Abstract
Virulence-associated genes in bacteria are often located on chromosomal regions, termed pathogenicity islands (PAIs). Several PAIs are found in Escherichia coli strains that cause extraintestinal infections, but their role in commensal bowel colonization is unknown. Resident strains are enriched in adhesins (P fimbriae and type 1 fimbriae), capsular antigens (K1 and K5), hemolysin, and aerobactin and mostly belong to phylogenetic group B2. Here, we investigated whether six pathogenicity islands and the virulence determinants malX and usp are associated with fitness of E. coli in the infant bowel microbiota. E. coli strains isolated from stools of 130 Swedish infants during the first year of life were examined for their carriage of PAI markers, malX, and usp by PCR. Carriage was related to strain persistence: long-term colonizers (≥12 months) carried significantly more of PAI II from strain CFT703 (IICFT703), IV536, and IIJ96 and malX and usp than intermediate colonizers (1 to 11 months) and transient strains (<3 weeks). The accumulation of PAI markers in each individual strain correlated positively with its time of persistence in the colon. Phylogenetic group B2 accounted for 69% of long-term colonizers, 46% of intermediate colonizers and 14% of transient strains. These results support the hypothesis that some bacterial traits contributing to extraintestinal infections have in fact evolved primarily because they increase the fitness of E. coli in its natural niche, the colon; accordingly, they may be regarded as fitness islands in the gut.
Escherichia coli is a normal inhabitant of the large intestine, but certain strains also cause extraintestinal infections when they spread from their primary niche. Such pathogenic E. coli strains express several virulence-associated traits that contribute to the disease process, including adhesins, certain O serotypes and capsular antigens, iron-trapping compounds (such as aerobactin), and cytolytic toxins (such as hemolysin) (17).
Pathogenicity-associated islands (PAIs) are particular regions on the bacterial chromosome where virulence genes have accumulated. PAIs, and their associated virulence genes, have spread among bacterial populations by horizontal transfer (15). Several PAIs were previously identified in uropathogenic E. coli strains such as E. coli 536, E. coli J96, and E. coli CFT073. PAIs I to IV from strain 536 (I536 to IV536) encode a range of virulence factors, including P fimbriae, P-related fimbriae, α-hemolysin, S fimbriae, and the yersiniabactin siderophore system. PAI IJ96 and IIJ96 encode P fimbriae, P-related fimbriae, and α-hemolysin; PAI ICFT073 and IICFT073 encode P fimbriae, α-hemolysin, and aerobactin (42) (Table 1). usp is prevalent among strains causing urinary tract infections (24). It enhances infectivity in a mouse ascending urinary tract infection (UTI) model (48), displays homology with S-type pyocin, and may function as a bacteriocin (34). malX codes for a phosphotransferase system enzyme II that recognizes maltose and glucose (37) and is enriched in strains causing extraintestinal infections (18, 19, 41).
TABLE 1.
The pathogenicity islands and the functions encoded
| Pathogenicity island or gene | Product(s) | Reference(s) |
|---|---|---|
| Pathogenicity islands | ||
| IICFT073 | P fimbriae, iron-regulated proteins | 36 |
| I536 | α-Hemolysin, F17-like fimbriae, CS12-like fimbriae | 12 |
| II536 | Hek adhesin, P-related fimbriae, α-hemolysin, hemagglutinin-like adhesin | 12 |
| III536 | S fimbriae, salmochelin, HmuR-like heme receptor, Sat toxin, Tsh-like hemoglobin protease, antigen 43 | 12 |
| IV536 | Yersiniabactin siderophore system | 12 |
| IIJ96 | α-Hemolysin, Prs fimbriae, cytotoxic, necrotizing factor | 3, 6 |
| Virulence-associated genes | ||
| malX | Maltose- and glucose-specific component IIa of a phosphoenolpyruvate-dependent phosphotransferase system | 14, 38 |
| usp | Putative bacteriocin | 24 |
E. coli segregate into four main phylogenetic groups, termed A, B1, B2, and D (16). Strains of group A and B1 seldom cause extraintestinal infections and carry few virulence factor genes (4, 7). Extraintestinal pathogenic E. coli strains belong mostly to group B2 (4, 18) and to a lesser extent to group D and usually carry several PAIs (5, 33, 40).
Intestinal E. coli strains differ widely in colonization capacity; resident strains have the ability to persist in the gut microbiota, while transient strains disappear from the microbiota within a short time (44-46). Previously, in several epidemiological studies, we demonstrated that genes for certain virulence-associated factors, such as P fimbriae, type 1 fimbriae, hemolysin, capsular polysaccharides (K1 and K5), and aerobactin, are significantly enriched in resident strains (27-29). Furthermore, strains of B2 origin have an enhanced ability to persist in the intestinal microbiotas of infants (32).
The present study was designed to investigate whether several markers for pathogenicity islands and the virulence-associated genes malX and usp are associated with an increased capacity of E. coli to persist in the infantile intestinal microbiota. In regard to this, 273 E. coli strains obtained from 130 healthy Swedish infants, monitored longitudinally over the first year of life, were assessed for carriage of markers for PAIs I536, II536, III536, IV536, IICFT073, and IIJ96 and for malX and usp. Carriage of PAI-associated markers was compared with a strain's persistence.
MATERIALS AND METHODS
E. coli strains.
The source population comprised 130 healthy Swedish infants born in 1998 to 2001 at the Sahlgrenska University Hospital, Göteborg, Sweden. They were part of a prospective birth cohort study examining the relation between the intestinal colonization pattern and allergy development, the ALLERGYFLORA study (1, 2). E. coli strains (n = 149) from 70 of these were previously described with regard to E. coli colonization pattern, association between certain virulence factor genes, phylogenetic group distribution, and persistence (28, 30). Here in a larger infant cohort, carriage of six PAI markers, including PAI I536, II536, III536, IV536, IIJ96, and IICFT073, as well as malX and usp, in the intestinal E. coli strains was identified. Phylogenetic group distribution was further determined.
E. coli strains were isolated and quantified in stools as previously described (1). In brief, a rectal swab was obtained at 3 days of age and cultured semiquantitatively under aerobic condition for facultative bacteria. Fecal samples were collected at 1, 2, 4, and 8 weeks and at 6 and 12 months of age. They were diluted serially and cultured on Drigalski's agar for isolation of Enterobacteriaceae. One to six colonies with different morphologies were isolated from each positive culture and enumerated, subcultured, identified to species level by using biotyping (API20E; API Systems SA, La Balme les Grottes, Montalieu-Vercieu, France), and stored at −70°C. CFU with the same morphology within each sample were regarded as the same strain. After species identification, E. coli isolates were analyzed by random amplified polymorphic DNA (RAPD) profiling. Isolates from a particular infant that exhibited RAPD patterns differing by no more than three weak bands were considered to be the same strain (28).
The strains were divided into three groups according to the time they persisted in the host: strains colonizing the microbiota for <3 weeks were termed transient strains, strains colonizing for 1 to 11 months were termed intermediate colonizers, and those colonizing for ≥12 months were termed long-term colonizers. Strains appearing only once in the last sample or in samples with long sampling intervals could not be classified according to these definitions and therefore were included only in characterization of the infant E. coli microbiota.
Detection of markers for pathogenicity islands and assignment to phylogenetic groups by PCR.
Phylogenetic group distribution was determined using triplex PCR as described by Clermont et al. (8) with slight modifications. Bacterial DNA was added to a PCR mixture containing HotStarTaq master mix (Qiagen, Spånga, Sweden) and 20 pmol of each primer pair amplifying the genes chuA and yjaA and the DNA fragment TspE4.C2.
All primers used for identification of the screened PAI markers have already been described (39). They were selected from PAI-associated sequences (11, 20, 23, 25), and their similarity to E. coli and specificity for the PAI sequences have also been screened using BLASTN (39).
To extract genomic DNA, a small amount of bacteria was suspended in Tris-EDTA buffer (Sigma-Aldrich Sweden AB, Stockholm, Sweden) and heated at 95°C for 10 min. The mixture was centrifuged for 5 min at room temperature, and supernatants were used for PCRs. The DNA extraction method was identical in all PCR assays.
PAI IV536 (high-pathogenicity island) was identified as previously described (9). The presence of malX was confirmed by a PCR assay described elsewhere (31) using the previously published primers (20).
PAI I and II in E. coli 536 and PAI IIJ96 were identified by a duplex and a single PCR, respectively. The following PCR procedure was used for both assays: 1.2 U Expand high-fidelity Taq DNA polymerase (Roche Diagnostics, Bromma, Sweden), 0.5 mM deoxynucleoside triphosphate s (dNTPs), 3 mM MgCl2, 0.5 mM each primer (primers 1.9, 1.10, orf1 up, and orf1 down for duplex PCR and primers hlyd and cnf for single PCR) in a final volume of 25 μl. The PCR program was 95°C for 5 min and then 30 cycles of 94°C for 1 min, 57°C for 1 min, and 72°C for 3 min, with a final extension step at 72°C for 10 min.
The presence of PAI IICFT073 and PAI III536 were verified in a duplex PCR using 12.5 μl HotStarTaq master mix (Qiagen, Spånga, Sweden), 0.1 μM primers cft073.2Ent1 and cft073.2Ent2, 0.5 μM primers sfaAI.1 and sfaAI.2, and 2.0 mM MgCl2, in a final volume of 25 μl. The PCR program was 95°C for 15 min and then 30 cycles of 94°C for 1 min, 55°C for 1 min, and 72°C for 1 min, with a final extension step at 72°C for 10 min. The PCR amplicons were separated on agarose gels and stained with ethidium bromide.
We screened all strains for the presence of the papG allele I, which is known as a marker for PAI IJ96 (39), using a PCR assay described elsewhere (27). However, we found no papGI-positive strain, and we therefore excluded this PAI in the present study.
A real-time PCR was developed to detect usp. Primers and a probe were designed using Primer Express 3.0 software (Applied Biosystems, Stockholm, Sweden) based on the sequence for usp in E. coli strain Z42 as retrieved from DDBJ, accession number AB056434 (26). The primers were USPf (5′-CGGCGCATATTGCTGATAAAT-3′) and USPr (5′-CGCCTCGCGAAACTCATC-3′), and the probe was 5′-ACGGGAGTTTAAAACC-3′. The PCR was performed by adding 2.5 μl of bacterial lysate to a mixture containing 12.5 μl TaqMan universal PCR master mix (Applied Biosystems) and 0.2 μM each primer and probe in a final volume of 25 μl. The PCR was carried out using Applied Biosystems 7500 PCR systems (Applied Biosystems, Stockholm, Sweden). The PCR program was 95°C for 10 min, followed by 95°C for 15 s and 60°C for 1 min for 35 cycles.
To verify the real-time PCR assay, 100 strains were assessed using the conventional primers described by Nakano et al. (26). There was a 99% accordance.
Strains CFT073, 536, J96, Z42, Z13, and C10 (26) served as positive controls for PAI markers.
Virulence score representing carriage of PAI markers and malX and usp.
Each strain was assigned a score (from 0 to 8) corresponding to the number of PAI markers, malX, and usp. This score was used as an X variable (an independent variable) in the multivariate models.
Statistical methods.
Proportions were compared using Fisher's exact test. The prevalence of each individual PAI marker, malX, and usp in transient strains, intermediate colonizers, and long-term colonizers was first analyzed in a 2-by-3 table. The bacterial determinants that showed a significant nonrandom distribution (P < 0.05) were further compared between each group in a series of 2-by-2 tables. The virulence score was compared by the Mann-Whitney U test. Multivariate data analysis was performed using Simca-P + 12.0 (Umetrics AB, Umeå, Sweden). Projections to latent structures by means of partial least square (PLS) were used to assess the relationship between carriage of the PAI markers, usp, and malX and persistence of E. coli strains in the gut flora. This method is a regression development of principal component analysis (PCA) in which clustering data are related to a Y variable, in this case, persistence in the gut flora, that was generated by a score setting 0 for transient strains, 1 for intermediate colonizers, and 2 for long-term colonizers. The studied PAI markers, malX, usp, and phylogenetic group origin were modeled as X variables (independent variables) and persistence as the Y variable (dependent variable). Analyses were made using the default settings in SIMCA-P+ 12.0.
Logistic regression analysis, using stepwise selection, was performed to predict which trait was independently associated with persistence.
RESULTS
Prevalence of PAI markers, malX, and usp among E. coli strains from the infantile intestinal microbiota.
Overall, 273 distinct fecal E. coli strains were identified in samples from the 130 Swedish infants monitored from 3 days to 1 year of age. The median number of E. coli strains per infant was 2 (range 0 to 6). Of these, 60% carried at least one of the investigated bacterial traits. The prevalence of the PAI markers, malX, and usp is shown in Table 2. Markers for PAI IV536 were the most abundant, while markers for PAI I536, II536, and III536 were the least frequent. The carriage rates of malX, usp, and PAI IICFT073 were roughly similar.
TABLE 2.
Prevalence of PAI markers, malX, and usp in the intestinal E. coli strains of healthy Swedish infantsa
| Marker or gene | % positive strains |
|
|---|---|---|
| All strains (n = 273) | Strains defined by persistence (n = 100) | |
| PAI markers | ||
| IV536 | 57 | 62 |
| IICFT073 | 34 | 37 |
| I536 | 18 | 14 |
| IIJ96 | 17 | 14 |
| II536 | 12 | 8 |
| III536 | 3 | 4 |
| Virulence-associated genes | ||
| malX | 44 | 46 |
| usp | 45 | 52 |
A total of 273 commensal E. coli strains obtained from 130 Swedish infants during the first year of life were assessed for carriage of six PAI markers, malX, and usp. Of these, 100 were defined according to the time they persisted in the infantile gut.
Presence of PAI markers, malX, and usp in relation to persistence of E. coli strains in the gut microbiota.
The time of persistence was determined for 100 of the 273 strains. It was not possible to classify strains (n = 173) that appeared only once in the last sample or in a sample with a long sampling interval as long-term colonizers, intermediate colonizers, or transient strains. A total of 32 E. coli strains were classified as long-term colonizers (>12 months), 47 as intermediate colonizers (1 to 11 months), and 21 as transient strains (<3 weeks).
Each marker showed a clear tendency to increase in frequency with the time the strains persisted in the microbiota (Fig. 1). There was a significant association between persistence and markers for PAI IV536, PAI IICFT073, PAI IIJ96, malX, and usp. Indeed, they were considerably enriched in long-term colonizers compared to intermediate or transient strains (Fig. 1). No significant differences regarding markers for PAI I536, II536, and III536 were observed.
FIG. 1.
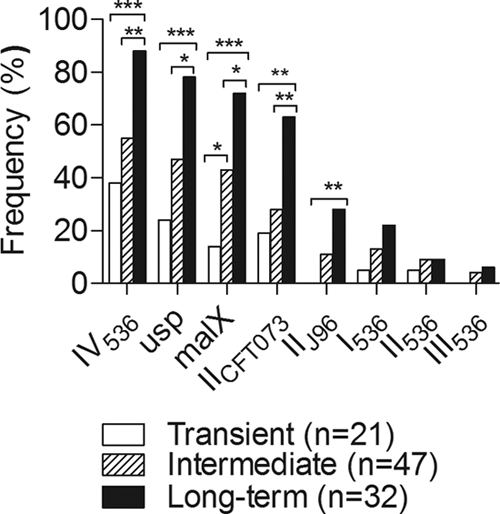
Prevalence of the PAI markers I536, II536, III536, IV536, IICFT073, and IIJ96 and the virulence determinants malX and usp in long-term-colonizer (n = 32), intermediate-colonizer (n = 47), and transient (n = 21) strains. Proportions were compared between groups using Fisher's exact test; asterisks indicate the level of significance: *, P < 0.05; **, P < 0.01; and ***, P < 0.001.
The relationship between virulence score and persistence is shown in Fig. 2. There was a dose-response relationship between the number of PAI markers, malX, and usp and persistence. Long-term colonizers had on average four virulence traits, compared with one trait in intermediate colonizers and no traits in transient strains (P = 0.0025 and P < 0.0001, respectively).
FIG. 2.
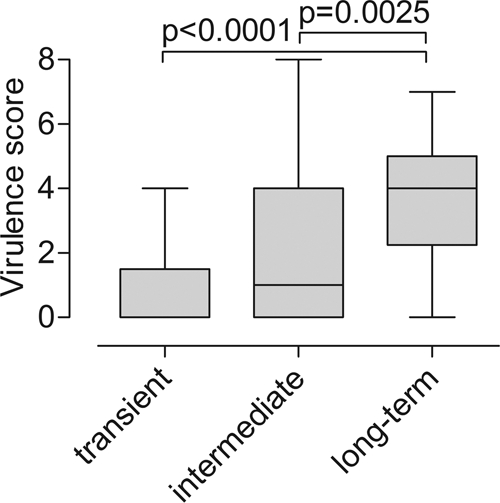
Virulence score (number of PAI markers or virulence determinants) in long-term-colonizer, intermediate-colonizer, and transient strains in relation to the time of persistence of E. coli strains in the microbiotas of Swedish infants. The median values were compared using the Mann-Whitney U test.
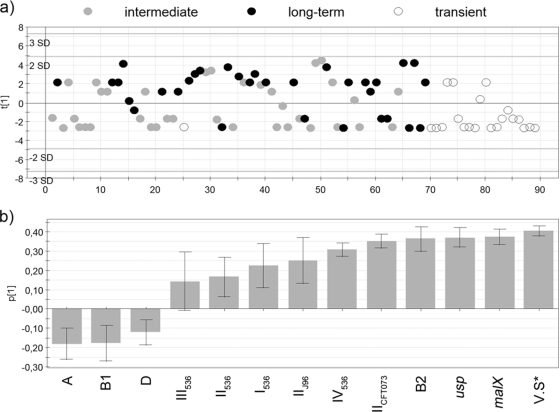
PLS regression analysis was used to investigate the relative strength of the associations of the PAI markers, malX, and usp and phylogenetic group with colonic persistence. The PLS analysis yielded a one-component model that revealed the clustering of E. coli strains based on these bacterial traits and phylogenetic group (Fig. 3a). Long-term colonizers were mainly positioned on the positive axis of the component, while transient strains were most often found on the negative axis of the component. In this model, intermediate colonizers were scattered across both axes.
FIG. 3.
Projections to latent structures by means of partial least square (PLS) was used to relate two data matrices, i.e., the X variables, including the virulence score and phylogenetic group, and the responder Y variable (persistence) to each other by a linear multivariate model. The model resulted in one component, which is on the y axis. (a) The score plot shows the position of observations (strains) along the principal component (y axis). Long-term colonizers (black dots) were mainly gathered on the positive axis and transient strains (white dots) on the negative axis. Intermediate colonizers (gray dots) appeared at either side. (b) Loadings, i.e., the variables generating the observed separation. PAI markers and virulence determinants, phylogenetic group, and the virulence score (V.S*) are included.
The bacterial determinants that generated the separation are shown in Fig. 3b. According to the model, variables located on the right side of diagram (with positive values on the y axis) were associated with long-term colonizers, while traits enriched in transient strains appear on the left (with negative values on the y axis). The taller the bar, the greater the trait's estimated contribution to persistence, and the smaller the error bar, the more reliable the estimate. The characteristics virulence score, malX, usp, group B2 origin, PAI IICFT073, PAI IV536, and PAI IIJ96 were obviously associated with persistence, while markers for PAI I536, II536, and III536 were less associated with persistence (Fig. 3b). Phylogenetic group origins A, B1, and D were linked to transient strains.
In addition, we used a logistic regression analysis with stepwise selection; in this model, transient strains and long term colonizers were included. Here we entered the screened PAI markers, malX, usp, and phylogenetic group B2 as independent variables and persistence as the dependent variable. malX was the only variable that yielded significance in this model (odds ratio, 16.2; 95% confidence interval, 3.8 to 68; P < 0.000).
Phylogenetic group in relation to PAI markers, malX, usp, and persistence.
Of the 273 distinct fecal strains, approximately half (45%) belonged to phylogenetic group B2, followed by groups A (28%), D (15%), and B1 (12%). The phylogenetic distribution of the 100 strains characterized regarding time of persistence were similar to that for the whole collection: B2, 46%; A, 23%; D, 18%; and B1, 13%.
Phylogenetic group B2 strains were significantly more common in long-term colonizers than intermediate colonizers (69% versus 46%; P = 0.01), and transient strains (69% versus 14%; P < 0.0001). In contrast, 32% of transient strains belonged to group B1, compared to 7% of intermediate strains (P = 0.01) and 9% of long-term colonizers (not significant).
The presence of PAI markers, malX, and usp varied by phylogenetic group. Group B2 strains were much more likely to carry each of the 8 investigated traits than all other strains (P = 0.002 for PAI III536; P < 0.0001 for all other markers). The only trait which was common in other phylogenetic groups was PAI IV536 (6% of B1, 25% of A, and 49% of D strains compared to 91% of B2 strains). malX was also relatively frequent among group D strains (17%).
DISCUSSION
Pathogenicity islands (PAIs) are enriched among E. coli strains causing extraintestinal infections, and extraintestinal pathogenic E. coli (ExPEC) strains mostly belong to phylogenetic groups B2 and D. Here, we examined whether carriage of six PAI markers, malX, and usp was associated with the capacity of E. coli strains to persist in the intestinal microbiota of 130 Swedish infants. The infants were monitored over the first year of life with regular quantitative cultures of stool samples. Individual E. coli strains were identified and characterized with respect to carriage of PAI markers, malX, and usp, phylogenetic group origin in each stool sample, and duration of persistence in the microbiota.
The most direct method of detecting PAIs is to determine the physical location of genetic elements representative of specific PAIs (5). Hence, the combination of pulsed-field gel electrophoresis analysis of macrorestricted genomic DNA and Southern hybridization is a reliable method. However, in the present study, for efficiency we used several PCR assays for detection of six PAI markers, malX, and usp.
We found that more than half of the intestinal commensal E. coli strains presumptively contained at least one of the studied bacterial determinants, including I536, II536, III536, IV536, IIJ96, I, malX, and usp, and that each of these was found in a higher frequency than generally reported among commensal strains (22, 24, 39, 43). The high prevalence of PAI markers is likely to reflect the dominance of phylogenetic group B2 strains in the intestinal microbiota of the Swedish infants examined here (32).
There was no significant association between carriage of PAIs I536, II536, and III536 and persistence. However, none of the transient strains carried markers for PAI III536, and few carried markers for PAIs I536 and II536. This indicates that PAIs I536, II536, and III536 possibly have an impact on persistence, although their low prevalence here as a single marker precluded statistical significance. In fact, 96% of PAIs I536, II536, and III536 appear with other common markers in combination, i.e., mostly together with 5 to 7 markers. Only two strains carried PAIs I536, II536, and III536 as single markers. Notably, acquisition of PAIs is not a random event (13), and here the rarest PAI markers were present only in strains already carrying the more common markers.
PLS analysis demonstrated the internal order of hierarchy in the association between accumulation of pathogenicity traits and persistence. The variable representing the virulence score exhibited the strongest association with persistence. Indeed, the greater the virulence score, the longer the duration of persistence. One may speculate that pathogenicity markers have an additive effect on intestinal colonization. It might be due to genes carried by different pathogenicity islands having an additive and/or synergistic effect. Or it may be that PAIs tend to occur in greater numbers in strains that have other unknown factors which promotes intestinal colonization. In support of the first interpretation, a recent experimental study by Diard et al. demonstrated an additive influence of seven PAIs (I536 to VII536) on persistence of E. coli in the intestines of mice. No single PAI showed a significant effect on intestinal colonization (10). Similarly, an additive function of the same seven PAIs for stabilizing extraintestinal virulence has been observed in a mouse septicemia model (47).
Many of the investigated markers probably do contribute to persistence, but as known virulence associated genes appear together and this multicollinearity leads to reduced precision in the prediction (35), it is difficult to show using logistic regression. Indeed, malX was the only marker that yielded significance, which is in accordance with the PLS results.
The association of phylogenetic group B2 and several extraintestinal virulence genes with persistence has previously been documented among E. coli strains from 70 of the 130 infants examined here (28, 32).
Our previous observations suggest that several well-known bacterial traits in E. coli, such as P fimbriae, type 1 fimbriae, aerobactin, capsular antigens (K1 and K5 capsule) and hemolysin, are associated with persistence of E. coli in its natural ecological niche, the large intestine (27-29).
Notably, hylA and papC may also be carried by PAI IIJ96, PAI I536, PAI II536, and PAI IICFT073, respectively (3, 6, 12, 36); these PAIs were associated with persistence in the present study. We also showed previously that resident E. coli strains possessed siderophores such as aerobactin (27, 29) and yersiniabactin (27). Interestingly, PAI IV536 codes for yersiniabactin, and a variety of other siderophore systems are also located on several PAIs (3, 6, 12, 36). The virulence determinants malX, also found on PAI ICFT073 (14), and usp are slightly associated with the B2 phylogenetic group (20, 21); strains of this group are, in general, excellent intestinal colonizers. One may speculate that the superior capacity of B2 strains to colonize the human gut is due partly to their accumulation of virulence factor genes, many of which are encoded by PAIs.
In summary, we have confirmed association between additional factors, i.e., six PAI markers, malX, and usp, with persistence of E. coli strains in the intestine in a large infant cohort. Further, this shows that the studied pathogenicity traits exhibit an accumulation effect promoting persistence of group B2 strains in the intestinal microbiota.
Taken together, our findings support the hypothesis that B2 strains may have evolved many of their so-called pathogenic characteristics in order to survive in the complex ecosystem in the human colon, with no reference to virulence. These pathogenicity markers likely cooperate in an additive manner for promoting the intestinal persistence of E. coli strains; thus, they may be regarded as “fitness islands.”
Acknowledgments
We thank James R. Johnson (Veterans Affairs Medical Centre and University of Minnesota, Minneapolis, MN) for critical manuscript review and S. Yamamoto (Department of Medical Technology, School of Health Sciences, Okayama University, Okayama, Japan) for provision of the E. coli PCR control strains.
The study was supported by grants from the Swedish Medical Research Council, the medical faculty of the University of Gothenburg (LUA/ALF grant), and the Wilhelm and Martina Lundgrens Foundation.
There is no conflict of interest.
Footnotes
Published ahead of print on 11 February 2011.
REFERENCES
- 1.Adlerberth, I., et al. 2006. Reduced enterobacterial and increased staphylococcal colonization of the infantile bowel: an effect of hygienic lifestyle? Pediatr. Res. 59:96-101. [DOI] [PubMed] [Google Scholar]
- 2.Adlerberth, I., et al. 2007. Gut microbiota and development of atopic eczema in 3 European birth cohorts. J. Allergy Clin. Immunol. 120:343-350. [DOI] [PubMed] [Google Scholar]
- 3.Bidet, P., et al. 2005. Multiple insertional events, restricted by the genetic background, have led to acquisition of pathogenicity island IIJ96-like domains among Escherichia coli strains of different clinical origins. Infect. Immun. 73:4081-4087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bingen, E., et al. 1998. Phylogenetic analysis of Escherichia coli strains causing neonatal meningitis suggests horizontal gene transfer from a predominant pool of highly virulent B2 group strains. J. Infect. Dis. 177:642-650. [DOI] [PubMed] [Google Scholar]
- 5.Bingen-Bidois, M., et al. 2002. Phylogenetic analysis and prevalence of urosepsis strains of Escherichia coli bearing pathogenicity island-like domains. Infect. Immun. 70:3216-3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blum, G., V. Falbo, A. Caprioli, and J. Hacker. 1995. Gene clusters encoding the cytotoxic necrotizing factor type 1, Prs-fimbriae and alpha-hemolysin form the pathogenicity island II of the uropathogenic Escherichia coli strain J96. FEMS Microbiol. Lett. 126:189-195. [DOI] [PubMed] [Google Scholar]
- 7.Boyd, E. F., and D. L. Hartl. 1998. Chromosomal regions specific to pathogenic isolates of Escherichia coli have a phylogenetically clustered distribution. J. Bacteriol. 180:1159-1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clermont, O., S. Bonacorsi, and E. Bingen. 2000. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl. Environ. Microbiol. 66:4555-4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clermont, O., S. Bonacorsi, and E. Bingen. 2001. The Yersinia high-pathogenicity island is highly predominant in virulence-associated phylogenetic groups of Escherichia coli. FEMS Microbiol. Lett. 196:153-157. [DOI] [PubMed] [Google Scholar]
- 10.Diard, M., et al. 2010. Pathogenicity-associated islands in extraintestinal pathogenic Escherichia coli are fitness elements involved in intestinal colonization. J. Bacteriol. 192:4885-4893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dobrindt, U., et al. 2001. S-fimbria-encoding determinant sfaI is located on pathogenicity island III536 of uropathogenic Escherichia coli strain 536. Infect. Immun. 69:4248-4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dobrindt, U., et al. 2002. Genetic structure and distribution of four pathogenicity islands (PAI I536 to PAI IV536) of uropathogenic Escherichia coli strain 536. Infect. Immun. 70:6365-6372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Escobar-Paramo, P., et al. 2004. A specific genetic background is required for acquisition and expression of virulence factors in Escherichia coli. Mol. Biol. Evol. 21:1085-1094. [DOI] [PubMed] [Google Scholar]
- 14.Guyer, D. M., J. S. Kao, and H. L. Mobley. 1998. Genomic analysis of a pathogenicity island in uropathogenic Escherichia coli CFT073: distribution of homologous sequences among isolates from patients with pyelonephritis, cystitis, and catheter-associated bacteriuria and from fecal samples. Infect. Immun. 66:4411-4417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hacker, J., G. Blum-Oehler, I. Muhldorfer, and H. Tschape. 1997. Pathogenicity islands of virulent bacteria: structure, function and impact on microbial evolution. Mol. Microbiol. 23:1089-1097. [DOI] [PubMed] [Google Scholar]
- 16.Herzer, P. J., S. Inouye, M. Inouye, and T. S. Whittam. 1990. Phylogenetic distribution of branched RNA-linked multicopy single-stranded DNA among natural isolates of Escherichia coli. J. Bacteriol. 172:6175-6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnson, J. R. 1991. Virulence factors in Escherichia coli urinary tract infection. Clin. Microbiol. Rev. 4:80-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson, J. R., E. Oswald, T. T. O'Bryan, M. A. Kuskowski, and L. Spanjaard. 2002. Phylogenetic distribution of virulence-associated genes among Escherichia coli isolates associated with neonatal bacterial meningitis in the Netherlands. J. Infect. Dis. 185:774-784. [DOI] [PubMed] [Google Scholar]
- 19.Johnson, J. R., K. Owens, A. Gajewski, and M. A. Kuskowski. 2005. Bacterial characteristics in relation to clinical source of Escherichia coli isolates from women with acute cystitis or pyelonephritis and uninfected women. J. Clin. Microbiol. 43:6064-6072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnson, J. R., and A. L. Stell. 2000. Extended virulence genotypes of Escherichia coli strains from patients with urosepsis in relation to phylogeny and host compromise. J. Infect. Dis. 181:261-272. [DOI] [PubMed] [Google Scholar]
- 21.Kanamaru, S., et al. 2006. Subtyping of uropathogenic Escherichia coli according to the pathogenicity island encoding uropathogenic-specific protein: comparison with phylogenetic groups. Int. J. Urol. 13:754-760. [DOI] [PubMed] [Google Scholar]
- 22.Kao, J. S., D. M. Stucker, J. W. Warren, and H. L. Mobley. 1997. Pathogenicity island sequences of pyelonephritogenic Escherichia coli CFT073 are associated with virulent uropathogenic strains. Infect. Immun. 65:2812-2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Karch, H., et al. 1999. A genomic island, termed high-pathogenicity island, is present in certain non-O157 Shiga toxin-producing Escherichia coli clonal lineages. Infect. Immun. 67:5994-6001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kurazono, H., et al. 2000. Characterization of a putative virulence island in the chromosome of uropathogenic Escherichia coli possessing a gene encoding a uropathogenic-specific protein. Microb. Pathog. 28:183-189. [DOI] [PubMed] [Google Scholar]
- 25.Landraud, L., M. Gibert, M. R. Popoff, P. Boquet, and M. Gauthier. 2003. Expression of cnf1 by Escherichia coli J96 involves a large upstream DNA region including the hlyCABD operon, and is regulated by the RfaH protein. Mol. Microbiol. 47:1653-1667. [DOI] [PubMed] [Google Scholar]
- 26.Nakano, M., et al. 2001. Structural and sequence diversity of the pathogenicity island of uropathogenic Escherichia coli which encodes the USP protein. FEMS Microbiol. Lett. 205:71-76. [DOI] [PubMed] [Google Scholar]
- 27.Nowrouzian, F., I. Adlerberth, and A. E. Wold. 2001. P fimbriae, capsule and aerobactin characterize colonic resident Escherichia coli. Epidemiol. Infect. 126:11-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nowrouzian, F., et al. 2003. Escherichia coli in infants' intestinal microflora: colonization rate, strain turnover, and virulence gene carriage. Pediatr. Res. 54:8-14. [DOI] [PubMed] [Google Scholar]
- 29.Nowrouzian, F., A. E. Wold, and I. Adlerberth. 2001. P fimbriae and aerobactin as intestinal colonization factors for Escherichia coli in Pakistani infants. Epidemiol. Infect. 126:19-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nowrouzian, F. L., I. Adlerberth, and A. E. Wold. 2006. Enhanced persistence in the colonic microbiota of Escherichia coli strains belonging to phylogenetic group B2: role of virulence factors and adherence to colonic cells. Microbes Infect. 8:834-840. [DOI] [PubMed] [Google Scholar]
- 31.Nowrouzian, F. L., A. E. Ostblom, A. E. Wold, and I. Adlerberth. 2009. Phylogenetic group B2 Escherichia coli strains from the bowel microbiota of Pakistani infants carry few virulence genes and lack the capacity for long-term persistence. Clin. Microbiol. Infect. 15:466-472. [DOI] [PubMed] [Google Scholar]
- 32.Nowrouzian, F. L., A. E. Wold, and I. Adlerberth. 2005. Escherichia coli strains belonging to phylogenetic group B2 have superior capacity to persist in the intestinal microflora of infants. J. Infect. Dis. 191:1078-1083. [DOI] [PubMed] [Google Scholar]
- 33.Parham, N. J., et al. 2005. Prevalence of pathogenicity island IICFT073 genes among extraintestinal clinical isolates of Escherichia coli. J. Clin. Microbiol. 43:2425-2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parret, A. H., and R. De Mot. 2002. Escherichia coli's uropathogenic-specific protein: a bacteriocin promoting infectivity? Microbiology 148:1604-1606. [DOI] [PubMed] [Google Scholar]
- 35.Ramette, A. 2007. Multivariate analyses in microbial ecology. FEMS Microbiol. Ecol. 62:142-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rasko, D. A., J. A. Phillips, X. Li, and H. L. Mobley. 2001. Identification of DNA sequences from a second pathogenicity island of uropathogenic Escherichia coli CFT073: probes specific for uropathogenic populations. J. Infect. Dis. 184:1041-1049. [DOI] [PubMed] [Google Scholar]
- 37.Reidl, J., and W. Boos. 1991. The malX malY operon of Escherichia coli encodes a novel enzyme II of the phosphotransferase system recognizing glucose and maltose and an enzyme abolishing the endogenous induction of the maltose system. J. Bacteriol. 173:4862-4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rouquet, G., et al. 2009. A metabolic operon in extraintestinal pathogenic Escherichia coli promotes fitness under stressful conditions and invasion of eukaryotic cells. J. Bacteriol. 191:4427-4440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sabate, M., E. Moreno, T. Perez, A. Andreu, and G. Prats. 2006. Pathogenicity island markers in commensal and uropathogenic Escherichia coli isolates. Clin. Microbiol. Infect. 12:880-886. [DOI] [PubMed] [Google Scholar]
- 40.Sabate, M., et al. 2008. Virulence and antimicrobial resistance profiles among Escherichia coli strains isolated from human and animal wastewater. Res. Microbiol. 159:288-293. [DOI] [PubMed] [Google Scholar]
- 41.Sannes, M. R., M. A. Kuskowski, K. Owens, A. Gajewski, and J. R. Johnson. 2004. Virulence factor profiles and phylogenetic background of Escherichia coli isolates from veterans with bacteremia and uninfected control subjects. J. Infect. Dis. 190:2121-2128. [DOI] [PubMed] [Google Scholar]
- 42.Schmidt, H., and M. Hensel. 2004. Pathogenicity islands in bacterial pathogenesis. Clin. Microbiol. Rev. 17:14-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schubert, S., A. Rakin, H. Karch, E. Carniel, and J. Heesemann. 1998. Prevalence of the “high-pathogenicity island” of Yersinia species among Escherichia coli strains that are pathogenic to humans. Infect. Immun. 66:480-485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sears, H. J., H. Janes, R. Saloum, I. Brownlee, and L. F. Lamoreaux. 1956. Persistence of individual strains of Escherichia coli in man and dog under varying conditions. J. Bacteriol. 71:370-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sears, H. J., and I. Brownlee. 1952. Further observations on the persistence of individual strains of Escherichia coli in the intestinal tract of man. J. Bacteriol. 63:47-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sears, H. J., I. Brownlee, and J. K. Uchiyama. 1950. Persistence of individual strains of Escherichia coli in the intestinal tract of man. J. Bacteriol. 59:299-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tourret, J., M. Diard, L. Garry, I. Matic, and E. Denamur. 2010. Effects of single and multiple pathogenicity island deletions on uropathogenic Escherichia coli strain 536 intrinsic extra-intestinal virulence. Int. J. Med. Microbiol. 300:435-439. [DOI] [PubMed] [Google Scholar]
- 48.Yamamoto, S., et al. 2001. The presence of the virulence island containing the usp gene in uropathogenic Escherichia coli is associated with urinary tract infection in an experimental mouse model. J. Urol. 165:1347-1351. [PubMed] [Google Scholar]