
| XM_001395339 | An12g02970 | 378 bp downstream | 3 |
| XM_001390831 | An06g00400 | 283 bp downstream | 8 |
| XM_001400379 | An02g12850 | Intronb | 4 |
| XM_001397721 | An16g04470 | Intronb | 5 |
| XM_001397940 | An16g06750 | 489 bp upstreamb | 5 |
| XM_001398488 | An18g00560 | Intronb | 8 |
| XM_001389142 | An01g07540 | Exonc | 2 |
| XM_001396557 | An15g00180 | 1,295 bp upstreamb | 3 |
| XM_001398918 | An18g05010 | Intronc | 8 |
| XM_001397938 | An16g06730 | Intron | 5 |
| XM_001397364 | An06g00880 | Intron | 8 |
| XM_001398449 | An18g00160 | Intron | 8 |
| XM_001397605 | An16g03290 | Intron | 5 |
| XM_001398419 | An17g02280 | 556 bp upstream | 5 |
| XM_001400930 | An14g03420 | Intron | 1 |
| XM_001388909 | An01g05210 | Intron | 2 |
| XM_001395706 | An12g06870 | 88 bp downstream | 3 |
| XM_001389436 | An01g10550 | 319 bp upstream | 2 |
| XM_001389142 | An01g07540 | Intronb | 2 |
Three other reintegration sites were characterized, but the sequences were not included in the database.
This reinsertion site was found twice.
This reinsertion site was found three times.
Finally, it should be noted that it seems feasible to employ this method in other fungi as well, provided the transactivating transposase can be expressed in new hosts. The construction of vectors carrying both Vader and its transposase will aid this purpose and provide an additional tool for fungi relevant in biotechnology.
Supplementary Material
Acknowledgments
Part of this study was funded by DSM Anti-Infectives, Delft, Netherlands. I.B. and E.H. received a grant from the Max-Buchner-Stiftung.
We thank Hanna Schmidt for excellent technical help.
Footnotes
Published ahead of print on 4 February 2011.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Amutan, M., E. Nyyssönen, J. Stubbs, M. R. Diaz-Torres, and N. Dunn-Coleman. 1996. Identification and cloning of a mobile transposon from Aspergillus niger var. awamori. Curr. Genet. 29:468-473. [DOI] [PubMed] [Google Scholar]
- 2.Braumann, I., M. van den Berg, and F. Kempken. 2008. Repeat induced point mutation in two asexual fungi, Aspergillus niger and Penicillium chrysogenum. Curr. Genet. 53:287-297. [DOI] [PubMed] [Google Scholar]
- 3.Braumann, I., M. van den Berg, and F. Kempken. 2007. Transposons in biotechnologically relevant strains of Aspergillus niger and Penicillium chrysogenum. Fungal Genet. Biol. 44:1399-1414. [DOI] [PubMed] [Google Scholar]
- 4.Braumann, I., M. A. van den Berg, and F. Kempken. 2008. Strain-specific retrotransposon-mediated recombination in a commercially used Aspergillus niger strain. Mol. Genet. Genomics 280:319-325. [DOI] [PubMed] [Google Scholar]
- 5.Bryan, G., D. Garza, and D. Hartl. 1990. Insertion and excision of the transposable element mariner in Drosophila. Genetics 125:103-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Daboussi, M. J. 1996. Fungal transposable elements: generators of diversity and genetic tools. J. Genet. 75:325-339. [Google Scholar]
- 7.Daboussi, M. J., and P. Capy. 2003. Transposable elements in filamentous fungi. Annu. Rev. Microbiol. 57:275-299. [DOI] [PubMed] [Google Scholar]
- 8.de Queiroz, M. V., and M. J. Daboussi. 2003. Impala, a transposon from Fusarium oxysporum, is active in the genome of Penicillium griseoroseum. FEMS Microbiol. Lett. 218:317-321. [DOI] [PubMed] [Google Scholar]
- 9.Dufresne, M., et al. 2008. Transposon-tagging identifies novel pathogenicity genes in Fusarium graminearum. Fungal Genet. Biol. 45:1552-1561. [DOI] [PubMed] [Google Scholar]
- 10.Firon, A., F. Villalba, R. Beffa, and C. d'Enfert. 2003. Identification of essential genes in the human fungal pathogen Aspergillus fumigatus by transposon mutagenesis. Eukaryot. Cell 2:247-255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kaster, K. R., S. G. Burgett, R. N. Rao, and T. D. Ingolia. 1983. Analysis of a bacterial hygromycin B resistance gene by transcriptional and translational fusions and by DNA sequencing. Nucleic Acids Res. 11:6895-6911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kempken, F. 2003. Fungal transposable elements: inducers of mutations and molecular tools, p. 83-99. In D. K. Arora and G. G. Khachatourians (ed.), Applied mycology and biotechnology: fungal genomics, vol. 3. Elsevier Science, Amsterdam, Netherlands. [Google Scholar]
- 13.Kempken, F. 1999. Fungal transposons: from mobile elements towards molecular tools. Appl. Microbiol. Biotechnol. 52:756-760. [Google Scholar]
- 14.Kempken, F., and U. Kück. 2000. Tagging of a nitrogen pathway-specific regulator gene in Tolypocladium inflatum by the transposon Restless. Mol. Gen. Genet. 263:302-308. [DOI] [PubMed] [Google Scholar]
- 15.Kempken, F., and U. Kück. 1998. Transposons in filamentous fungi-facts and perspectives. Bioessays 20:652-659. [DOI] [PubMed] [Google Scholar]
- 16.Li Destri Nicosia, M. G., et al. 2001. Heterologous transposition in Aspergillus nidulans. Mol. Microbiol. 39:1330-1344. [PubMed] [Google Scholar]
- 17.Liu, Y. G., and R. F. Whittier. 1995. Thermal asymmetric interlaced PCR: automatable amplification and sequencing of insert end fragments from P1 and YAC clones for chromosome walking. Genomics 25:674-681. [DOI] [PubMed] [Google Scholar]
- 18.Mullaney, E. J., J. E. Hamer, K. A. Roberti, M. M. Yelton, and W. E. Timberlake. 1985. Primary structure of the trpC gene from Aspergillus nidulans. Mol. Gen. Genet. 199:37-45. [DOI] [PubMed] [Google Scholar]
- 19.Nyyssönen, E., M. Amutan, L. Enfield, J. Stubbs, and N. S. Dunn-Coleman. 1996. The transposable element Tan1 of Aspergillus niger var. awamori, a new member of the Fot1 family. Mol. Gen. Genet. 253:50-56. [DOI] [PubMed] [Google Scholar]
- 20.Pel, H. J., et al. 2007. Genome sequencing and analysis of the versatile cell factory Aspergillus niger CBS 513.88. Nat. Biotechnol. 25:221-231. [DOI] [PubMed] [Google Scholar]
- 21.Pöggeler, S., and F. Kempken. 2004. Mobile genetic elements in mycelial fungi. In U. Kück (ed.), The mycota II, genetics and biotechnology, 2nd ed., vol. 165-198. Springer Verlag, Heidelberg, Germany.
- 22.Punt, P., et al. 1990. Functional elements in the promoter region of the Aspergillus nidulans gpdA gene encoding glyceraldehyde-3-phosphate dehydrogenase. Gene 93:101-109. [DOI] [PubMed] [Google Scholar]
- 23.van den Brink, J. M., G. C. M. Selten, and J. P. T. W. van den Hombergh. July 1999. Expression cloning in filamentous fungi. U.S. patent WO 99/32617.
- 24.van Luenen, H. G., S. D. Colloms, and R. H. Plasterk. 1994. The mechanism of transposition of Tc3 in C. elegans. Cell 79:293-301. [DOI] [PubMed] [Google Scholar]
- 25.Villalba, F., M. H. Lebrun, A. Hua-Van, M. J. Daboussi, and M. C. Grosjean-Cournoyer. 2001. Transposon impala, a novel tool for gene tagging in the rice blast fungus Magnaporthe grisea. Mol. Plant Microbe Interact. 14:308-315. [DOI] [PubMed] [Google Scholar]
- 26.Windhofer, F., K. Hauck, D. E. A. Catcheside, U. Kück, and F. Kempken. 2002. Ds-like Restless deletion derivatives occur in Tolypocladium inflatum and two foreign hosts, Neurospora crassa and Penicillium chrysogenum. Fungal Genet. Biol. 35:171-182. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.