Abstract
Ammonia-oxidizing bacteria (AOB) in nitrifying biofilters degrading four regulated trihalomethanes—trichloromethane, bromodichloromethane, dibromochloromethane, and tribromomethane—were related to Nitrosomonas oligotropha. N. oligotropha is associated with chloraminated drinking water systems, and its presence in the biofilters might indicate that trihalomethane tolerance is another reason that this bacterium is dominant in chloraminated systems.
No evidence indicates that trihalomethanes (THMs) support microbial growth, but previous research demonstrated that four regulated THMs—trichloromethane (TCM), bromodichloromethane (BDCM), dibromochloromethane (DBCM), and tribromomethane (TBM)—were cometabolized in batch culture (15) by Nitrosomonas europaea (an ammonia oxidizer) and in batch culture (14) and laboratory-scale biofilters (16, 17) by mixed-culture nitrifiers. The current research extended such process evaluation by analyzing the ammonia-oxidizing bacteria (AOB) present in biofilters through quantitative real-time PCR (qPCR) and amoA gene sequence analysis.
Duplicate anthracite biofilters (1-min empty-bed contact time; 1.3 gallons min−1 ft−2 surface loading rate) were seeded with one of two mixed cultures (14). Trains A and B were seeded with a sample collected from Lake Austin in Austin, TX, and trains C and D were seeded with an enriched nitrifier culture initially dominated by Nitrosomonas oligotropha (N. oligotropha enrichment, provided by D. R. Noguera, University of Wisconsin). The trains were fed 4 mg N/liter TOTNH3 (sum of ammonia-nitrogen [NH3-N] and ammonium-nitrogen [NH4+-N]). The three operational periods were based on influent THM concentrations as follows: period I, TCM-DBCM (50 μg/liter TCM and 25 μg/liter DBCM); period II, DBCM (25 μg/liter DBCM); and period III, all THMs (15 μg/liter each of TCM, DBCM, BDCM, and TBM). THM removals are summarized in Table 1 for each period; all biofilters removed THMs with removals ranging from 7 to 24% (Table 1).
TABLE 1.
Biofilter performancea
| Train | Period | No. of samples | Δ0-1TOTNH3 (mg N/liter)b | % Removal (mean ± standard deviation)c |
|||
|---|---|---|---|---|---|---|---|
| TCM | BDCM | DBCM | TBM | ||||
| A | I | 6 | 2.7 ± 0.29 | 9.7 ± 6.4 | 12 ± 5.3 | ||
| II | 3 | 3.4 ± 0.26 | 24 ± 2.3 | ||||
| III | 4 | 3.1 ± 0.38 | 16 ± 7.6 | 18 ± 6.5 | 19 ± 6.3 | 16 ± 6.2 | |
| B | I | 7 | 2.1 ± 0.36 | 7.7 ± 5.2 | 13 ± 8.3 | ||
| II | 3 | 3.0 ± 0.23 | 14 ± 2.0 | ||||
| III | 4 | 2.5 ± 0.37 | 13 ± 6.2 | 14 ± 6.0 | 15 ± 5.2 | 13 ± 3.8 | |
| C | I | 6 | 2.6 ± 0.74 | 6.9 ± 6.0 | 12 ± 5.0 | ||
| II | 2 | 2.5 ± 0.62 | 17 ± 2.0 | ||||
| III | 4 | 2.8 ± 0.67 | 14 ± 5.7 | 21 ± 8.1 | 23 ± 6.0 | 18 ± 3.8 | |
| D | I | 7 | 2.0 ± 0.56 | 6.5 ± 6.5 | 13 ± 7.3 | ||
| II | 3 | 1.7 ± 0.22 | 10 ± 3.2 | ||||
| III | 4 | 2.4 ± 1.1 | 16 ± 6.9 | 16 ± 7.6 | 17 ± 8.1 | 15 ± 7.6 | |
Trains were nominally fed 4 mg N/liter TOTNH3 (sum of ammonia-nitrogen [NH3-N] and ammonium-nitrogen [NH4+-N]). The three operational periods were based on influent THM concentrations as follows: period I, TCM-DBCM (50 μg/liter TCM and 25 μg/liter DBCM); period II, DBCM (25 μg/liter DBCM); and period III, all THMs (15 μg/liter each of TCM, BDCM, DBCM, and TBM).
Δ0-1TOTNH3, TOTNH3 removed through biofilter (mean ± standard deviation).
TCM, trichloromethane; BDCM, bromodichloromethane; DBCM, dibromochloromethane; TBM, tribromomethane.
At the end of period II, backwash water was collected for batch kinetic tests conducted as described previously (16). Because backwash water may have contained extracellular material, attachment media, or nonnitrifying organisms, a biomass-independent analysis was required. To accomplish this, a previously described (16) simplified THM cometabolism model (equation 1) was used, as follows:
 |
(1) |
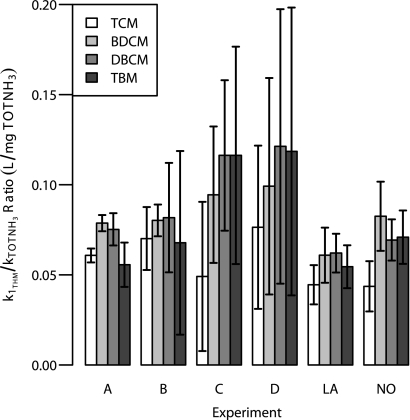
where STHM0 is the initial THM concentration (μg/liter THM); STHM1 is the final THM concentration (μg/liter THM); ΔTOTNH3 is initial TOTNH3 minus final TOTNH3 (mg N/liter); and k1THM/kTOTNH3 is the ratio of THM and TOTNH3 rate constants (liter/mg TOTNH3). The k1THM/kTOTNH3 ratio represents the THM removal efficiency, relating THM to ammonia removal. For purposes of comparison, the biofilter k1THM/kTOTNH3 ratios were calculated by pooling operational data for duplicate trains (where biofilter THM removal was not statistically different between duplicate trains, based on Tukey's paired comparison method with a two-sided 95% confidence interval of the studentized range statistic [2, 4]). The backwash batch kinetic test k1THM/kTOTNH3 ratios (Fig. 1) provided another confirmation that the biofilters were able to degrade THMs (initial concentrations, 70 to 120 μg/liter each THM and 5 mg N/liter TOTNH3).
FIG. 1.
Backwash batch kinetic test k1THM/kTOTNH3 ratios and associated 95% confidence limits. A, B, C, and D correspond to experiments conducted with backwash from trains A, B, C, and D, respectively. For comparison purposes, k1THM/kTOTNH3 ratios were also calculated using equation 1 and biofilter performance data. LA, Lake Austin; NO, N. oligotropha enrichment.
At the end of period III, biofilter DNA was isolated using an UltraClean soil DNA isolation kit (Mo Bio Laboratories, Inc., Carlsbad, CA). The biofilters were divided into 4 equally spaced vertical sections (influent end is designated section 1, and effluent end is designated section 4). The amoA qPCR method of Regan et al. (7) was used to assess the relative abundance of AOB in three general classes: (i) non-Oligotropha nitrosomonas, (ii) Nitrosospira, and (iii) N. oligotropha. Only N. oligotropha was detected (data not shown), indicating that this is the dominant AOB present.
Subsequently, amoA genes from trains A and C were cloned and sequenced to supplement the qPCR results. For cloning, amoA was amplified (7), and the amplicon purified using a QIAquick PCR purification kit (Qiagen, Valencia, CA) and cloned using a TOPO TA cloning kit for sequencing (Invitrogen, Carlsbad, CA). The clones were screened for inserts by PCR with M13 forward and reverse primers. The amplicon was sequenced using BigDye sequencing chemistry (Applied Biosystems, Foster City, CA).
Using MEGA 4 (12), 452-bp sequences (primer sequences removed) were generated, and DOTUR was used to sort amoA gene sequences with more than 98% similarity (11) into operational taxonomic units (OTUs). Analyses of cloned amoA gene sequences found two OTUs (GenBank accession numbers HQ399455 and HQ399456) that were present in both trains, hereinafter designated Biofilter OTU 1 and 2 (Table 2). These OTUs were incorporated into a phylogenetic tree (Fig. 2) constructed in MEGA4 by the neighbor-joining method using 10,000 bootstrap replicates. The sequence analysis confirmed the qPCR results, as these two OTUs were related to N. oligotropha whether the phylogenetic tree was generated based on amoA gene sequences (data not shown) or on deduced amino acid sequences from the amoA gene sequences (Fig. 2). Considering deduced amino acid sequences, the Biofilter OTU 1 consensus sequence was a 100% match to samples taken from a wastewater treatment plant activated sludge and membrane bioreactor and to Nitrosomonas NL7 and NM47, and the Biofilter OTU 2 consensus sequence was a 100% match to an environmental sample taken from a chloraminated drinking water distribution system. Taken together, the qPCR and gene sequence analyses show that the dominant AOB in the biofilters used for THM removal were related to N. oligotropha. AOB related to N. oligotropha have been reported as the dominant AOB in chloraminated drinking water systems (7-9). Because these systems contain THMs at levels similar to those used in our biofilter experiments, it is possible that one reason for the presence of AOB related to N. oligotropha in chloraminated drinking water systems is their ability to tolerate THMs. This is supported by the results of Bayer and Speitel (1), who found that the transformation capacities (including those of an N. oligotropha-dominated culture) were greater than those for N. europaea (15).
TABLE 2.
amoA gene sequences and OTU biofilter sample summary
| Samplea | No. of amoA gene sequences in: |
|
|---|---|---|
| Biofilter OTU 1 | Biofilter OTU 2 | |
| A1 (influent end) | 105 | 25 |
| A2 | 67 | 3 |
| A3 | 8 | 1 |
| A4 (effluent end) | 14 | 0 |
| Total for A | 194 | 29 |
| C1 (influent end) | 73 | 0 |
| C2 | 74 | 0 |
| C3 | 73 | 1 |
| C4 (effluent end) | 65 | 0 |
| Total for C | 285 | 1 |
| Total for A and C | 479 | 30 |
The influent end of the biofilter was designated section 1, with the numbering progressing down the length of the biofilter in equal lengths such that the effluent end was designated section 4. Each section was divided into three subsamples that were used for DNA extraction, resulting in triplicate extractions for each section. For the extractions, the mass of dry anthracite used was 0.60 ± 0.06 g (mean ± standard deviation) per extraction (wet anthracite moisture content, 32% ± 0.74% [mean ± standard deviation]).
FIG. 2.
Phylogenetic tree based on deduced amino acid sequences of the amoA gene sequences of AOB. The evolutionary history was inferred using the neighbor-joining method (10). The percentage of replicate trees (those >50%) in which the associated taxa clustered together in the bootstrap test (10,000 replicates) are shown next to the branches (3). The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the JTT matrix-based method (5), and the unit of measure is the number of amino acid substitutions per site, shown by the bar. The number of sequences belonging to the OTU/total number of sequences is given in parentheses.
Overall, four regulated THMs found in treated drinking water were degraded in biofilters seeded with Lake Austin and N. oligotropha enrichment cultures. The ability of the biofilter biomass to remove THMs was verified in batch kinetic tests. amoA qPCR and gene sequence analyses demonstrated that the biofilter AOB were related to N. oligotropha. Their presence in biofilters removing THMs suggests that one potential reason for their dominance in these systems is their THM tolerance. Future research should broaden the nitrifier community analysis to include ammonia-oxidizing archaea, since they are being reported in drinking water systems (6, 13).
Acknowledgments
We thank John M. Regan for plasmids used in constructing qPCR standard curves and Daniel R. Noguera for a mixed culture source.
This research was funded by AwwaRF.
Any opinions expressed are those of the authors and do not necessarily reflect the views of U.S. Environmental Protection Agency or AwwaRF; therefore, no official endorsement should be inferred. Any mention of trade names or commercial products does not constitute endorsement or recommendation for use.
Footnotes
Published ahead of print on 28 January 2011.
REFERENCES
- 1.Bayer, B. M., and G. E. Speitel, Jr. 2007. Significance of trihalomethanes in preventing distribution system nitrification. 2007 Am. Water Works Assoc. Annu. Conf. Proc. American Water Works Association, Denver, CO.
- 2.Berthouex, P. M., and L. C. Brown. 2002. Statistics for environmental engineers. Lewis Publishers, Boca Raton, FL.
- 3.Felsenstein, J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783-791. [DOI] [PubMed] [Google Scholar]
- 4.Harter, H. L. 1960. Tables of range and studentized range. Ann. Math. Stat. 31:1122-1147. [Google Scholar]
- 5.Jones, D. T., W. R. Taylor, and J. M. Thornton. 1992. The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 8:275-282. [DOI] [PubMed] [Google Scholar]
- 6.Kasuga, I., H. Nakagaki, F. Kurisu, and H. Furumai. 2010. Predominance of ammonia-oxidizing archaea on granular activated carbon used in a full-scale advanced drinking water treatment plant. Water Res. 44:5039-5049. [DOI] [PubMed] [Google Scholar]
- 7.Regan, J. M., A.-Y. Cho, S. Kim, and C. D. Smith. 2007. Monitoring ammonia-oxidizing bacteria in chloraminated distribution systems. AwwaRF Report 91162. Water Research Foundation, Denver, CO.
- 8.Regan, J. M., G. W. Harrington, H. Baribeau, R. D. Leon, and D. R. Noguera. 2003. Diversity of nitrifying bacteria in full-scale chloraminated distribution systems. Water Res. 37:197-205. [DOI] [PubMed] [Google Scholar]
- 9.Regan, J. M., G. W. Harrington, and D. R. Noguera. 2002. Ammonia- and nitrite-oxidizing bacterial communities in a pilot-scale chloraminated drinking water distribution system. Appl. Environ. Microbiol. 68:73-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 11.Schloss, P. D., and J. Handelsman. 2005. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl. Environ. Microbiol. 71:1501-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596-1599. [DOI] [PubMed] [Google Scholar]
- 13.van der Wielen, P. W. J. J., S. Voost, and D. van der Kooij. 2009. Ammonia-oxidizing bacteria and archaea in groundwater treatment and drinking water distribution systems. Appl. Environ. Microbiol. 75:4687-4695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wahman, D. G., A. E. Henry, L. E. Katz, and G. E. Speitel, Jr. 2006. Cometabolism of trihalomethanes by mixed culture nitrifiers. Water Res. 40:3349-3358. [DOI] [PubMed] [Google Scholar]
- 15.Wahman, D. G., L. E. Katz, and G. E. Speitel, Jr. 2005. Cometabolism of trihalomethanes by Nitrosomonas europaea. Appl. Environ. Microbiol. 71:7980-7986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wahman, D. G., L. E. Katz, and G. E. Speitel, Jr. 2011. Performance and biofilm activity of nitrifying biofilters removing trihalomethanes. Water Res. 45:1669-1680. [DOI] [PubMed] [Google Scholar]
- 17.Wahman, D. G., L. E. Katz, and G. E. Speitel, Jr. 2006. Trihalomethane cometabolism by a mixed-culture nitrifying biofilter. J. Am. Water Works Assoc. 98:2-20. [Google Scholar]