Abstract
Taking advantage of the gene manipulation system developed in Thermococcus kodakarensis, here, we developed a system for gene expression and efficient protein secretion using this hyperthermophilic archaeon as a host cell. DNA fragments encoding the C-terminal domain of chitinase (ChiAΔ4), which exhibits endochitinase activity, and the putative signal sequence of a subtilisin-like protease (TK1675) were fused and positioned under the control of the strong constitutive promoter of the cell surface glycoprotein gene. This gene cassette was introduced into T. kodakarensis, and secretion of the ChiAΔ4 protein was examined. ChiAΔ4 was found exclusively in the culture supernatant and was not detected in the soluble and membrane fractions of the cell extract. The signal peptide was specifically cleaved at the C-terminal peptide bond following the Ala-Ser-Ala sequence. Efficient secretion of the orotidine-5′-monophosphate decarboxylase protein was also achieved with the same strategy. We next individually overexpressed two genes (TK1675 and TK1689) encoding proteases with putative signal sequences. By comparing protein degradation activities in the host cells and transformants in both solid and liquid media, as well as measuring peptidase activity using synthetic peptide substrates, we observed dramatic increases in protein degradation activity in the two transformants. This study displays an initial demonstration of cell engineering in hyperthermophiles.
Hyperthermophiles are organisms that exhibit optimal growth at temperatures above 80°C (33). Due to the fact that they occupy the deepest lineages within the phylogenies of both Archaea and Bacteria based on rRNA sequences, the organisms have attracted much attention in terms of biological evolution (11, 33). Hyperthermophiles have also been focused upon as a source for (thermo)stable enzymes, which can be expected to be applicable in a broad range of enzyme-based technologies (3, 10, 35). Besides the thermostable DNA polymerase used in PCR, a vast range of enzymes from hyperthermophiles have been examined, including various lipases/esterases, proteases, sugar-modifying enzymes, and dehydrogenases. In contrast to the large number of studies on the enzymes from hyperthermophiles, attempts to utilize the hyperthermophile cells themselves are limited (9, 23). This may be in part due to the fact that genetic manipulation systems have not been developed in most of the hyperthermophiles, which makes it difficult to utilize metabolic and cell-engineering strategies that are commonplace in mesophilic microorganisms.
Thermococcus kodakarensis (previously called Thermococcus kodakaraensis) is a sulfur-reducing hyperthermophilic archaeon isolated near the coast of Kodakara Island, Kagoshima, Japan (4, 18). The strain is an obligate heterotroph and grows on a variety of carbon sources, such as maltooligosaccharides, amino acids, and pyruvate. The entire genome sequence of T. kodakarensis has been determined (12), and gene manipulation systems based on homologous recombination and shuttle vectors have been developed (16, 27, 28, 30, 31). This allows us to rationally design and engineer T. kodakarensis cells for use in various aspects of biotechnology, as well as to study gene function in vivo (5, 13, 21, 26, 27, 29, 32).
In recent studies, we have shown that T. kodakarensis can be utilized as a host cell for protein expression. By positioning genes under the control of strong, constitutive promoters, such as those of the glutamate dehydrogenase gene (TK1431) or the cell surface glycoprotein gene (TK0895), the 3-hydroxy-3-methylglutaryl coenzyme A (HMG-CoA) reductase genes from T. kodakarensis and Pyrococcus furiosus (16), the endogenous pantoate kinase gene (38), and the α-1,4-glucan phosphorylase gene from Sulfolobus solfataricus (19) have been expressed in T. kodakarensis, leading to active proteins. Functional expression of the last two genes was not possible in conventional mesophilic host cells, such as Escherichia coli, indicating that gene expression in T. kodakarensis can be regarded as a practical alternative for the preparation of active recombinant proteins from hyperthermophilic archaea.
In this study, we examined the possibilities of secreting proteins using T. kodakarensis as a host strain. We also examined whether the overexpression/secretion system could be applied to engineer T. kodakarensis cells. In this case, we overexpressed putative protease genes (TK1675 and TK1689) with predicted secretion signals and found that these engineered cells have significantly greater protein degradation capabilities than the wild-type strain, T. kodakarensis KOD1.
MATERIALS AND METHODS
Strains, media, and plasmids.
T. kodakarensis strains were cultivated under anaerobic conditions at 85°C in a nutrient-rich medium (ASW-YT) or a synthetic medium (ASW-AA) supplemented with various organic substrates or elemental sulfur (24, 31). ASW-YT medium was composed of 0.8× artificial seawater (ASW), 5.0 g liter−1 of yeast extract, and 5.0 g liter−1 of tryptone. Resazurin was added at a concentration of 0.8 mg liter−1, and prior to inoculation, Na2S 9H2O was added to the medium until it became colorless. In the case of plate culture, elemental sulfur and Na2S 9H2O were replaced with 2 ml of a polysulfide solution (10 g of Na2S 9H2O and 3 g of sulfur flowers in 15 ml of H2O) per liter, and Gelrite (10 g liter−1) was added to solidify the medium. The composition of ASW-AA medium is described elsewhere (31). Unless otherwise indicated, all medium components were purchased from Wako Pure Chemical Industries (Osaka, Japan) or Nacalai Tesque (Kyoto, Japan). E. coli strain DH5α was cultivated in Luria-Bertani medium at 37°C with 100 μg ml−1 ampicillin.
Construction of expression/secretion plasmids.
The plasmid pSecChiA was constructed to insert the ChiAΔ4 expression/secretion cassette into the genome through double-crossover homologous recombination. The 5′-flanking region (998 bp) of the chiA gene of T. kodakarensis, which contains 758 bp of the TK1764 gene, and the 3′ portion of the chiA gene itself (920 bp) were amplified from genomic DNA with the primer sets chiAF1/chiAR1 and chiAF2/chiAR2, respectively (see Table S1 in the supplemental material). The two fragments were digested with SpeI and ligated with pUC118, which had been digested with EcoRI/SalI and treated with exonuclease to obtain blunt ends (pChiA1). The trpE marker gene cassette in pUMT2 (30) was amplified with the primer set trpEF/trpER, digested with XbaI/SpeI, and inserted in the SpeI site of pChiA1. Inverse PCR and self-ligation were first performed with the primer set CsgF/CsgR to introduce the cell surface glycoprotein gene (TK0895) promoter (csg promoter) and then with SigF/SigR to introduce a fragment corresponding to the signal sequence. To construct the PyrF expression/secretion cassette (pSecPyrF), the coding region of the pyrF gene (TK2276) and the 3′-flanking region (300 bp) of the glutamate dehydrogenase gene (TK1431) were inserted between the signal peptide sequence and the ChiAΔ4 coding region of pSecChiA. The plasmid (pSecPyrF) was designed so that the second amino acid of PyrF follows the signal peptide and a STREP tag (Trp-Ser-His-Pro-Gln-Phe-Glu-Lys) sequence is added at the C terminus.
Plasmids for overexpression/secretion of TK1675 and TK1689 were designed so that the csg promoter would be inserted directly upstream of the TK1675 and TK1689 genes at their native loci via single-crossover insertion/pop-out recombination. For TK1675, the coding region, along with a 1,000-bp region of the TK1675 5′-flanking region, was amplified with the primer set (1675F1-X/1675R-B), digested with BamHI/XbaI, and inserted into pUD2 digested with BamHI/XbaI (pUD2-1675). pUD2 contains the pyrF marker gene cassette inserted at the HincII site of pUC118. Additionally, the TK1675 coding region alone was amplified with the primer set (1675F2/1675R2) and ligated to the 187-bp csg promoter region amplified with the primers 0895PF/0895PR to construct the csg-TK1675 fusion cassette. An inverse PCR of pUD2-1675 was performed with the primer set 1675PI/1675F2, and after the amplified product was digested with BamHI, it was ligated to the BamHI-digested csg-TK1675 fusion cassette. In the case of TK1689, a fragment containing 1,000 bp of the 5′-flanking and coding regions of TK1689 was amplified with primer set 1689F1-B/1689R1-E, digested with BamHI/EcoRI, and inserted into pUD2 at the respective sites. Inverse PCR was performed with the primer set 1689F2-csg/1689R2-csg, and the obtained fragment was self-ligated. The sequences of all plasmids were confirmed.
Transformation of T. kodakarensis.
Transformation of T. kodakarensis with the trpE marker was performed using T. kodakarensis KUW1 (ΔpyrF ΔtrpE), which displays tryptophan and uracil auxotrophy (30). Cells at late log phase were harvested, resuspended in 200 μl of 0.8× ASW, and kept on ice for 30 min. After the addition of 3.0 μg plasmid and further incubation on ice for 1 h, the cells were spread on ASW-AA plate medium with 10 μg ml−1 uracil but without tryptophan, and transformants displaying tryptophan prototrophy were isolated.
Transformation of T. kodakarensis KUW1 with the pyrF marker via single-crossover insertion and pop-out recombination was performed as described previously (38). Cells transformed with the plasmids were cultivated twice in ASW-AA medium for 24 h at 85°C to enrich transformants that displayed uracil prototrophy due to single-crossover insertion. The cells were then diluted with 0.8× ASW and spread onto solid medium supplemented with 10 g liter−1 of 5-fluoroorotic acid (5-FOA) and 60 mM NaOH. As cells with an intact pyrimidine biosynthesis pathway convert 5-FOA to the toxic 5-fluoroorotidine-5′-phosphate, only cells that have lost the pyrF gene via a second pop-out recombination can grow under these conditions. Cells were grown for 2 days at 85°C. Individual transformants were selected, and their genotypes were examined by PCR using primer sets 1675F3/1675R3 and 1689F3/1689R3 and DNA sequencing. All loci were analyzed by PCR and sequenced to confirm the absence of unintended mutations.
Preparation of culture supernatant and soluble/membrane fractions of T. kodakarensis.
T. kodakarensis strains were cultivated in ASW-YT liquid medium supplemented with 5 g liter−1 sodium pyruvate. Cells were separated by centrifugation (5,000 × g; 15 min). The supernatant was centrifuged again at 10,000 × g for 15 min, and the supernatant was used as the culture supernatant fraction. The fraction was concentrated and desalted using Amicon Ultra-15 (Millipore, Bedford, MA). The cell pellet was suspended in 50 mM Tris-HCl (pH 7.5) to promote cell disruption and centrifuged (20,400 × g; 15 min). The supernatant was used as the soluble fraction of the cell extracts. The pellet was suspended in 50 mM Tris-HCl (pH 7.5), centrifuged (20,400 × g; 15 min), and resuspended in 50 mM Tris-HCl (pH 7.5) and was used as the membrane fraction.
Examination of ChiAΔ4 and PyrF proteins in the culture supernatant.
For Western blot analysis, aliquots of the culture supernatant, soluble fraction, and membrane fraction were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (12.5% acrylamide), followed by blotting to a polyvinylidene difluoride membrane (Hybond-P; GE Healthcare, Little Chalfont, United Kingdom). In the case of ChiAΔ4, rabbit anti-ChiAΔ4 antiserum was used, along with horseradish peroxidase (HRP)-rec-protein G (1:100,000 dilution) (Zymed Laboratories, San Francisco, CA). STREP-Tag II antibody HRP conjugate (Merck KGaA, Darmstadt, Germany) was used for the STREP-tagged PyrF protein. For signal detection, the ECL Advance Western Blotting Detection System (GE Healthcare) and Lumi Vision PRO 400EX (Aisin, Kariya, Japan) were used. For N-terminal amino acid sequence analysis, the culture supernatant of OSC1 was applied to anion-exchange chromatography (ResourceQ; GE Healthcare), and the ChiAΔ4 protein was eluted with a linear gradient of NaCl (0 to 1 M) in Tris-HCl (pH 7.5). After SDS-PAGE, proteins were electroblotted onto a polyvinylidene difluoride membrane (Mini ProBlott Membranes; Applied Biosystems). The membrane was washed with double-distilled H2O (ddH2O) for 5 min and rinsed with 100% methanol, stained with CBB solution (0.2% Coomassie brilliant blue, 10% acetic acid, 40% methanol) for 15 s, and destained with 60% methanol. Bands were excised and analyzed with a protein sequencer (Procise 491HT; Applied Biosystems). A chitinase activity assay was performed using a fluorometric substrate, 4-methylumbelliferyl β-d-N,N′-diacetyl chitobioside (Sigma, St. Louis, MO) (34). After a 30-min reaction at 80°C, the fluorescence of liberated 4-methylumbelliferone was measured with excitation and emission wavelengths (λex and λem) at 350 and 440 nm, respectively.
Measurements of protein-degrading activity.
In order to examine halo formation on solid media, T. kodakarensis strains were cultivated at 85°C for 12 h in ASW-YT liquid medium. Cells from 100 μl culture were suspended in 0.8× ASW (100 μl). After dilution (1:1,000), 10 μl was inoculated onto solidified ASW-YT medium supplemented with 3.5% skim milk. The cells were incubated at 85°C for 3 days. Activity measurements with peptide substrates were performed at 60°C with the substrate succinyl (Suc)-Ala-Ala-Pro-Phe-4-methyl-coumaryl-7-amide (MCA) (Peptide Institute, Osaka, Japan). The reaction mixture (1 ml) contained culture supernatant fractions corresponding to 100 μl culture, 600 μM substrate, and 5 mM CaCl2 in 50 mM Tris-HCl (pH 7.5). The final concentration of Me2SO used to dissolve the substrate was constant at 6%. Release of 7-amino-4-methyl-coumarin was monitored consecutively with λex and λem at 380 and 460 nm, respectively. Other methods are described in the figure legends.
RESULTS
Gene design for overexpression and secretion.
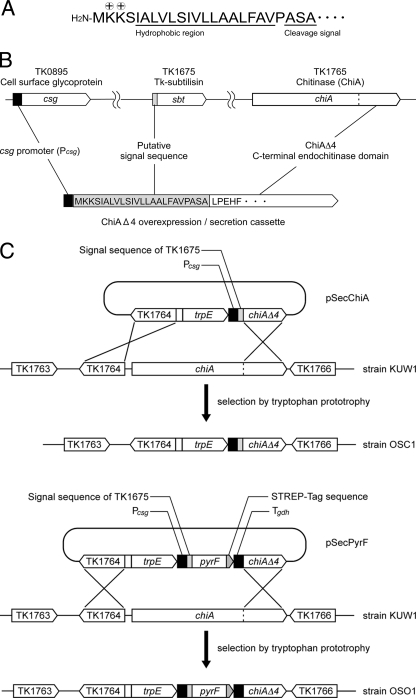
We first designed a gene cassette for overexpression and secretion of proteins in T. kodakarensis (Fig. 1). For use as a reporter protein, we selected the ChiAΔ4 protein, the C-terminal region (amino acid residues 911 to 1215) of chitinase (TK1765) from T. kodakarensis. The ChiAΔ4 protein displays endochitinase activity and is extremely thermostable, with a half-life of over 7 h at 100°C (34). We can also quantify protein levels by Western blot analysis using antibodies specific to ChiAΔ4. As a signal sequence for secretion, we selected the N-terminal region of Tk-subtilisin (14) (the product of TK1675) (Fig. 1A). The putative signal peptide region includes two basic Lys residues following the initial Met residue; a hydrophobic region consisting of 16 residues; and a putative cleavage signal, Ala-Ser-Ala. As for the promoter, we used the promoter of TK0895, annotated to encode a cell surface glycoprotein. Judging from DNA microarray experiments, the TK0895 transcript is abundant in T. kodakarensis cells under various growth conditions, suggesting that the promoter is strong and constitutive. The promoter has also been used to express the endogenous pantoate kinase gene (38) and the α-1,4-glucan phosphorylase gene from S. solfataricus (19) using T. kodakarensis as a host cell.
FIG. 1.
Strategies for constructing protein expression and secretion strains of T. kodakarensis. (A) Amino acid sequence of the putative secretion signal in the N-terminal region of TK1675. Basic residues following the initial Met residue are indicated with + symbols, and the hydrophobic region and the cleavage signal Ala-Ser-Ala are underlined. (B) Strategy for constructing the gene cassette for overexpression and secretion of ChiAΔ4. (C) Strategies for integrating the ChiAΔ4 and PyrF expression/secretion gene cassettes into the T. kodakarensis KUW1 genome with pSecChiA and pSecPyrF, respectively. Tgdh represents the 3′-flanking region of the glutamate dehydrogenase gene (TK1431), inserted to promote transcription termination.
A DNA fragment corresponding to the putative signal sequence of Tk-subtilisin was fused to the chiAΔ4 gene (Fig. 1B). The fusion gene was placed downstream of the cell surface glycoprotein gene promoter (csg promoter). This ChiAΔ4 expression/secretion cassette was inserted into a plasmid harboring the trpE gene used for transformant selection, resulting in the plasmid pSecChiA. As shown in Fig. 1C, pSecChiA was introduced into the chiA locus of T. kodakarensis KUW1 via homologous double-crossover recombination. T. kodakarensis KUW1 displays tryptophan and uracil auxotrophy, and transformants exhibiting tryptophan prototrophy were selected. The genotypes of 16 transformants were examined with PCR, and DNA sequencing was performed to confirm the occurrence of the expected recombination and the absence of unintended mutations. One strain was designated T. kodakarensis OSC1 and subjected to further analyses.
Secretion of ChiAΔ4 from T. kodakarensis OSC1.
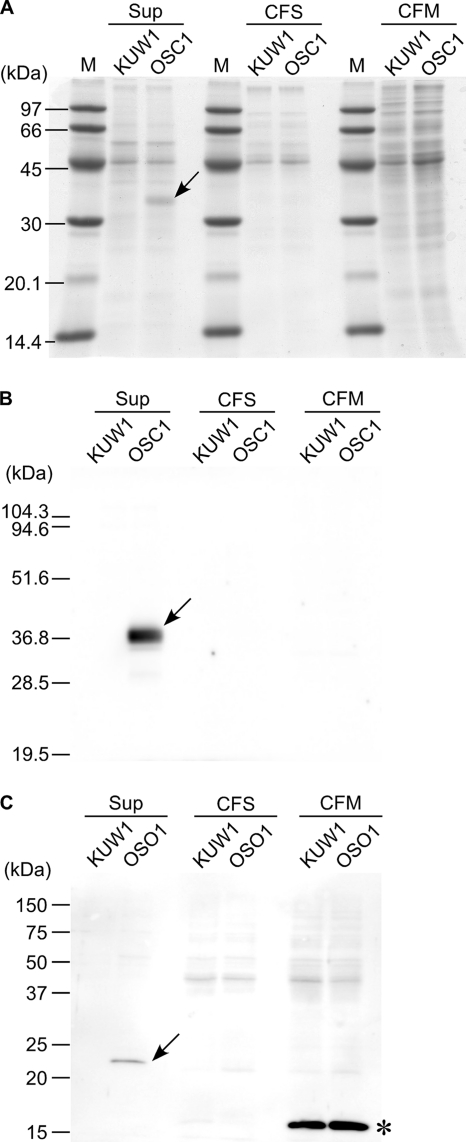
The host strain T. kodakarensis KUW1 and the isolated strain OSC1 were cultivated in nutrient-rich ASW-YT medium, and the cells and culture supernatant were separated. The cells were disrupted and further separated to obtain soluble and membrane fractions of the cell extracts. The three fractions were subjected to SDS-PAGE, with the amounts of proteins in each lane corresponding to those obtained from an equivalent volume (100 μl) of culture. After being stained with Coomassie brilliant blue, a band with a molecular mass corresponding to that of the ChiAΔ4 protein was specifically observed in the culture supernatant fraction of OSC1 cells (Fig. 2A). Using antibodies specific to ChiAΔ4, Western blot analysis was carried out in order to examine the ChiAΔ4 protein levels in further detail (Fig. 2B). We observed a strong signal in the culture supernatant of OSC1 cells corresponding to the band observed in Fig. 2A. Furthermore, we found that almost all of the ChiAΔ4 protein produced was secreted; no signals were detected in the soluble and membrane fractions of OSC1 cells.
FIG. 2.
Protein secretion from T. kodakarensis OSC1 and OSO1. Cultures of T. kodakarensis KUW1, OSC1, and OSO1 were separated into culture supernatant (SUP) and soluble (CFS) and membrane (CFM) fractions of the cell extracts as described in the text and subjected to SDS-PAGE. Each lane contained proteins obtained from an equivalent volume (100 μl) of culture. Bands specific to strains OSC1 and OSO1 are indicated by arrows. (A) Gels stained with Coomassie brilliant blue (KUW1 and OSC1). Lanes M contain molecular mass markers. (B) Western blot analysis using antisera against ChiAΔ4 (KUW1 and OSC1). (C) Western blot analysis with STREP-Tag II antibody HRP conjugate (KUW1 and OSO1). The band indicated with an asterisk is an endogenous protein in the membrane fraction that is recognized by the STREP-Tag II antibody.
With a similar strategy, we also attempted to secrete the cytosolic enzyme orotidine-5′-monophosphate decarboxylase, encoded by the pyrF gene (TK2276). The plasmid (pSecPyrF) was constructed so that the second amino acid residue of PyrF was connected to the signal sequence, and a STREP tag (Trp-Ser-His-Pro-Gln-Phe-Glu-Lys) sequence was fused at its C terminus (Fig. 1C). The obtained recombinant strain, T. kodakarensis OSO1, was examined for PyrF secretion. Using STREP-Tag II antibody HRP conjugate, we observed a signal in the culture supernatant of strain OSO1 that agreed with the calculated molecular mass of PyrF (24.2 kDa) (Fig. 2C). The signal was not observed in the host strain, KUW1, indicating that it was not due to an endogenous protein reacting with the STREP antibodies. As the signal was not observed in the soluble and membrane fractions of OSO1, this indicates that the PyrF protein was also efficiently secreted from the cells into the culture supernatant.
The results with ChiAΔ4 and PyrF indicate that the strategies described above are applicable for the efficient secretion of proteins from T. kodakarensis. We stress, however, that secretion of another cytosolic protein, β-glycosidase, the product of TK1761, was not successful. The protein was detected in the soluble fraction in the cells, but not in the culture supernatant (data not shown).
Examination of the secreted ChiAΔ4 protein.
We quantified the secreted ChiAΔ4 protein in the culture supernatants of OSC1 cells by Western blot analysis and enzyme assay. According to the results of Western blot analysis using purified ChiAΔ4 protein produced in E. coli as a standard, the amount of secreted ChiAΔ4 protein was estimated to be 2.1 mg liter culture−1. In order to examine chitinase activity, GlcNac2-4MU was used as a fluorescent substrate. Using the specific activity value of purified, recombinant ChiAΔ4 produced in E. coli, chitinase activity levels in the culture supernatant of OSC1 cells corresponded to a production level of 4.2 mg liter culture−1.
We also analyzed the N-terminal amino acid sequence of the secreted ChiAΔ4 protein. The culture supernatant of OSC1 cells was subjected to anion-exchange chromatography, and the band corresponding to ChiAΔ4 (33.8 kDa) was examined. We obtained the sequence Leu-Pro-Glu-His-Phe, which corresponds exactly to the N-terminal region of ChiAΔ4. This indicates that the signal peptide of TK1675 was cleaved specifically after the Ala-Ser-Ala sequence (Fig. 1B).
Application of the secretion system.
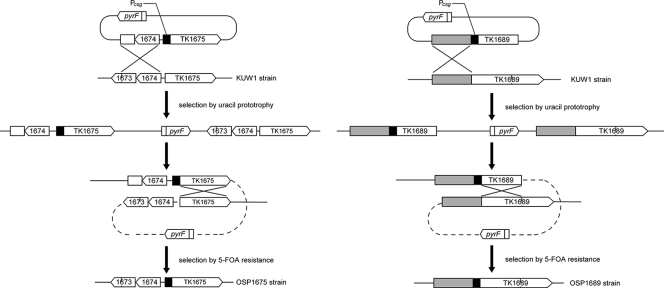
We applied the expression/secretion system and tried to enhance the protein degradation capacity of T. kodakarensis. Two genes encoding putative proteases with predicted signal sequences in their N-terminal regions were selected for overexpression and secretion (TK1675 and TK1689). The signal sequence of TK1675 has been verified to function in the experiments described above. We therefore only exchanged the native promoters of these two genes with the csg promoter to enhance expression levels. Gene manipulation for the insertion of the csg promoter was performed via single-crossover insertion and pop-out recombination (Fig. 3). By examining 16 colonies for each gene insertion, we were able to isolate promoter insertion strains for both TK1675 (1/16) and TK1689 (3/16). The two strains were designated T. kodakarensis OSP1675 and OSP1689, respectively.
FIG. 3.
Construction of protease overexpression strains of T. kodakarensis. Shown are strategies applied in the construction of T. kodakarensis OSP1675 and OSP1689 via single-crossover insertion/pop-out recombination.
Analysis of the protease oversecretion strain.
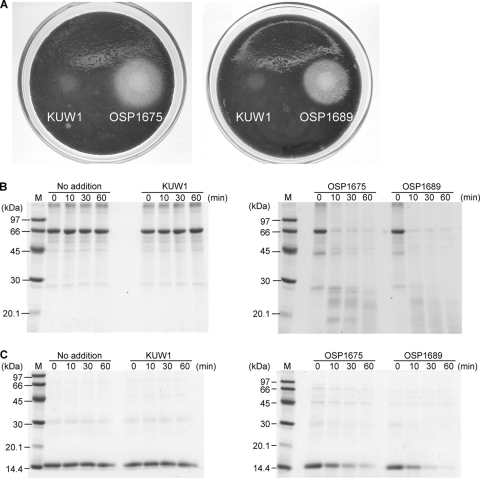
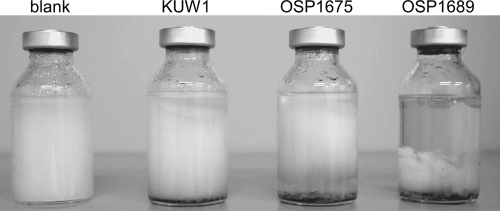
In order to examine whether the two recombinant strains OSP1675 and OSP1689 displayed enhanced protease activity due to the overexpression of TK1675 and TK1689, we first performed a plate assay using solid medium containing skim milk. Aliquots of OSP1675, OSP1689, and KUW1 cells were added to the plates and incubated at 85°C (Fig. 4A). As a result, halos brought about by protein degradation were observed only with the OSP1675 and OSP1689 cells, suggesting that the expression and secretion of proteases are enhanced in the two strains. We next grew the strains in liquid culture and examined their culture supernatants. SDS-PAGE analysis revealed significant differences in the protein bands when the culture supernatants of OSP1675, OSP1689, and KUW1 cells were compared. A large portion of high-molecular-weight proteins that were observed in the host strain cultures could not be observed in the cultures of OSP1675 and OSP1689 cells (data not shown). As a significant increase in low-molecular-weight proteins could also be observed, this is most likely a result of enhanced protein degradation activity in the culture supernatants of OSP1675 and OSP1689 cells.
FIG. 4.
Protein degradation by strains OSP1675 and OSP1689 in solidified and liquid media. (A) Formation of halos in solid media containing 3.5% skim milk. Cell suspensions were inoculated as described in the text and incubated at 85°C for 3 days. (B and C) Degradation of bovine serum albumin (B) and hemoglobin (C) with incubation in the culture supernatants of strains OSP1675 and OSP1689. Reaction mixtures (100 μl) contained culture supernatants from 40 μl culture and 30 μg of bovine serum albumin or hemoglobin in 50 mM Tris-HCl (pH 7.5). Incubation was carried out at 60°C for the indicated periods. Lanes M contain molecular mass markers.
In order to confirm whether the protein degradation activity resides in the culture supernatant, cells were removed and aliquots of the supernatant were added to a reaction mixture containing bovine serum albumin or hemoglobin. After incubation at 60°C for various periods, protein degradation was examined by SDS-PAGE (Fig. 4B and C). While no notable degradation of the two proteins was observed with the culture supernatant of KUW1 cells, those of OSP1675 and OSP1689 cells displayed significant protease activity. Activity measurements with the fluorescent peptide substrate (Suc)-Ala-Ala-Pro-Phe-MCA indicated that the hydrolytic activity levels in the supernatants of OSP1675 and OSP1689 cultures were 8.0 nmol min−1 ml culture−1 and 2.0 nmol min−1 ml culture−1, respectively. Activity in the supernatants of KUW1 culture was 0.2 nmol min−1 ml culture−1. We next examined the protein degradation capacities of the cells in a liquid culture supplemented with 5 g liter−1 ovalbumin. After inoculation of KUW1, OSP1675, and OSP1689 cells, the cultures were incubated at 85°C for 12 h. A major portion of the ovalbumin was degraded in the OSP1689 culture and, to a lesser extent, in the OSP1675 culture (Fig. 5). No notable degradation was observed in the KUW1 cell culture compared to a control experiment in which no cells were inoculated.
FIG. 5.
Protein degradation capacities of strains OSP1675 and OSP1689 in liquid medium. KUW1, OSP1675, and OSP1689 cells were inoculated in a liquid medium containing 0.8× ASW, 10 mg liter−1 of yeast extract, and 2.0 g liter−1 of elemental sulfur supplemented with 5 g liter−1 ovalbumin and incubated at 85°C for 12 h.
DISCUSSION
In this study, we examined the possibilities of utilizing T. kodakarensis cells as host cells for protein secretion. The use of the signal peptide from TK1675 led to efficient secretion and specific processing of the reporter proteins ChiAΔ4 and PyrF. Furthermore, the overexpression of two protease genes with putative signal sequences resulted in significant increases in protease activity in the culture supernatants, and the capacities of the transformants to degrade proteins were clearly higher than that of the host strain. However, a fifth protein, β-glycosidase, was not secreted with our strategy. At present, we cannot provide an explanation as to why one cytosolic protein (PyrF) is secreted while another (β-glycosidase) is not. Increasing the number of examinations with different proteins will be necessary to determine what governs the efficiency of protein secretion.
Although the efficiency of secretion seems to depend on the protein, our results suggest that T. kodakarensis can be used as a host cell to functionally produce and secrete proteins derived from hyperthermophiles. A present drawback that is now being addressed is the low cell yield of T. kodakarensis cells in batch cultures. This is the major reason why protein production levels per volume of culture were not high. The amount of protein produced per gram dry cell weight (DCW) of T. kodakarensis was ∼16 mg protein g DCW−1 with the ChiAΔ4 protein and is comparable to those of other microbial host cells. Protein production levels of 7 to over 150 mg protein g DCW−1 have been reported for E. coli (8, 15, 17), while 40 to over 140 mg protein g DCW−1 have been reported for Bacillus subtilis (36). With yeast host cells, 25 to 30 mg protein g DCW−1 have been reported for Saccharomyces cerevisiae (1, 7, 25), while 2 to 4 mg protein g DCW−1 have been reported for Pichia pastoris (6, 20). It is often the case that large differences in production levels are observed using the same host cell, presumably due to differences in promoters, culture conditions, and the protein being expressed.
Taking advantage of halo formation on casein-supplemented solid media, the protease secretion system developed in this study can also be expected to be useful as a tool for further biological studies of T. kodakarensis. Obviously, it can be applied directly as a reporter system for studies on the protein secretion mechanism in T. kodakarensis. By fusing the signal peptide/protease gene to a promoter of choice, one may also be able to carry out in vivo promoter analysis and identify cis-acting elements or to screen for transcriptional regulators.
This study displays an initial demonstration of cell engineering in hyperthermophiles, which is now very practical, considering the accumulation of genetic-manipulation techniques developed for T. kodakarensis and Sulfolobus species (2, 22, 37). Cell engineering and metabolic engineering of hyperthermophiles should open up new alternatives in various aspects of biotechnology, particularly in microbial whole-cell biocatalysis and fermentation. Besides the acceleration of the reaction rate, elevation of the reaction temperature will in many cases provide advantages in the solubility of substrates, such as poly- and oligosaccharides, and may also contribute to the downstream processing of volatile products, such as ethanol.
Supplementary Material
Acknowledgments
This work was supported by the Japan Society for the Promotion of Science under a Grant-in-Aid for Creative Scientific Research (no. 18GS0421) (to T.I.) and under a Grant-in-Aid for Scientific Research (no. 21350092) and a Grant-in-Aid for Scientific Research on Priority Areas “Applied Genomics” (no. 20018013) (to H.A.).
Footnotes
Published ahead of print on 28 January 2011.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Ahn, J. O., et al. 2004. Enhanced secretion of Bacillus stearothermophilus L1 lipase in Saccharomyces cerevisiae by translational fusion to cellulose-binding domain. Appl. Microbiol. Biotechnol. 64:833-839. [DOI] [PubMed] [Google Scholar]
- 2.Albers, S. V., and A. J. Driessen. 2008. Conditions for gene disruption by homologous recombination of exogenous DNA into the Sulfolobus solfataricus genome. Archaea 2:145-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Atomi, H. 2005. Recent progress towards the application of hyperthermophiles and their enzymes. Curr. Opin. Chem. Biol. 9:166-173. [DOI] [PubMed] [Google Scholar]
- 4.Atomi, H., T. Fukui, T. Kanai, M. Morikawa, and T. Imanaka. 2004. Description of Thermococcus kodakaraensis sp. nov., a well studied hyperthermophilic archaeon previously reported as Pyrococcus sp. KOD1. Archaea 1:263-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Atomi, H., R. Matsumi, and T. Imanaka. 2004. Reverse gyrase is not a prerequisite for hyperthermophilic life. J. Bacteriol. 186:4829-4833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bhatacharya, P., G. Pandey, and K. J. Mukherjee. 2007. Production and purification of recombinant human granulocyte-macrophage colony stimulating factor (GM-CSF) from high cell density cultures of Pichia pastoris. Bioprocess. Biosyst. Eng. 30:305-312. [DOI] [PubMed] [Google Scholar]
- 7.Brown, R., R. D. O'Kennedy, J. Helwigh, E. Madden, and M. Hoare. 2000. Accelerated prediction of recombinant protein production in Saccharomyces cerevisiae by using rapid monitoring techniques. Enzyme Microb. Technol. 26:801-807. [DOI] [PubMed] [Google Scholar]
- 8.Cheng, S., Q. Song, D. Wei, and B. Gao. 2007. High-level production of penicillin G acylase from Alcaligenes faecalis in recombinant Escherichia coli with optimization of carbon sources. Enzyme Microb. Technol. 41:326-330. [Google Scholar]
- 9.Chou, C. J., et al. 2007. Impact of substrate glycoside linkage and elemental sulfur on bioenergetics of and hydrogen production by the hyperthermophilic archaeon Pyrococcus furiosus. Appl. Environ. Microbiol. 73:6842-6853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Egorova, K., and G. Antranikian. 2005. Industrial relevance of thermophilic Archaea. Curr. Opin. Microbiol. 8:649-655. [DOI] [PubMed] [Google Scholar]
- 11.Forterre, P. 1996. A hot topic: the origin of hyperthermophiles. Cell 85:789-792. [DOI] [PubMed] [Google Scholar]
- 12.Fukui, T., et al. 2005. Complete genome sequence of the hyperthermophilic archaeon Thermococcus kodakaraensis KOD1 and comparison with Pyrococcus genomes. Genome Res. 15:352-363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Imanaka, H., A. Yamatsu, T. Fukui, H. Atomi, and T. Imanaka. 2006. Phosphoenolpyruvate synthase plays an essential role for glycolysis in the modified Embden-Meyerhof pathway in Thermococcus kodakaraensis. Mol. Microbiol. 61:898-909. [DOI] [PubMed] [Google Scholar]
- 14.Kannan, Y., et al. 2001. Active subtilisin-like protease from a hyperthermophilic archaeon in a form with a putative prosequence. Appl. Environ. Microbiol. 67:2445-2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kasche, V., et al. 2005. Ca2+ is a cofactor required for membrane transport and maturation and is a yield-determining factor in high cell density penicillin amidase production. Biotechnol. Prog. 21:432-438. [DOI] [PubMed] [Google Scholar]
- 16.Matsumi, R., K. Manabe, T. Fukui, H. Atomi, and T. Imanaka. 2007. Disruption of a sugar transporter gene cluster in a hyperthermophilic archaeon using a host-marker system based on antibiotic resistance. J. Bacteriol. 189:2683-2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mergulhão, F. J. M., M. A. Taipa, J. M. S. Cabral, and G. A. Monteiro. 2004. Evaluation of bottlenecks in proinsulin secretion by Escherichia coli. J. Biotechnol. 109:31-43. [DOI] [PubMed] [Google Scholar]
- 18.Morikawa, M., Y. Izawa, N. Rashid, T. Hoaki, and T. Imanaka. 1994. Purification and characterization of a thermostable thiol protease from a newly isolated hyperthermophilic Pyrococcus sp. Appl. Environ. Microbiol. 60:4559-4566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mueller, M., R. Takemasa, A. Schwarz, H. Atomi, and B. Nidetzky. 2009. “Short-chain” alpha-1,4-glucan phosphorylase having a truncated N-terminal domain: functional expression and characterization of the enzyme from Sulfolobus solfataricus. Biochim. Biophys. Acta 1794:1709-1714. [DOI] [PubMed] [Google Scholar]
- 20.Murasugi, A., Y. Tohma-Aiba, and Y. Asami. 2000. Production of recombinant human midkine in yeast, Pichia pastoris. J. Biosci. Bioeng. 90:395-399. [DOI] [PubMed] [Google Scholar]
- 21.Orita, I., et al. 2006. The ribulose monophosphate pathway substitutes for the missing pentose phosphate pathway in the archaeon Thermococcus kodakaraensis. J. Bacteriol. 188:4698-4704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peeters, E., S. V. Albers, A. Vassart, A. J. Driessen, and D. Charlier. 2009. Ss-LrpB, a transcriptional regulator from Sulfolobus solfataricus, regulates a gene cluster with a pyruvate ferredoxin oxidoreductase-encoding operon and permease genes. Mol. Microbiol. 71:972-988. [DOI] [PubMed] [Google Scholar]
- 23.Rinker, K. D., and R. M. Kelly. 2000. Effect of carbon and nitrogen sources on growth dynamics and exopolysaccharide production for the hyperthermophilic archaeon Thermococcus litoralis and bacterium Thermotoga maritima. Biotechnol. Bioeng. 69:537-547. [DOI] [PubMed] [Google Scholar]
- 24.Robb, F. T., and A. R. Place. 1995. Media for thermophiles, p. 167-168. In F. T. Robb and A. R. Place (ed.), Archaea: a laboratory manual-thermophiles. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 25.Sagt, C. M., W. H. Muller, J. Boonstra, A. J. Verkleij, and C. T. Verrips. 1998. Impaired secretion of a hydrophobic cutinase by Saccharomyces cerevisiae correlates with an increased association with immunoglobulin heavy-chain binding protein (BiP). Appl. Environ. Microbiol. 64:316-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Santangelo, T. J., L. Cubonova, C. L. James, and J. N. Reeve. 2007. TFB1 or TFB2 is sufficient for Thermococcus kodakaraensis viability and for basal transcription in vitro. J. Mol. Biol. 367:344-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Santangelo, T. J., et al. 2008. Polarity in archaeal operon transcription in Thermococcus kodakaraensis. J. Bacteriol. 190:2244-2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Santangelo, T. J., L. Cubonova, and J. N. Reeve. 2010. Thermococcus kodakarensis genetics: TK1827-encoded b-glycosidase, new positive-selection protocol, and targeted and repetitive deletion technology. Appl. Environ. Microbiol. 76:1044-1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Santangelo, T. J., L. Cubonova, K. M. Skinner, and J. N. Reeve. 2009. Archaeal intrinsic transcription termination in vivo. J. Bacteriol. 191:7102-7108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sato, T., T. Fukui, H. Atomi, and T. Imanaka. 2005. Improved and versatile transformation system allowing multiple genetic manipulations of the hyperthermophilic archaeon Thermococcus kodakaraensis. Appl. Environ. Microbiol. 71:3889-3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sato, T., T. Fukui, H. Atomi, and T. Imanaka. 2003. Targeted gene disruption by homologous recombination in the hyperthermophilic archaeon Thermococcus kodakaraensis KOD1. J. Bacteriol. 185:210-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sato, T., et al. 2004. Genetic evidence identifying the true gluconeogenic fructose-1,6-bisphosphatase in Thermococcus kodakaraensis and other hyperthermophiles. J. Bacteriol. 186:5799-5807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stetter, K. O. 1996. Hyperthermophilic procaryotes. FEMS Microbiol. Rev. 18:149-158. [Google Scholar]
- 34.Tanaka, T., et al. 1999. A unique chitinase with dual active sites and triple substrate binding sites from the hyperthermophilic archaeon Pyrococcus kodakaraensis KOD1. Appl. Environ. Microbiol. 65:5338-5344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vieille, C., and G. J. Zeikus. 2001. Hyperthermophilic enzymes: sources, uses, and molecular mechanisms for thermostability. Microbiol. Mol. Biol. Rev. 65:1-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vuolanto, A., N. von Weymarn, J. Kerovuo, H. Ojamo, and M. Leisola. 2001. Phytase production by high cell density culture of recombinant Bacillus subtilis. Biotechnol. Lett. 23:761-766. [Google Scholar]
- 37.Worthington, P., V. Hoang, F. Perez-Pomares, and P. Blum. 2003. Targeted disruption of the a-amylase gene in the hyperthermophilic archaeon Sulfolobus solfataricus. J. Bacteriol. 185:482-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yokooji, Y., H. Tomita, H. Atomi, and T. Imanaka. 2009. Pantoate kinase and phosphopantothenate synthetase, two novel enzymes necessary for CoA biosynthesis in the Archaea. J. Biol. Chem. 284:28137-28145. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.