Abstract
O-antigen biosynthetic (wbf) regions for Vibrio cholerae serogroups O5, O8, and O108 were isolated and sequenced. Sequences were compared to those of other published V. cholerae O-antigen regions. These wbf regions showed a high degree of heterogeneity both in gene content and in gene order. Genes identified frequently showed greater similarities to polysaccharide biosynthesis genes from species other than V. cholerae. Our results demonstrate the plasticity of O-antigen genes in V. cholerae, the diversity of the genetic pool from which they are drawn, and the likelihood that new pandemic serogroups will emerge.
Cholera is a pandemic diarrheal disease that continues to be an important cause of morbidity and mortality worldwide. Cholera is associated with a clonally related subset of Vibrio cholerae strains, which carry ctxAB (cholera toxin subunits A and B), the vibrio pathogenicity island (VPI), and other cholera-associated genes (15, 40). Until 1992, only serogroup O1 (out of more than 206 serogroups currently described) was recognized as a cause of cholera. In 1992, a new, non-O1 V. cholerae strain (subsequently designated V. cholerae O139) appeared in India and rapidly spread across much of Asia (32). Extensive studies (5, 12, 31, 41) demonstrated that the O139 strain was closely related to the O1 El Tor strains of the 7th pandemic, except that the genes responsible for O1-antigen biosynthesis were deleted and replaced with DNA that encodes the O139 antigen. Since the O antigen is the major protective epitope, its alteration was sufficient to allow O139 strains to move in epidemic form through populations previously immune to cholera caused by O1. Thus, adults were more commonly affected than children (1). Recent studies suggest that the pandemic cluster carries other serogroups as well, including O37, O27, O53, O65, and O75 (31, 40, 43). These observations are consistent with the hypothesis that pathogenic V. cholerae strains are able to easily acquire and/or exchange O-antigen genes, with the new O antigen allowing strains to evade preexisting immunity to cholera.
Changes in O-antigen structure also may provide selective advantages in the environment. During epidemics, bacteriophage may play a crucial role in controlling the number of V. cholerae in the environment (19), and since the O-antigen may serve as bacteriophage receptors, serogroup conversion also may be beneficial for evading phage predation (33). It is well documented that mobile genetic elements, bacteriophage, and the competence of V. cholerae to take up and assimilate free DNA from the environment significantly contribute to genetic diversity in V. cholerae (18). Serogroup conversion was demonstrated while V. cholerae was growing on a chitin substrate (6). This transfer may have been facilitated by the JUMPstart sequence, which has nucleotide similarity to a DNA uptake signal that causes the preferential uptake of free DNA containing that sequence in the Pasteurellaceae and is present in front of a laterally transferred O-antigen region in V. cholerae (20). DNA transfer in V. cholerae also occurs by phage transduction (7, 21).
Numerous studies have focused on the variability and origins of O-antigen genes in enteric bacteria. Reeves et al. (35, 36) have undertaken extensive studies of the genetic basis of O-antigen variation in Salmonella enterica and Escherichia coli. They showed that some O-antigen DNA originated in species other than S. enterica or E. coli and had been captured by lateral transfer. By sequencing and comparison of O-antigen gene clusters, they found evidence that DNA recombination events between E. coli and Klebsiella played a role in the formation of new O-antigen forms (25). Despite the extensive variation in E. coli (186) serotypes, the variation can be organized into groups of similar serotypes. In E. coli, the O-antigen-processing genes pairs wzm-wzt (together an ABC transport system) and wzx (flippase)-wzy (polymerase) have been used to separate distinct classes of serogroups (37, 45). A similar system was applied to V. cholerae serogroups based on a limited number of strains (46). In a nonenteric bacterial species, Streptococcus pneumoniae, 90 serotypes have been sequenced, and all serotypes have the same translocation system wzx and wzy and can be classified based on shared sugar synthesis genes (4).
Several V. cholerae lipopolysaccharide (LPS) regions, primarily from toxigenic isolates, have been sequenced. These include O1, O139, O22, O37, O31, O12, O14, O39, O141, and three of unknown serogroups (5, 9, 11, 12, 13, 14, 30, 31, 41). These regions vary in size from the 18 open reading frames (ORFs) of O1 LPS to the 56 ORFs identified in the unknown serotype of the 623-39 strain. Vibrio cholerae O antigens include a number of sugars that are somewhat unusual, including quinovosamine (O139 and O108) and fucosamine. 2,4-Diacetamido-2,4,6-trideoxyglucose (QuiNAc4NAc) has been reported in both O8 (26) and O5 serogroups (22). In addition, glycero-d-manno-heptose (O5, O8, and O108), glucose (O5 and O8), fructose (O5 and O8), glucosamine (O108), galactosamine (O108), and fucosamine (O108) had been reported in these serogroups (28). In both O139 and O31 serogroups, the wbf region encodes both O antigen and a high-molecular-weight capsule (9, 44). To expand our understanding of this region and in hopes of defining classes of O antigens, we sequenced the wbf region of three additional strains.
MATERIALS AND METHODS
Three strains, CO545, CO845, and CO603B (from serogroups O5, O8, and O108, respectively), were selected and sequenced. The strains were collected from patients admitted with a diagnosis of diarrhea to the Infectious Disease Hospital by the National Institute of Enteric Disease in Kolkata (formerly known as Calcutta), India, in 1994 and 1995. These isolates were collected as part of routine surveillance (26, 34). The strains were selected based on overlapping sugar composition (see Introduction) and ease of long-range PCR amplification.
V. cholerae O-antigen genes studied to date have been localized in the wbf region flanked by two genes: gmhD (which encodes d-glycero-d-manno-heptose 1-phosphate guanosyltransferase) and the right junction gene rjg (hypothetical protein with similarities to mRNA 3′-end processing factor) (38). Primer sequences were derived from gmhD and rjg (12), and the wbf regions in the three strains were isolated by long-range PCR using the GeneAmp XL PCR kit (Perkin-Elmer). Gel-purified (QIAquick gel extraction kit; Qiagen, CA) products, ∼25 kb for O5, ∼20 kb for O8, and ∼30 kb for O108, were sonicated, and the sheared DNA was used to create individual libraries for each strain. These shotgun libraries were constructed with the TOPO shotgun subcloning kit (Invitrogen, CA) according to the manufacturer's protocol. Approximately 300 to 400 clones per strain grown on LB agar were picked and subjected to PCR amplification. The PCR was carried out with the following steps: an initial denaturing at 94°C for 2 min; 12 cycles of 94°C for 15 s and 68°C for 15 min; 12 cycles of 94°C for 15 s and 15 min (increased by 15 s with each cycle) at 68°C; and finally, 72°C for 30 min. Amplified products from the libraries were purified with a Microcon YM-100 column (Millipore) and used to initiate sequencing PCR using the BigDye Terminator cycle sequencing kit (Applied Biosystems). Sequencing was performed with an Applied Biosystems 3700 automated DNA sequencer. Sequence data were assembled by using the Phred/Phrap package (16, 17), and the sequence annotation was done by using the program Artemis from the Sanger Centre (http://www.sanger.ac.uk/Software/Artemis/).
The nucleotide and amino acid sequences of each gene were used to search available databases for an indication of function. The PFAM (3) database was searched by BLASTP and BLASTX (2), and only hits with an E score of ≤0.02 or lower were considered matches for potential ORFs. In addition, the GenBank database (www.ncbi.nlm.nih.gov/BLAST) was searched by BLASTP and BLASTX (2), and only hits with P values of 10−6 or lower were considered matches for potential ORFs. A representative subset of BLASTP hits to each strain, together with the respective sequences from the strains, were aligned with ClustalX (10), and genetic relatedness was determined with neighbor-joining (NJ) analysis as implemented in the PAUP 4.0 package (42) and with the Splitstree method (24). Bootstrap analysis was done to statistically confirm the robustness of the phylogenetic analyses.
Nucleotide sequence accession numbers.
The sequences determined in the course of this work have been deposited in GenBank under accession numbers GU576497 to GU576499.
RESULTS AND DISCUSSION
Long-range PCR was attempted on nine strains from different serogroups based on carbohydrate analysis, suggesting that they have a number of sugars in common (27). Three strains were amplified: CO545, CO845, and CO603B, representing serogroups O5, O8, and O108, respectively. These were compared to the wbf regions from an additional 13 sequences available in hopes of being able to identify classes, as was done for E. coli and S. pneumoniae.
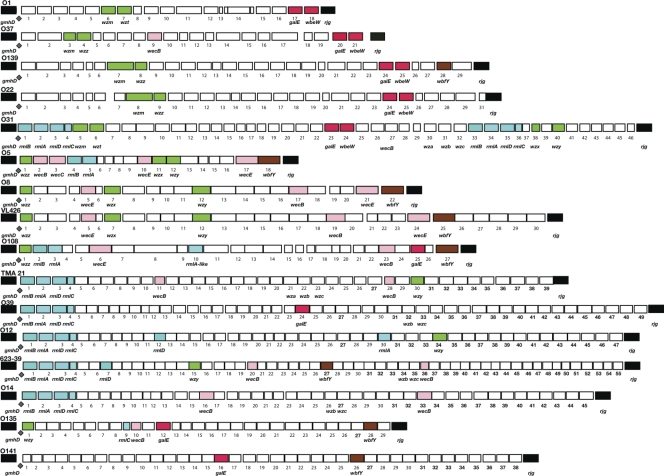
Overall, the wbf regions from 16 different serogroups ranged from 18 ORFs in serogroups O1 and O5 (CO545) to 56 ORFs for strain 623-39 (Fig. 1). The O139 and O22 serogroup sequences were shown previously to have three stretches (∼2, 12, and 16 kb) of sequence with greater than 91% similarity interrupted by two blocks of genes with low (50%) or no sequence similarity (46). Like O22 and O139, the LPS region of serogroup O8 (CO845) (Table 1) was similar to the LPS region of V. cholerae bv. albensis strain VL426 (11). Both strains have a similar order and high sequence similarity (>97% for 6,577 amino acids covering ORFs 1 to 12, and 16 to 22 of O8 and 1 to 12, 18 to 22, 24, and 25 of VL426) of the genes. The interrupting blocks consisted of three consecutive ORFS of O8 (CO845), 13, 14, and 15, that had low amino acid similarities. ORF 13 was 63% similar to Psychrobacter cryohalolentis glutamine amidotransferase, while ORFs 14 and 15 were only 37 and 79% similar to V. cholerae bv. albensis glycosyl transferase and d-glucuronyl C5-epimerase, respectively. Furthermore, there were nine additional genes, one (V. cholerae bv. albensis ORF 23) between ORFs 20 and 21 of O8, three (V. cholerae bv. albensis ORFs 14 to 16) between ORFs 14 and 15 in O8, and five additional genes (V. cholerae bv. albensis ORFs 26 to 30) found between the epimerase (wbfY; ORF 22) and rjg of O8 (CO845) (Fig. 1). The serogroup O5 (CO545) strain had 18 ORFs, and O108 (CO845) had 27 ORFs (Table 1). Each region had genes at the 3′ and 5′ end that had high similarities to another Vibrio sequence as well as internal sequences that did not match any known Vibrio sequence.
FIG. 1.
Pictorial representation of the genes in the LPS/capsule polysaccharide regions of Vibrio cholerae serogroups O1, O37, O22, O139, O31, O5, O8, O108, O12, O14, O39, O135, and O141, as well as those of strains 623-39, TMA 21, and VL426. We selected three strains, CO545, CO845, and CO603B, from serogroups O5, O8, and O108, respectively, for analysis based on carbohydrate analysis, which suggested that these serogroups have at least two sugars, glycero-d-manno-heptose and glucose, in common in their LPS (28). The sequences of the other serogroups were taken from the literature for comparison, with the expectation that there would be identifiable subgroups. Flanking genes gmhD and rjg, which delineate the region, are marked in black. ORFs between these loci are numbered. JUMPstart sites are indicated by a diamond. Similar genes present in more than two rfb regions are color coded. Transport genes are marked in green; galE and wbeW genes are red; rhamnose pathway genes are marked in blue; wecB, wecE, and wecC are pink; and wbfY is brown. The unlabeled genes represented by white boxes are not found in common across the regions.
TABLE 1.
Identification of open reading frames in O5 (CO545), O8 (CO845), and O108 (CO603B)a
| Serogroup and ORF no. | Putative function of product | Name | Organism | Accession no. | % Similarity (no./total no.) |
|---|---|---|---|---|---|
| O5 | ADP-l-glycero-d-mannoheptose-6-epimerase | gmhD | |||
| 1 | Chain length determinant | wzz | Vibrio cholerae | AAO88947.1 | 90 (318/350) |
| 2 | UDP-N-acetylglucosamine 2-epimerase | wecB | Vibrio vulnificus YJ016 | NP_933134.1 | 96 (361/374) |
| 3 | UDP-N-acetyl-d-manno-saminuronate dehydrogenase | wecC | Vibrio vulnificus YJ016 | NP_759756.1 | 90 (382/422) |
| 4 | Dtdp-glucose 4,6-dehydratase | rmlB | Vibrio cholerae O141 | YP_002072336.1 | 90 (321/355) |
| 5 | Glucose-1-phosphate thymidylyltransferase | rmlA | Listonella anguillarum | AAZ66343.1 | 82 (237/288) |
| 6 | Lipopolysaccharide biosynthesis protein | wblP | Vibrio harveyi HY01 | ZP_01987019.1 | 64 (84/130) |
| 7 | Transferase, putative | Xanthomonas oryzae | YP_201636.1 | 61 (135/220) | |
| 8 | Hypothetical protein | Francisella philomiragia | YP_001677984.1 | 59 (182/304) | |
| 9 | Hypothetical protein | Burkholderia phymatum | YP_001858531.1 | 39 (121/303) | |
| 10 | DegT/DnrJ/EryC1/StrS aminotransferase | wecE | Shewanella denitrificans | YP_563660.1 | 62 (220/366) |
| 11 | O-antigen flippase | wzx | Pectobacterium carotovorum | ZP_03830717.1 | 25 (38/151) |
| 12 | O-antigen polymerase | wzy | Bacillus cereus | ZP_04173869.1 | 37 (44/117) |
| 13 | WblR protein | Rhodoferax ferrireducens | YP_522520.1 | 26 (111/414) | |
| 14 | Glycosyl transferase | Vibrio vulnificus YJ016 | NP_933153.1 | 84 (354/418) | |
| 15 | Putative UDP-galactose phosphate transferase | wbfU-wcaJ | Vibrio cholerae VL426 | ZP_04414194.1 | 93 (190/204) |
| 16 | Putative acetyltransferase | Vibrio cholerae VL426 | ZP_04414193.1 | 99 (232/233) | |
| 17 | DegT/DnrJ/EryC1/StrS aminotransferase | wecE | Vibrio cholerae O1 | ZP_01982757.1 | 99 (389/391) |
| 18 | Putative epimerase/dehydratase | wbfY | Vibrio cholerae | BAA33644.1 | 99 (561/562) |
| O8 | ADP-l-glycero-d-mannoheptose-6-epimerase | gmhD | |||
| 1 | Chain length determinant | wzz | Vibrio cholerae VL426 | ZP_04414215.1 | 95 (356/374) |
| 2 | Myo-inositol 2-dehydrogenase | Vibrio cholerae VL426 | ZP_04414214.1 | 100 (349/349) | |
| 3 | UDP-glucose/GDP-mannose dehydrogenase family | Vibrio cholerae VL426 | ZP_04414213.1 | 98 (416/424) | |
| 4 | Bacterial transferase | Vibrio cholerae VL426 | ZP_04414212.1 | 100 (196/196) | |
| 5 | DegT/DnrJ/EryC1/StrS aminotransferase | wecE | Vibrio cholerae VL426 | ZP_04414211.1 | 98 (352/359) |
| 6 | Acyltransferase family | Vibrio cholerae VL426 | ZP_04414210.1 | 96 (86/89) | |
| 7 | Flippase | wzx | Vibrio cholerae VL426 | ZP_04414209.1 | 97 (342/351) |
| 8 | Asparagine synthetase | Vibrio cholerae VL426 | ZP_04414208.1 | 99 (592/596) | |
| 9 | Formyl transferase | Vibrio cholerae VL426 | ZP_04414207.1 | 100 (318/318) | |
| 10 | Conserved protein | Vibrio cholerae VL426 | ZP_04414206.1 | 83 (270/324) | |
| 11 | Polysaccharide deacetylase | Vibrio cholerae VL426 | ZP_04414205.1 | 93 (422/455) | |
| 12 | O-antigen polymerase | wzy | Vibrio cholerae VL426 | ZP_04414204.1 | 96 (369/382) |
| 13 | Glutamine amidotransferase | Psychrobacter cryohalolentis | YP_579889.1 | 63 (400/627) | |
| 14 | Glycosyl transferase | Vibrio cholerae VL426 | ZP_04414203.1 | 37 (143/379) | |
| 15 | d-Glucuronyl C5-epimerase | Vibrio cholerae VL426 | ZP_04414199.1 | 79 (246/310) | |
| 16 | Glycosyl transferases group I | wbpH | Vibrio cholerae VL426 | ZP_04414198.1 | 98 (362/367) |
| 17 | UDP-N-acetylglucosamine 2-epimerase | wecB | Vibrio cholerae VL426 | ZP_04414197.1 | 99 (356/359) |
| 18 | Glycosyl transferase | Vibrio cholerae VL426 | ZP_04414195.1 | 98 (240/244) | |
| 19 | Bacterial sugar transferase | wbfU-wcaJ | Vibrio cholerae VL426 | ZP_04414194.1 | 100 (204/204) |
| 20 | Putative acetyltransferase | wbfU-wcaJ | Vibrio cholerae VL426 | ZP_04414193.1 | 100 (233/233) |
| 21 | DegT/DnrJ/EryC1/StrS aminotransferase | wecE | Vibrio cholerae VL426 | ZP_04414192.1 | 98 (386/391) |
| 22 | Putative epimerase/dehydratase | wbfY | Vibrio cholerae | BAA33644.1 | 99 (559/562) |
| O108 | ADP-l-glycero-d-manno-heptose-6-epimerase | gmhD | |||
| 1 | Chain length determinant | wzz | Vibrio cholerae VL426 | ZP_04414215.1 | 87 (329/375) |
| 2 | dTDP-glucose 4,6-dehydratase | rmlB | Vibrio cholerae TMA21 | ZP_04402237.1 | 98 (348/354) |
| 3 | Glucose-1-phosphate thymidylyltransferase | rmlA | Shewanella sp. | YP_733455.1 | 81 (235/288) |
| 4 | Hypothetical protein | wblP | Photorhabdus luminiscens | NP_931970.1 | 72 (92/127) |
| 5 | Hypothetical protein | Shewanella pealeana | YP_001501263.1 | 70 (217/308) | |
| 6 | Aminotransferase | wecE | Vibrio cholerae O141 | YP_002072334.1 | 92 (352/379) |
| 7 | CDP-glycerol:poly(glycerophosphate) glycerophosphotransferase | Providencia rettgeri | ZP_03638641.1 | 46 (168/360) | |
| 8 | Pyruvate phosphate dikinase | Providencia rettgeri | ZP_03638640.1 | 51 (525/1016) | |
| 9 | Glutamine amidotransferase class I | Providencia rettgeri | ZP_03638639.1 | 53 (106/198) | |
| 10 | Putative sugar nucleotidyltransferase | rmlA | Providencia rettgeri | ZP_03638638.1 | 64% (160/250) |
| 11 | Lipopolysaccharide biosynthesis protein WzxC | Providencia rettgeri | ZP_03638637.1 | 58% (270/460) | |
| 12 | Beta-1,3-glucosyltransferase | Pseudomonas fluorescens | YP_002870154.1 | 30% (94/305) | |
| 13 | Polysaccharide polymerase | Lactobacillus plantarum | ZP_04012602.1 | 26% (64/239) | |
| 14 | Hypothetical protein | Vibrio cholerae VL426 | ZP_04414203.1 | 24% (39/158) | |
| 15 | Transposase | Vibrio cholerae | AAA76604.1 | 97% (215/221) | |
| 16 | Colanic acid biosynthesis glycosyl transferase | Bryantella formatexigens | ZP_03688482.1 | 45% (100/218) | |
| 17 | Glycosyl transferase, group 1/2 family protein | Bryantella formatexigens | ZP_03688482.1 | 38% (63/164) | |
| 18 | Putative LPS biosynthesis protein | Escherichia coli | AAV74532.1 | 76% (290/380) | |
| 19 | Imidazole glycerol phosphate synthase | Escherichia coli | AAX58765.1 | 59% (125/210) | |
| 20 | Imidazole glycerol phosphate synthase cyclase | Escherichia coli | AAX58766.1 | 69% (177/255) | |
| 21 | UDP-N-acetylglucosamine 4,6-dehydratase | Vibrio cholerae O141 | ZP_01982240.1 | 94% (325/345) | |
| 22 | Capsular polysaccharide synthesis enzyme Cap5F | Vibrio cholerae O1 | ZP_01982224.1 | 99% (365/368) | |
| 23 | UDP-N-acetylglucosamine 2-epimerase | wecB | Vibrio cholerae O1 | ZP_01982220.1 | 96% (363/376) |
| 24 | Putative l-fucosamine transferase | Vibrio vulnificus | ABD38629.1 | 73% (279/380) | |
| 25 | UDP-galactose 4-epimerase | galE | Vibrio cholerae | BAA33640.1 | 68% (217/317) |
| 26 | Lipid carrier:UDP-N-acetylgalactosaminyltransferase | wbfU | Vibrio cholerae | YP_002069533.1 | 94% (173/184) |
| 27 | Putative epimerase/dehydratase | wbfY | Vibrio cholerae | BAA33644.1 | 99% (557/562) |
The putative function of product, gene name, and organism were based on the function and name assigned to the gene sequence most similar to the ORF. The accession number is that of the gene most similar to the ORF. Similarity is given as a percentage, and the number of matches and length of the ORF and its closest match in GenBank are in parentheses.
Otherwise, there was a conspicuous absence of conserved genes and little conserved gene order in the V. cholerae wbf regions from the 16 serogroups. In O1 strains, the perosamine pathway genes are clustered at the beginning of the region, and tetronate genes also are clustered together (8). This type of arrangement also is seen in many enteric bacteria (37). All of the regions had a JUMPstart sequence adjacent to the gmhD-flanking gene (23). Nine serogroups had at least one gene from the dTDP-sugar biosynthesis pathways leading to l-rhamnose or other 6-deoxyhexose sugars. l-Rhamnose requires four rml genes that generally appear in the order rmlBADC (37). Li et al. (29) reported that these genes often are involved in recombination within V. cholerae if both donor and recipient have rml genes. In six V. cholerae isolates (O31, TMA21, O39, O12, 623-39, and O14), rmlBADC appear at the beginning of the region (Fig. 1), as was true in 9 of 11 partially sequenced regions (29). Of note, these authors did not report duplicated rml regions or isolated rml genes. However, the serogroup O31 has a second copy of rmlBADC comprising ORFs 33 to 36. This region has a total of 46 ORFs and could be formed by the incorporation of a second wbf cluster into the region. In addition, two other isolates (O5 and O108) have rmlAB, which could lead to a 6-deoxyhexose sugar other than l-rhamnose. Single rml genes also appear, apparently randomly, in O108, O12, 623-39, and O135. A pair of genes from the rjg end of the O1 biosynthetic pathway, galE and wbeW (red in Fig. 1), occupy a similar position in O37 and also are found in O139, O22, and O31. Four additional isolates have galE alone. Also primarily at the 3′ end, there is an epimerase/dehydratase/UDP-d-quinovosamine 4-dehydrogenase (wbfY; brown in Fig. 1) exhibiting 98% amino acid similarity between O5 (CO545), O8 (CO845), and O108 (CO603B); it is found more centrally in O139, O135, O141, 623-39, and VL426 strains. Finally, four serogroups start with wzz, but this gene also appears in the middle of O37, O139, and O22 serogroups.
E. coli and S. enterica use either the wzm-wzt or wzx-wzy-(wzz) O-antigen export system (36, 37). The export of heteropolymers generally uses the wzx system, with wzx encoding a flippase that transports O-subunits across the inner membrane, wzy is a polymerase, and wzz determines the chain length and is optional (37). Homopolymers use the wzm-wzt system. Serogroup O1 has wzm-wzt, as would be expected with the perosamine homopolymer backbone of O1. However, the mechanisms of O-antigen transport for other serogroups is much less clear. Both export systems appear in O31, again suggesting that this is a fusion of two regions. wzm is found in five strains but is accompanied by a recognizable wzt in only O1 and O31. wzm is accompanied by a recognizable wzz gene in O37, O139, and O22. wzx was found in O31, O5 (CO545), O8 (CO845), and VL426, while its common partner wzy was found in O31, O5 (CO545), O8 (CO845), O12, O135, VL426, and TMA 21. That more strains have wzy than wzx was surprising, because wzy often is recognized by predicted structure rather than sequence similarity (9). The absence of common patterns of genes inhibits the identification of classes, as has been done for E. coli, Salmonella, and S. pneumoniae (4, 36).
The heterogeneity and gene duplications seen in V. cholerae O-antigen regions may be due to frequent recombination within the region, resulting in random assortments of genes. Of note, in O108 (CO603B), there was a block of five genes (ORFs 7 to 11) that had best-hit similarities of 46 to 64% to genes from Providencia rettgeri. A boundary in similarities between regions is a defining observation for recombination or horizontal gene transfer (39). In addition, this same boundary phenomena, which is indicative of recombination, was found in five other blocks of contiguous genes in O5 (CO545) (ORFs 1-2 and 15-16) and O108 (CO603B) (ORFs 16, 17, 18, 19, and 20, and 23-24); in each the contiguous genes have the highest similarity to genes from a single source that is different from those genes outside the block.
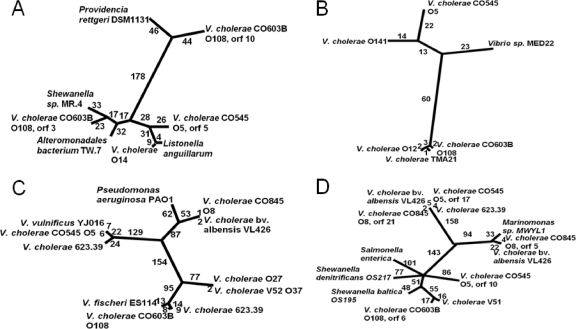
To look further at gene recombination, we examined the phylogenetic relationship of gene homologs. The rml loci not only had the most conserved location but also showed higher levels of similarity than others, so trees were constructed for rmlA and rmlB. Interestingly, O108 (CO603B) had two genes, rmlA (ORF 3) and one paralog, an rmlA-like (ORF 10) ORF, that share only 29% amino acid similarity (Fig. 1, Table 1). In the NJ tree (Fig. 2A), the O5 (CO545) rmlA amino acid sequence was more closely related to its V. cholerae O14 and Listonella anguillarum orthologs than to O108 (CO603B). The closest neighbor to O108 (CO603B) is a Shewanella sp. sequence. O108 (CO603B) rmlA-like ORF 10 and its most closely related sequences formed a branch separated by 368 amino acid differences from O5 (CO545) ORF 13 and by 363 amino acid differences from O108 (CO603B) ORF 3 clades, a result not unexpected with the low percent identity. Li et al. (29) showed that homologous recombination occurring within the rml genes maintained the entire rml cassette structure at its location in the beginning of the wbf region; we showed that single rml genes may be found elsewhere in the region.
FIG. 2.
Unrooted phylogram showing the relatedness of similar genes from different LPS regions. The number of amino acid changes is indicated along each branch. (A) rmlA; (B) rmlB; (C) UDP-N-acetylglucosamine 2-epimerase; (D) DegT/DnrJ/EryC1/StrS aminotransferase. In each case, sequences from V. cholerae isolates were closer to homologous sequences from other species than to those from other V. cholerae isolates. Furthermore, each pair of paralogs was separated by more than 270 amino acid changes.
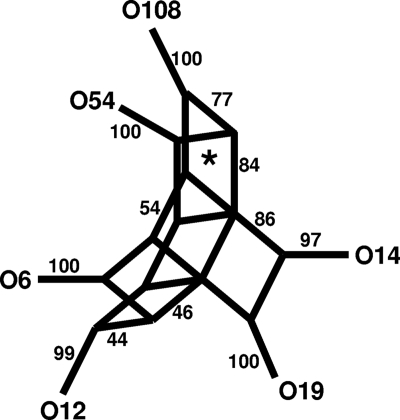
The two rmlB sequences in O108 (CO603B) and O5 (CO545) are highly divergent (Fig. 2B). However, rmlB from O108 (CO603B) was closely related to rmlB in V. cholerae TMA 21, a non-O1/O139 strain, and V. cholerae O12. Additional V. cholerae loci with a high degree of similarity to the O108 (CO603B) and O12 rmlB have been described (29). These loci had sufficient nucleotide similarity to permit split decomposition analysis. Split decomposition was not used for other alleles and loci, because they were too divergent and similarities were apparent only at the amino acid level. The split decomposition method was applied to test for the presence of recombination within the gene between alleles. The computed split graphs based on the analysis of six rmlB sequences (Fig. 3) shows seven parallelograms between the strains, which is indicative of recombination events. The analysis was sound because the fit was high, and the bootstrapping values were 76.9 and 83.6% on two sides of one parallelogram. Thus, at least one of the parallelograms is supported statistically. Since Li et al. (29) identified the rmlABCD genes as a hot spot for recombination, additional support for recombination might have been expected from our analysis. However, in their study, 50% (5 of 10) of the recombination events were in rmlC and none in the rmlB gene, the gene we examined, perhaps accounting for the perceived difference.
FIG. 3.
Splitstree cladogram of selected rmlB genes. The numbers along the edges of the cladogram indicate the proportion of times that an edge occurred in 1,000 bootstrap replicates. The asterisk identified the parallel change support by bootstrapping in more than 84% replicates on one edge and 77% on the other.
Sequences for UDP-N-acetylglucosamine 2-epimerase (wecB; pink in Fig. 1) were also most similar to homologous genes from other species. UDP-N-acetylglucosamine 2-epimerase was found in all three strains (ORF 16 in O5 [CO545], ORF 6 in O8 [CO845], and ORF 5 in O108 [CO603B]), but the amino acid similarity is only in the range of 28 to 32%. The unrooted NJ phylogram of UDP-N-acetylglucosamine 2-epimerase (Fig. 2C) revealed that the O5 (CO545) sequence was closely related to an ortholog from V. vulnificus and to one of two paralogs from a non-O1/O139 strain, V. cholerae 623-39. These three sequences were only distantly related to the O8 (CO845) and O108 (CO603B) sequences. The O8 (CO845) sequence was 99% similar to V. cholerae bv. albensis VL426 and related to Pseudomonas aeruginosa, while the O108 (CO603B) sequence was closer to the other paralog from the non-O1/O139 strain V. cholerae 623-39 and the O37 sequence from V. cholerae strain V52.
Extensive diversity was noted in the five putative aminotransferase genes (Fig. 2D). ORFs 10 and 17 in O5 (CO545), ORFs 5 and 21 in strain O8 (CO845), and ORF 6 in O108 (CO603B) were identified as members of the DegT/DnrJ/EryC1/StrS aminotransferase family of proteins. ORF 17 from O5 (CO545) and ORF 21 from O8 (CO845) had 98% similarity and were closely related to genes from one paralog of V. cholerae non-O1/O139 strain 623-39 and an ortholog in V. cholerae bv. albensis VL426. In contrast, the other ORFs all were less than 50% similar to each other. ORFs 10, 5, and 6 from O5 (CO545), O8 (CO845), and O108 (CO603B), respectively, matched aminotransferase-encoding genes from Shewanella denitrificans (YP_563660.1), the other paralog of V. cholerae bv. albensis (ZP_04414211.1), and V. cholerae V51 (YP_002072334.1), with amino acid similarity in the range of 62 to 96%. The paralogs (ORFs 10 and 17) found in O5 (CO545) have only 28% amino acid similarity, while those in O8 (CO845), ORF 21 and ORF 5, have only 25% similarity. These paralogs were expected to belong to different pathways, but as yet our data cannot delineate the pathway involved in the production of a particular sugar.
The diversity shown by V. cholerae was greater than that seen in either S. pneumoniae or E. coli. In contrast to V. cholerae, where no genes were found in all 16 sequenced wbf regions, all 90 known serotypes of S. pneumoniae had six genes in common: four (wzg, wzh, wzd, and wze) were almost always at the 5′ end, and two genes, wzy and wzx, were always present together downstream (4). In V. cholerae, despite having 16 sequences for the wbf region, we could not define subtypes based on the wzm-wzt or wzx-wzy transport system, as was done for E. coli, because the pairs of transport genes were not always found together and sometimes were absent entirely.
In summary, our data show that V. cholerae shares at least portions of pathways identified in other Gram-negative bacteria, and genes from both the wzm-wzt and wzx-wzy transport systems were found either together or at random in the wbf regions. There is minimal identity between nucleotide sequences of some homologs, and there frequently is greater similarity to polysaccharide biosynthesis genes from species other than V. cholerae. There also are gene duplications in several strains. These data highlight the diversity of V. cholerae O-antigen genes, in keeping with the relatively large number of serogroups identified to date, and suggest that future antigenic changes in the epidemic lineage are likely to occur.
Acknowledgments
This research was supported in part by an NIH grant (RO1 GM060791) to J. G. Morris and the University of Maryland Clinical Research Unit of the Food and Waterborne Diseases Integrated Research Network, which is funded by the National Institute of Allergy and Infectious Diseases, National Institutes of Health, under contract number N01-AI-40014.
Footnotes
Published ahead of print on 11 February 2011.
REFERENCES
- 1.Albert, M. J., et al. 1993. Large outbreak of clinical cholera due to Vibrio cholerae non-O1 in Bangladesh. Lancet 341:704. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., et al. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bateman, A., E. Birney, et al. 2002. The Pfam protein families database. Nucleic Acids Res. 30:276-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bentley, S. D., et al. 2006. Genetic analysis of the capsular biosynthetic locus from all 90 pneumococcal serotypes. PLoS Genet 2:e31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bik, E. M., et al. 1995. Genesis of the novel epidemic Vibrio cholerae O139 strain: evidence for horizontal transfer of genes involved in polysaccharide synthesis. EMBO J. 14:209-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blokesch, M., and G. K. Schoolnik. 2007. Serogroup conversion of Vibrio cholerae in aquatic reservoirs. PLoS Pathog. 3:e81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Campos, J., E. Martinez, et al. 2010. VEJφ, a novel filamentous phage of Vibrio cholerae able to transduce the cholera toxin genes. Microbiology 156(Pt 1):108-115. [DOI] [PubMed] [Google Scholar]
- 8.Chatterjee, S. N., and K. Chaudhuri. 2004. Lipopolysaccharides of Vibrio cholerae II. Genetics of biosynthesis. Biochim. Biophys. Acta 1690:93-109. [DOI] [PubMed] [Google Scholar]
- 9.Chen, Y., et al. 2007. The capsule polysaccharide structure and biogenesis for non-O1 Vibrio cholerae NRT36S: genes are embedded in the LPS region. BMC Microbiol. 7:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chenna, R., et al. 2003. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 31:3497-3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chun, J., et al. 2009. Comparative genomics reveals mechanism for short-term and long-term clonal transitions in pandemic Vibrio cholerae. Proc. Natl. Acad. Sci. U. S. A. 106:15442-15447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Comstock, L. E., et al. 1996. Cloning and sequence of a region encoding a surface polysaccharide of Vibrio cholerae O139 and characterization of the insertion site in the chromosome of Vibrio cholerae O1. Mol. Microbiol. 19:815-826. [DOI] [PubMed] [Google Scholar]
- 13.Cox, A. D., et al. 1997. Structural analysis of the lipopolysaccharide from Vibrio cholerae serotype O22. Carbohydr. Res. 304:191-208. [DOI] [PubMed] [Google Scholar]
- 14.Dumontier, S., and P. Berche. 1998. Vibrio cholerae O22 might be a putative source of exogenous DNA resulting in the emergence of the new strain of Vibrio cholerae O139. FEMS Microbiol. Lett. 164:91-98. [DOI] [PubMed] [Google Scholar]
- 15.Dziejman, M., et al. 2002. Comparative genomic analysis of Vibrio cholerae: genes that correlate with cholera endemic and pandemic disease. Proc. Natl. Acad. Sci. U. S. A. 99:1556-1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ewing, B., and P. Green. 1998. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 8:186-194. [PubMed] [Google Scholar]
- 17.Ewing, B., et al. 1998. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 8:175-185. [DOI] [PubMed] [Google Scholar]
- 18.Faruque, S. M., et al. 1998. Epidemiology, genetics, and ecology of toxigenic Vibrio cholerae. Microbiol. Mol. Biol. Rev. 62:1301-1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Faruque, S. M., et al. 2005. Seasonal epidemics of cholera inversely correlate with the prevalence of environmental cholera phages. Proc. Natl. Acad. Sci. U. S. A. 102:1702-1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.González-Fraga, S., et al. 2008. Lateral gene transfer of O1 serogroup encoding genes of Vibrio cholerae. FEMS Microbiol. Lett. 286:32-38. [DOI] [PubMed] [Google Scholar]
- 21.Hava, D. L., and A. Camilli. 2001. Isolation and characterization of a temperature-sensitive generalized transducing bacteriophage for Vibrio cholerae. J. Microbiol. Methods 46:217-225. [DOI] [PubMed] [Google Scholar]
- 22.Hermansson, K., P.-E. Jansson, T. Holme, and B. Gustavsson. 1993. Structural studies of the Vibrio cholerae O:5 O-antigen polysaccharide. Carbohydr. Res. 248:199-211. [DOI] [PubMed] [Google Scholar]
- 23.Hobbs, M., and P. R. Reeves. 1994. The JUMPstart sequence: a 39 bp element common to several polysaccharide gene clusters. Mol. Microbiol. 12:855-856. [DOI] [PubMed] [Google Scholar]
- 24.Huson, D. H., and D. Bryant. 2006. Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 23:254-267. [DOI] [PubMed] [Google Scholar]
- 25.Jiang, X. M., et al. 1991. Structure and sequence of the rfb (O antigen) gene cluster of Salmonella serovar typhimurium (strain LT2). Mol. Microbiol. 5:695-713. [DOI] [PubMed] [Google Scholar]
- 26.Kocharova, N. A., et al. 2001. Structural studies of the O-specific polysaccharide of Vibrio cholerae O8 using solvolysis with triflic acid. Carbohydr. Res. 330:83-92. [DOI] [PubMed] [Google Scholar]
- 27.Kondo, S., and K. Hisatsune. 1989. Sugar composition of the polysaccharide portion of lipopolysaccharides isolated from non-O1 Vibrio cholerae O2 to O41, O44, and O68. Microbiol. Immunol. 33:641-648. [DOI] [PubMed] [Google Scholar]
- 28.Kondo, S., Y. Kawamata, Y. Sano, T. Iguchi, and K. Hisatsune. 1997. A chemical study of the sugar composition of the polysaccharide portion of lipopolysaccharides isolated from Vibrio cholerae non-01 from O2 to O155. Syst. Appl. Microbiol. 20:1-11. [Google Scholar]
- 29.Li, Q., M. Hobbs, et al. 2003. The variation of dTDP-l-rhamnose pathway genes in Vibrio cholerae. Microbiology 149(Pt 9):2463-2474. [DOI] [PubMed] [Google Scholar]
- 30.Manning, P. A., et al. 1995. Putative O-antigen transport genes within the rfb region of Vibrio cholerae O1 are homologous to those for capsule transport. Gene 158:1-7. [DOI] [PubMed] [Google Scholar]
- 31.Mooi, F. R., and E. M. Bik. 1997. The evolution of epidemic Vibrio cholerae strains. Trends Microbiol. 5:161-165. [DOI] [PubMed] [Google Scholar]
- 32.Nair, G. B., et al. 1994. Spread of Vibrio cholerae O139 Bengal in India. J. Infect. Dis. 169:1029-1034. [DOI] [PubMed] [Google Scholar]
- 33.Nesper, J., et al. 2000. Characterization of Vibrio cholerae O1 antigen as the bacteriophage K139 receptor and identification of IS1004 insertions aborting O1 antigen biosynthesis. J. Bacteriol. 182:5097-5104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ramamurthy, T., et al. 1993. Virulence patterns of Vibrio cholerae non-O1 strains isolated from hospitalised patients with acute diarrhoea in Calcutta, India. J. Med. Microbiol. 39:310-317. [DOI] [PubMed] [Google Scholar]
- 35.Reeves, P. 1993. Evolution of Salmonella O antigen variation by interspecific gene transfer on a large scale. Trends Genet. 9:17-22. [DOI] [PubMed] [Google Scholar]
- 36.Reeves, P. P., and L. Wang. 2002. Genomic organization of LPS-specific loci. Curr. Top. Microbiol. Immunol. 264:109-135. [PubMed] [Google Scholar]
- 37.Samuel, G., and P. Reeves. 2003. Biosynthesis of O-antigens: genes and pathways involved in nucleotide sugar precursor synthesis and O-antigen assembly. Carbohydr. Res. 338:2503-2519. [DOI] [PubMed] [Google Scholar]
- 38.Sozhamannan, S., et al. 1999. Cloning and sequencing of the genes downstream of the wbf gene cluster of Vibrio cholerae serogroup O139 and analysis of the junction genes in other serogroups. Infect. Immun. 67:5033-5040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stephens, J. C. 1985. Statistical methods of DNA sequence analysis: detection of intragenic recombination or gene conversion. Mol. Biol. Evol. 2:539-556. [DOI] [PubMed] [Google Scholar]
- 40.Stine, O. C., et al. 2000. Phylogeny of Vibrio cholerae based on recA sequence. Infect. Immun. 68:7180-7185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stroeher, U. H., et al. 1995. Genetic rearrangements in the rfb regions of Vibrio cholerae O1 and O139. Proc. Natl. Acad. Sci. U. S. A. 92:10374-10378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Swofford, D. 2003. PAUP. Sinauer Associates, Sutherland, MA.
- 43.Tobin-D'Angelo, M., et al. 2008. Severe diarrhea caused by cholera toxin-producing Vibrio cholerae serogroup O75 infections acquired in the southeastern United States. Clin. Infect. Dis. 47:1035-1040. [DOI] [PubMed] [Google Scholar]
- 44.Waldor, M. K., et al. 1994. The Vibrio cholerae O139 serogroup antigen includes an O-antigen capsule and lipopolysaccharide virulence determinants. Proc. Natl. Acad. Sci. U. S. A. 91:11388-11392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Whitfield, C., and I. S. Roberts. 1999. Structure, assembly and regulation of expression of capsules in Escherichia coli. Mol. Microbiol. 31:1307-1319. [DOI] [PubMed] [Google Scholar]
- 46.Yamasaki, S., et al. 1999. The genes responsible for O-antigen synthesis of vibrio cholerae O139 are closely related to those of Vibrio cholerae O22. Gene 237:321-332. [DOI] [PubMed] [Google Scholar]