Abstract
Kimchi, a traditional food in the Korean culture, is made from vegetables by fermentation. In this study, metagenomic approaches were used to monitor changes in bacterial populations, metabolic potential, and overall genetic features of the microbial community during the 29-day fermentation process. Metagenomic DNA was extracted from kimchi samples obtained periodically and was sequenced using a 454 GS FLX Titanium system, which yielded a total of 701,556 reads, with an average read length of 438 bp. Phylogenetic analysis based on 16S rRNA genes from the metagenome indicated that the kimchi microbiome was dominated by members of three genera: Leuconostoc, Lactobacillus, and Weissella. Assignment of metagenomic sequences to SEED categories of the Metagenome Rapid Annotation using Subsystem Technology (MG-RAST) server revealed a genetic profile characteristic of heterotrophic lactic acid fermentation of carbohydrates, which was supported by the detection of mannitol, lactate, acetate, and ethanol as fermentation products. When the metagenomic reads were mapped onto the database of completed genomes, the Leuconostoc mesenteroides subsp. mesenteroides ATCC 8293 and Lactobacillus sakei subsp. sakei 23K genomes were highly represented. These same two genera were confirmed to be important in kimchi fermentation when the majority of kimchi metagenomic sequences showed very high identity to Leuconostoc mesenteroides and Lactobacillus genes. Besides microbial genome sequences, a surprisingly large number of phage DNA sequences were identified from the cellular fractions, possibly indicating that a high proportion of cells were infected by bacteriophages during fermentation. Overall, these results provide insights into the kimchi microbial community and also shed light on fermentation processes carried out broadly by complex microbial communities.
Kimchi is a traditional food, emblematic of Korean culture, that is fermented from vegetables such as Chinese cabbage and radish. In recent years, kimchi's health-promoting characteristics have been recognized (40, 54). Kimchi is usually processed with various seasonings, such as red pepper powder, garlic, ginger, green onion, fermented seafood (jeotgal), and salts, at low temperatures to ensure proper ripening and preservation. Spontaneous fermentation without the use of starter cultures or sterilization leads to the growth of various microorganisms during kimchi preparation. Moreover, the influence of ingredients and fermentation conditions on the microbial community is unexplored, and a rational approach to the control of the microbial community for the improvement of kimchi flavor is currently almost impossible.
Microbes involved in kimchi fermentation have been studied previously (25, 26). Taxonomic studies of bacteria isolated and purified from kimchi fermenting cultures have shown that lactic acid bacteria (LAB), including the Leuconostoc species Le. mesenteroides, Le. kimchii, Le. citreum, Le. gasicomitatum, and Le. gelidum, the Lactobacillus species Lb. brevis, Lb. curvatus, Lb. plantarum, and Lb. sakei, Lactococcus lactis, Pediococcus pentosaceus, Weissella confusa, Weissella kimchii, and Weissella koreensis, are likely to be key players responsible for kimchi fermentation (2, 21, 23, 27, 42, 51). However, culture-based approaches have known limitations in terms of reproducibility and challenges in the culturability of some bacteria. Therefore, the set of cultured isolates may not reflect the true microbial composition of fermented kimchi.
The use of 16S rRNA genes as a phylogenetic marker that does not require isolation and cultivation of microorganisms has resulted in a tremendous amount of information about naturally occurring microbial communities in various habitats (8, 48). When applied to kimchi, the construction of 16S rRNA clone libraries has extended information about kimchi microbiology (42). Denaturing gradient gel electrophoresis, amplified ribosomal DNA restriction analysis, and the subsequent sequencing of 16S rRNA genes were additional techniques used to elucidate the microbial community involved in kimchi fermentation (23, 27). Although these culture-independent approaches can estimate species richness within a kimchi microbial community, they are unable to resolve the true genotypic diversity, because diversity of 16S rRNA gene sequences may not correlate well with genotypic and phenotypic diversity (60). Thus, although progress made using 16S RNA gene analysis and culture-based methods has been significant, current understanding of kimchi microbiology is piecemeal and has significant limitations regarding community dynamics and function during fermentation (12, 28, 38).
Metagenomic approaches based on the random sequencing of environmental DNA can provide a wealth of information (free of PCR bias) about gene content, metabolic potential, and the function of microbial communities (9). Recently, the development of next-generation DNA sequencing technologies (50), such as 454 pyrosequencing, has led to the large-scale metagenomic sequencing of environmental habitats, such as activated sludge (15, 49), the coastal ocean (36), glacier ice (52), and the human distal gut (45).
It is well known that kimchi fermentation is generally characterized by the production of metabolites, such as organic acids (lactic and acetic acids) and other flavoring compounds, which are important components in defining kimchi quality (19). The presence of various metabolites can reflect the presence of related genes in the microbial population and give a more direct collective phenotypic view of the kimchi microbiome as a consequence of the end products of gene expression. The fermentation rate is also very important to kimchi flavor development during fermentation (24). 1H nuclear magnetic resonance (1H-NMR) is one of the most comprehensive, easy, and nondestructive multinuclear techniques to simultaneously monitor several substances present in one sample (14, 29).
Here, we applied metagenomic approaches using pyrosequencing technology developed by 454 Life Sciences to kimchi. In addition, we conducted a metabolite analysis of fermenting kimchi using 1H-NMR to collectively understand the overall features of the kimchi microbiome and to investigate relationships between metabolite production, metabolic potential, and composition of the kimchi microbial community during fermentation.
MATERIALS AND METHODS
Preparation of kimchi and sampling.
An industrial-scale batch of kimchi was prepared by Daesang F&F Corporation using their production line. Briefly, Chinese cabbages (Brassica rapa subsp. pekinensis) were soaked in a solution of 15% (wt/vol) solar salt for 5 to 6 h. The soaked Chinese cabbages were washed with tap water three times and drained of excessive water. These salted cabbages were then mixed with various seasonings, including red pepper powder, radish, garlic, ginger, green onion, sugar, and fermented seafood (jeotgal), and were packaged into airtight plastic bags in 1-kg portions. Thirty 1-kg kimchi bags were stored at the standard fermentation temperature (4°C). Periodically over the 29-day fermentation period, three bags were sacrificed, the contents were filtered through three layers of coarse gauze (Daehan, Republic of Korea) to remove only large kimchi particles, and the filtrates were pooled. The pH of each filtrate was measured immediately, and the filtrates were centrifuged (8,000 rpm for 20 min at 4°C) to harvest microorganisms. The pellets and supernatants were stored at −80°C for extraction of bulk genomic DNA and metabolite analysis, respectively. One ml of each sample was centrifuged separately for the measurement of 16S rRNA gene copies using quantitative real time-PCR (qRT-PCR). The kimchi samples were identified as Jx, where “x” designates the sample day during fermentation.
qRT-PCR to determine 16S rRNA gene copy number.
To estimate the total number of bacteria during the kimchi fermentation, total genomic DNA from pellets derived from 1.0 ml of kimchi filtrate by centrifugation was extracted using a FastDNA Spin kit (MPbio, Solon, OH) according to the manufacturer's instructions. Genomic DNA concentrations were measured using an enzyme-linked immunosorbent assay (ELISA) reader equipped with a Take3 multivolume plate (SynergyMx; BioTek, CA). The genomic DNA was serially diluted in a range of from 50 μg/ml to 200 μg/ml and was used as the template to amplify 16S rRNA genes from each group of bacteria using primers 340F (5′-CCTACGGGAGGCAGCAG-3′) and 758R (5′-CTACCAGGGTATCTAATCC-3′) (20). The PCR mixture consisted of 2 μl of genomic DNA, 0.3 μM primer concentrations (for 340F and 758R, each), 10 μl of 2× iQ SYBR Green Supermix (Bio-Rad, Hercules, CA), and water for a total volume of 20 μl. The quantitative real-time PCR (qRT-PCR) amplifications were conducted using a CFX96 real-time PCR system (Bio-Rad, Hercules, CA) with the following qRT-PCR conditions: initial denaturation at 95°C for 3 min, followed by 40 cycles of 95°C for 45 s, 59°C for 45 s, and 72°C for 50 s. Bacterial standard curves were generated on the basis of the concentration of a pCR2.1 vector (Invitrogen, CA) carrying the 16S rRNA gene of Leuconostoc mesenteroides subsp. mesenteroides ATCC 8293T. The 16S rRNA gene copy number of each sample was calculated as described previously (41, 46).
1H-NMR spectroscopic analysis of kimchi metabolites.
The analysis of metabolites, including organic acids and monosaccharides, in kimchi supernatants during fermentation was performed using 1H-NMR spectroscopy according to a modification of the methods described previously (29, 53). Briefly, 15 ml of kimchi supernatant without cells was adjusted to pH 6.0 and then lyophilized. The freeze-dried powders were dissolved in 600 μl of 10% (wt/vol) D2O with 0.5 mM sodium 2,2-dimethyl-2-silapentane-5-sulfonate (DSS) and centrifuged at 13,000 rpm for 5 min. The resulting supernatants were transferred into 5-mm NMR tubes, and 1H-NMR spectra were acquired using a Varian Inova 600-MHz NMR spectrometer (Varian, Inc., Palo Alto, CA). Identification and quantification of individual metabolites was performed using the Profiler module of the Chenomx NMR suite, v. 6.1 (Chenomx, Inc., Edmonton, Alberta, Canada).
Genomic DNA extraction and pyrosequencing.
Total genomic DNA from pellets was extracted using a FastDNA Spin kit (MPbio) according to the manufacturer's instructions. The extracted crude DNA was loaded on 1% (wt/vol) low-melting agarose (Promega, WI) and electrophoresed at 100 V for 1 h to remove impurities. A strip from each side of the gel was removed and stained to localize the genomic DNA. DNA-containing regions from the rest of the unstained gel were removed and digested with β-agarase I, and DNA recovery was conducted as described by the manufacturer (NEB, MA). The concentration of purified genomic DNA was measured with an ELISA reader equipped with a Take3 multivolume plate (SynergyMx; BioTek, CA), and the DNA samples were concentrated to a minimum of 100 ng/μl using a speed vacuum centrifuge (Eyela CVE-2000; Japan). The total genomic DNA was nebulized to obtain a mean fragment size of ∼680 bp, confirmed by electrophoresis, for pyrosequencing. The fragmented genomic DNA was individually tagged using multiplex identifier (MID) adaptors according to the manufacturer's instructions (Roche, Mannheim, Germany), which allowed for the automatic sorting of the pyrosequencing-derived sequencing reads based on MID adaptors. Pyrosequencing of the 10 tagged genomic DNA samples was performed by Macrogen (Korea) using a 454 GS FLX Titanium system (Roche, Mannheim, Germany).
Sequencing data sets were subjected to systematic checks to remove replicates, duplicates, and low-quality reads. Briefly, replicate and duplicate sequences, which are known artifacts of the pyrosequencing approach, were first checked through a Web-based program (http://microbiomes.msu.edu/replicates/) (17) using the default settings (sequence identity cutoff, 0.9; length identity requirement, 0; number of beginning base pairs to check, 3) of the open-source program Cd-hit (30). Replicates and duplicates were removed, and the remaining sequence data were then trimmed using the LUCY program (6) with a stringency similar to that for Phred scores of 20 or higher (error, 0.01) and with long reads being those over 100 bp. Only high-quality sequence data obtained after LUCY trimming were used for further subtraction with SeqClean (http://compbio.dfci.harvard.edu/tgi/software/). Resulting clean sequence data sets were then used for further analysis.
Comparative analysis of metagenomic data.
Unassembled clean reads were annotated by Metagenome Rapid Annotation using Subsystem Technology (MG-RAST) server (http://metagenomics.nmpdr.org/), an open-access metagenome curation and analysis platform (35). To analyze bacterial community changes during kimchi fermentation, 16S rRNA gene sequences from all sequences were identified by comparing the read data set against the Ribosomal Data Project II (RDP II) database (7), using recommended parameters (minimum alignment length, ≥50 bp; E-value cutoffs, 0.01). 16S rRNA gene phylotypes were classified at the genus, family, order, class, and phylum levels based on the respective taxonomic categorizations of the MG-RAST server. To identify putative protein-encoding sequences, the sequences were also analyzed using the SEED platform (http://www.theseed.org/), which comprises all available genomic data from genome sequencing centers (1). Kimchi microbial community metabolic potential was determined by assigning functional annotation to metagenomic sequences with subsequent sequence assignment to SEED subsystems using the recommended parameters (minimum alignment length, ≥50 bp; E-value cutoffs, 0.01). Statistical analyses were conducted using the number of sequencing reads within each subsystem rather than using the number of total metagenomic reads. Therefore, the abundance of metabolic genes in the MG-RAST server was calculated based on the relative proportion of sequencing reads within each subsystem. The metabolic potential of the kimchi microbiome was compared with that in other metagenomic data sets on the MG-RAST server.
Individual genome recruitment plots.
The degree to which completed, whole-genome sequences were represented in metagenomic reads was assessed using the “recruitment plot” procedure described by Ghai et al. (16). Sequence fragments from unassembled clean reads derived from a 454 GS FLX Titanium system were compared to all complete and draft microbial genomes using the BLASTN algorithm. Whole clean metagenomic reads were matched to multiple reference genomes and were matched multiple times to the same genome (16). The criteria for counting a given sequence were a minimum identity of 95% and a minimum alignment of 90 bp. Whole clean reads were multiply matched to the four highest-scoring genomes: Leuconostoc mesenteroides subsp. mesenteroides ATCC 8293T (NC_008531), Lactobacillus sakei 23K sakei (NC_007576), the partially assembled genome of Weissella paramesenteroides ATCC 33313, and Leuconostoc citreum KM20 (NC_010471). The data were plotted using the gnuplot program (http://www.gnuplot.info/).
Assembly of pyrosequencing-derived reads and analysis of assembled contigs.
Pyrosequencing-derived sequencing reads were assembled by Macrogen (Korea) using 454 Newbler Assembler software (ver. 2.0.01.14; Roche). Low-complexity sequence regions (simple sequence repeats) were identified and excluded from consideration during initial pair-wise comparison but were included during final alignment and consensus building. Assembled contigs longer than 10 kb were assigned to their taxonomic affiliations by BLASTN comparisons to the GenBank nonredundant nucleotide (nr/nt) database and selections of the top BLASTN hits for phylogenetic affiliation as described previously (43, 44). Clustered regularly interspaced short palindromic repeats (CRISPR) elements were searched from assembled contigs longer than 10 kb by using the Web-based tool CRISPRFinder (18).
Phylogenetic analysis of phage-related contigs and analysis of their abundance.
Four contigs (contigs 01645, 27827, 02778, and 03545), assigned as putative bacteriophage genome fragments among the assembled contigs, were analyzed in detail. Molecular phylogenetic analysis of the putative phage contigs was performed using bacteriophage major capsid proteins (MCPs). Putative MCP sequences were aligned with homologous MCP sequences and retrieved from the NCBI database using the MUSCLE program (11). Phylogenetic trees were constructed using MEGA software (ver. 4.0) based on deduced amino acid sequences (55). The abundance of putative phage reads in the cellular fractions of kimchi supernatants during fermentation was estimated by multiple matching of the metagenomic data sets against the four putative phage contigs, with the criteria of a minimum percent identity of 95% and a minimum overlap of 90 bp as individual genome recruitment plots, which was performed as described above.
Nucleotide sequence accession numbers.
The kimchi metagenomic data sets are publicly available in the MG-RAST system under project identifiers 4450209.3 to 4450218.3 and in the NCBI Short Read Archive under accession no. SRA023444. Assembled contig sequences with more than 10 kb have been deposited in GenBank under accession numbers AEKF01000001 to AEKF01000127.
RESULTS AND DISCUSSION
Metagenomics makes it possible to study microbial communities by deciphering genetic information from DNA extracted directly from naturally occurring habitats. Various metagenomic approaches have been applied to environmental samples to characterize the microbial and viral communities present in an ecosystem, thereby elucidating metabolic capabilities and native taxa (10, 47, 56, 57, 59). Advantages of metagenomic studies include avoiding the need to culture individual microorganisms, eliminating cloning and PCR biases, and reducing the cost per sequencing read. At present, the 454 GS FLX Titanium platform generates relatively long read lengths (400 to 500 bp), which make it possible to reliably assemble metagenomic reads. Here, we applied pyrosequencing technology to analyze the microbiome of kimchi, the most popular fermented food in Korea. In addition, we analyzed the metabolites of fermenting kimchi using 1H-NMR to investigate the link between the kimchi microbial community metagenome and the phenotype.
General features of kimchi fermentation.
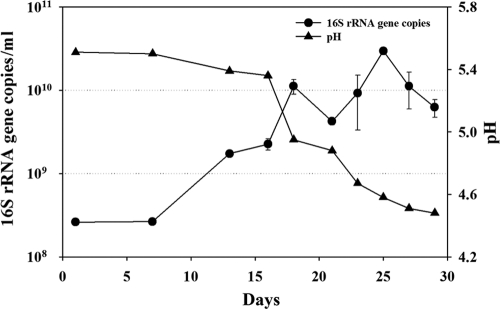
Kimchi fermentation is an anaerobic process, and sampling kimchi supernatants from one large fermentation batch can cause oxygen infiltration, disturbing the normal fermentation process and thereby influencing the microbial community and metabolite production. Therefore, 30 sealed bags, each containing 1 kg of kimchi that was manufactured by a one-batch process (incubated at the standard temperature of 4°C), were prepared for metagenome and metabolite analysis. The pH profile of kimchi supernatants over the 30 day-fermentation process (Fig. 1) was similar to those of typical kimchi fermentation (41). In this study, the pH was about 5.5 at the beginning and decreased sharply after 16 days. After 27 days of fermentation, the pH reached below 4.5 and became stable.
FIG. 1.
Change in pH and 16S rRNA gene copy number for total bacterial numbers during kimchi fermentation. Measurements of 16S rRNA gene copy numbers were performed independently in triplicate. Error bars represent the standard deviations.
A qRT-PCR approach was used to enumerate the total number of bacteria present during the kimchi fermentation process. The exact determination of bacterial cell numbers by qRT-PCR in a microbial community is impossible because the copy numbers of chromosomal 16S rRNA gene operons vary with species type (13); however, the data do allow for an estimation of the cell numbers. In our study, the total number of bacteria in kimchi samples was estimated using a standard curve (R2, 0.999) prepared from the 16S rRNA gene of Leuconostoc mesenteroides subsp. mesenteroides ATCC 8293. The 16S rRNA gene copy numbers of kimchi samples increased from an initial value (sample J1, day 1) of 2.6 × 108 copies/ml to the highest value (sample J25, day 25) of 3.0 × 1010 copies/ml during the kimchi fermentation. After day 25, the bacterial population decreased steadily. The increase of bacterial abundance correlated well with a decrease in pH value (Fig. 1).
Identification and quantification of metabolites during the kimchi fermentation process.
It is known that kimchi flavor is dependent principally on free sugars, amino acids, and organic acids, production of which are surely influenced by the microbial community during fermentation (19). Therefore, we analyzed the change of metabolites and microbial communities during kimchi fermentation using 1H-NMR spectroscopy to investigate their relationships. Identification and quantification of metabolites using 1H-NMR peaks were performed based on comparisons with chemical shifts of standard compounds using Chenomx NMR suite software. 1H-NMR spectra of the kimchi supernatants were associated with the presence of various metabolites, including carbohydrates, amino acids, and organic acids, as expected. In particular, intense parts of the 1H-NMR spectra were in the 0.5- to 2.0-ppm and 3.0- to 5.0-ppm ranges, corresponding to organic acids and carbohydrates, respectively (data not shown).
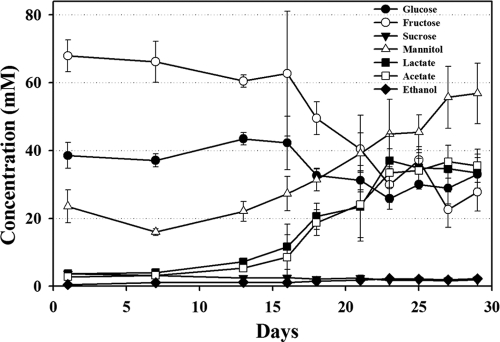
In this study, fructose and glucose were detected as major free sugars (Fig. 2). Free sugars play important roles in the taste of kimchi because free sugars are not only sweeteners but also serve as carbon sources for microorganisms, including LAB, to produce various products. The level of free sugars decreased slowly in the early phases; between days 16 and 23, their concentrations decreased relatively quickly. After day 23, carbohydrate levels were relatively constant (Fig. 2). With the decrease in free sugars, lactate, acetate, and ethanol were detected as fermentation products, indicating that heterotrophic lactic acid fermentation occurred during fermentation. It was also found that a considerable amount of mannitol (Fig. 2), a naturally occurring six-carbon polyol produced from the reduction of fructose by LAB in vegetable fermentations (34, 61), accumulated during the fermentation. The presence of mannitol in foods results in a refreshing taste, and mannitol has noncariogenic properties and is a good replacement for sugars in diabetic foods (61). In general, the profile of fermentation products inversely correlated with the level of free sugars. However, the level of fermentation products, especially mannitol, increased continually at the early and late phases, and free sugars were relatively constant; this constant level might be due to the continuous liberation of free sugar from cabbage. The change in free sugars and fermentation products were closely correlated with pH changes and growth of the bacterial populations (Fig. 1). However, ethanol levels were very low, which might be explained by the loss of ethanol during sample preparation (freeze drying) for NMR analysis.
FIG. 2.
Quantification of identified key metabolites in kimchi supernatants during fermentation by 1H-NMR. Data are given as means ± standard errors, calculated from three replicates. Quantification was determined using the Chenomx NMR suite, v. 6.1, with 2,2-dimethyl-2-silapentane-5-sulfonate (DSS) as the internal standard.
Statistics of sequences generated by 454 pyrosequencing.
Pyrosequencing of 10 kimchi samples during fermentation yielded a total of 701,566 reads by a single run of 10 tagged samples (Table 1). However, there was considerable variation in the number of sequences obtained from each 454 sequencing sample, which could be explained by a difference in the amount of genomic DNA from each sample, even though the total amount of DNA was adjusted before the pooling of the samples for sequencing. It is well known that the 454 sequencers produce artificial replicated and duplicated reads, which might produce misleading conclusions (17). Therefore, replicates, duplicates, and sequences of low quality or with short read lengths (<100 bp) were removed from the raw data set through Web-based approaches. Here, these culled sequences constituted 18.9 to 21.3% of the total sequences, depending on the kimchi sample, and this was a typical output for the Roche GS FLX system (39). Finally, 560,907 clean sequencing reads (79.95% of total reads) with an average sequence length of 438 bp were obtained. This allowed clear assembly and annotation analysis. The GC content (%) of the sequences in the data sets ranged from 30 to 40%, which reflects the predominance of low-GC-content Gram-positive LAB. Table 1 summarizes the statistics for pyrosequencing-derived data sets from the kimchi samples.
TABLE 1.
Summary of the pyrosequencing-derived data sets for and statistical analysis of the kimchi fermentation samples
| Kimchi sample characteristic | Result for indicated sample |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| J1 | J7 | J13 | J16 | J18 | J21 | J23 | J25 | J27 | J29 | Total | |
| Total size of sequences (bp) | 11,417,220 | 15,705,531 | 9,806,465 | 13,411,871 | 8,009,837 | 27,622,094 | 28,190,450 | 104,312,483 | 49,419,903 | 25,183,023 | 281,661,657 |
| No. of reads | 27,337 | 38,388 | 23,287 | 31,647 | 18,846 | 66,096 | 67,771 | 249,849 | 118,313 | 60,022 | 701,556 |
| Size of clean sequences (bp) | 9,646,351 | 13,090,858 | 8,301,534 | 11,390,298 | 6,765,854 | 23,238,193 | 23,565,573 | 86,509,803 | 41,595,359 | 21,345,546 | 245,449,369 |
| No. of clean reads | 22,006 | 30,202 | 18,885 | 25,737 | 15,208 | 53,106 | 53,919 | 197,709 | 95,322 | 48,813 | 560,907 |
| Avg length of clean reads (bp) | 438 | 433 | 439 | 442 | 444 | 437 | 437 | 437 | 436 | 437 | 438 |
| Avg GC content (%) of clean readsa | 37.53 | 37.25 | 37.54 | 38.62 | 38.03 | 37.38 | 37.49 | 38.13 | 37.81 | 37.26 | 37.70 |
| No. (%) of predicted 16S rRNA genesb | 38 (0.17) | 57 (0.19) | 90 (0.48) | 199 (0.77) | 101 (0.66) | 300 (0.56) | 334 (0.62) | 1,316 (0.67) | 592 (0.62) | 252 (0.52) | 3,279 (0.59) |
| No. (%) of reads categorized by SEED subsystemb | 732 (3.33) | 2,249 (7.45) | 8,059 (42.67) | 12,419 (48.25) | 8,865 (58.29) | 23,849 (44.91) | 24,486 (46.11) | 102,770 (51.98) | 43,808 (45.96) | 23,406 (47.95) | 252,936 (45.09) |
Fractional GC contents of DNA sequence was calculated by the GEECEE program (http://emboss.bioinformatics.nl/cgi-bin/emboss/geecee).
An E value of less than 0.01 and a minimum alignment length (≥50 bp) were used to predict 16S rRNA and functional genes using the MG-RAST server. The numbers in parentheses indicate the percentages of respective read numbers for clean read numbers.
Phylogenetic compositions of the bacterial communities.
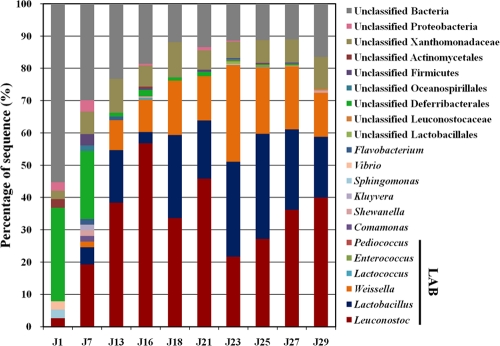
Among the pyrosequencing-derived data sets, 16S rRNA genes accounted for 0.17 to 0.67%, with an average of 0.54% (Table 1). The number of 16S rRNA genes increased as the fermentation progressed, and the 16S rRNA gene frequencies were slightly higher than those in cultured genome prokaryotic genomes (36), perhaps reflecting kimchi's enrichment in LAB, which contain several copies of 16S rRNA genes within relatively small genomes (∼2 Mb). Sequences were classified at each level using the MG-RAST server RDP II (16S rRNA gene) database (E value of 0.01 using a minimum alignment length of ≥50 bp). Phylogenetic classifications of the 16S rRNA gene sequences are shown in Fig. 3.
FIG. 3.
Phylogenetic taxonomic composition of the kimchi microbiome during fermentation. 16S rRNA gene sequences were classified to the genus level using the MG-RAST sever based on the RDP II (16S rRNA gene) database (E value, 0.01; minimum alignment length, ≥50 bp).
Genomic DNA from kimchi substrates such as Chinese cabbage, red pepper, garlic, and fermented seafood could be a reason for the relatively low frequency of 16S rRNA genes in the early fermentation phases (J1, 0.17%; J7, 0.19%). In fact, in early fermentation, the proportion of unclassified phylotypic groups, including unclassified Deferribacterales and other unclassified Bacteria, was highest (J1, ∼97%), and these bacteria might have originated from Chinese cabbage and kimchi additives. However, the proportion of unclassified phylotypic groups decreased gradually as the fermentation progressed, due to an increase in bacterial abundance. Over the entire kimchi fermentation, somewhat greater than 20% of the 16S rRNA gene sequences for all samples fell into unclassified phylotypic groups. Archaeal 16S rRNA gene sequences were not detected.
Figure 3 shows that the kimchi fermentation was governed by the distinct population dynamics of three genera, Leuconostoc, Lactobacillus, and Weissella. Among them, the genus Leuconostoc was most abundant during the kimchi fermentation, followed by Lactobacillus and Weissella. Leuconostoc dominated at the beginning stages of kimchi fermentation, but as the fermentation progressed, the abundance of Lactobacillus and Weissella increased. After the middle stages of fermentation, Leuconostoc, Lactobacillus, and Weissella became the predominant bacterial groups in the microbial community, and their cumulative abundance reached approximately 80% after day 23 (sample J23), and when the unclassified phylotypic groups were excluded from analysis, the three predominant bacterial groups constituted more than 98% of all bacterial groups. However, after day 23, the abundance of Leuconostoc increased again, and that of Lactobacillus and Weissella gradually decreased.
Previous studies reported that in kimchi fermentation, Leuconostoc species typically dominate during the beginning of fermentation with low acidity at high temperatures, whereas Weissella and Lactobacillus species have the capacity to grow under high acidity and low temperatures, indicating that fermentation temperature and acidity are important determinants of the microbial population (5, 27, 28). However, our results showed that Leuconostoc species dominated, even at a low temperature (4°C) and high acidity. Although Leuconostoc species were more dominant than Weissella and Lactobacillus species at the beginning of fermentation, Leuconostoc species increased again and Lactobacillus and Weissella decreased at the end of the fermentation. These results suggest that there could be another important determinant of microbial communities in addition to temperature and acidity. Data shown in Fig. 2 and 3 strongly indicate that the metabolism of free sugars and the production of lactate, acetate, and mannitol were closely correlated with the growth of Leuconostoc, Lactobacillus, and Weissella.
Comparative functional analysis of the kimchi microbiome.
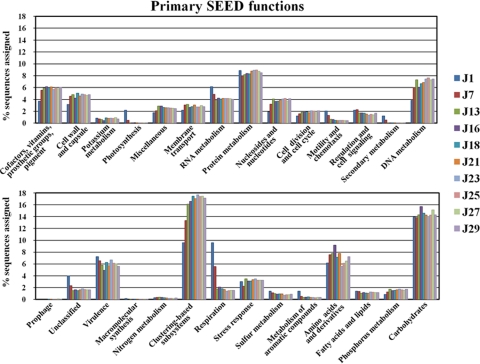
To explore the metabolic potential of the kimchi microbiome during fermentation, all sequences were analyzed using the SEED subsystem publicly available on the MG-RAST server. Assignments of orthologous sequences in the metagenome by comparison with both protein and nucleotide databases were categorized (28 metabolic categories, 519 subsystems). An average 45.09% of total sequences were matched to SEED functional categories of organisms present in the MG-RAST database at an E-value cutoff of 0.01 (Table 1). The level of reads categorized by SEED was very low for the early fermentation, as only 3.33% and 7.45% of reads from J1 and J7 samples were categorized into the SEED functional categories, respectively. This might be explained by a contamination of genomic DNA by cabbage and other kimchi ingredients. As the kimchi fermentation progressed, matching levels of the metagenomic sequence reads to SEED functional categories increased sharply due to the increase in bacterial abundance (Fig. 1).
Results of assigning these metagenomic sequences to functional categories are shown in Fig. 4 and 5. As expected, patterns in the data confirmed that kimchi fermentation is accomplished predominantly by heterofermentative LAB; therefore, carbohydrate categories, including metabolism of mono-, di-, and oligosaccharides and fermentation, were key categories for kimchi fermentation. Thus, among the many gene categories in the kimchi metagenome, our analyses focused on the carbohydrate categories. The comparative representation with several other environmental metagenomic data sets showed that genes involved in the fermentation process were enriched (Table 2). Carbohydrate metabolism yielded an average of 14.49% of all matches (Fig. 4). The fraction of the kimchi microbiome reads in the carbohydrate category was higher than those of microbiomes derived from the termite gut (12.89%), acid mine drainage (AMD) biofilm (12.17%), enhanced biological phosphorus removal (EBPR) sludge (10.72%), farm soil (11.25%), and the Sargasso Sea (12.23%). Analysis of metabolic potential within the carbohydrate category indicated that the kimchi microbiome was enriched for the genes involved and for mono-, di-, and oligosaccharide fermentation. The metagenomic read fractions of monosaccharide (28.70%), di- and oligosaccharide (28.38%), and fermentation (14.38%) within the carbohydrate category in the kimchi microbiome were also higher than those in the microbiomes derived from the termite gut (24.35%, 21.64%, 5.24%), AMD biofilm (7.12%, 24.1%, 10.51%), EBPR sludge (11.55%, 14.32%, 6.77%), farm soil (18.41%, 15.3%, 7.52%), and the Sargasso Sea (18.04%, 13.9%, 9.18%). The fermentation metabolism subcategory was associated mostly with various lactate fermentations (65.93%) and acetoin and butanediol metabolism (28.10%). In particular, lactate fermentation genes in the kimchi microbiome were predominantly enriched compared to those in the termite gut (33.33%), AMD biofilm (29.29%), EBPR sludge (27.19%), farm soil (25.66%), and the Sargasso Sea (16.07%). Moreover, metabolic genes involved in carbohydrate metabolism and fermentation generally increased as the kimchi fermentation progressed (Fig. 4 and 5). In summary, the above-mentioned results showed that the kimchi microbiome had high metabolic versatility with respect to lactic acid fermentations; these results were also supported by the profiles of free sugars and fermentation products (Fig. 2).
FIG. 4.
Predicted metabolic profiles of kimchi metagenomic data sets obtained from SEED subsystems. The MG-RAST server based on SEED subsystems provided categories for the metagenomic data sets. The MG-RAST recommended parameters (E value, 0.01; minimum sequence length, ≥50 bp) were used.
FIG. 5.
Predicted metabolic profiles of kimchi metagenomic data sets related to carbohydrate metabolism and fermentation. Categories for the metagenomic data sets were provided by the MG-RAST server based on SEED subsystems. The MG-RAST recommended parameters (E value, 0.01; minimum sequence length, ≥50 bp) were used. Acetyl-CoA, acetyl coenzyme A.
TABLE 2.
Comparative representation of the major metabolic groups of the kimchi microbiome versus several other environmental metagenomic data sets publicly available in MG-RAST
| SEED class | SEED category | Relative abundancea (%) in: |
|||||
|---|---|---|---|---|---|---|---|
| Kimchi | Termite gut (4442701.3)b | AMD biofilm (4441137.3)b | Farm soil (4441091.3)b | EBPR sludge (4441092.3)b | Sargasso Sea (4441571.3)b | ||
| Subsystem hierarchy 1 | Carbohydrate | 14.49 | 12.89 | 12.17 | 11.25 | 10.72 | 12.23 |
| Subsystem hierarchy 2 | Uptake system | 0.07 | 0.64 | 0.11 | 1.80 | 0.51 | 0.51 |
| Di- and oligosaccharides | 28.38 | 21.64 | 24.10 | 15.30 | 14.33 | 13.90 | |
| Central carbohydrate metabolism | 18.05 | 26.16 | 39.86 | 26.67 | 30.47 | 28.63 | |
| Amino sugars | 1.66 | 7.90 | 1.88 | 4.71 | 2.65 | 2.47 | |
| One-carbon metabolism | 0.69 | 2.60 | 3.28 | 8.15 | 11.09 | 8.76 | |
| Organic acids | 2.82 | 3.22 | 6.12 | 5.34 | 8.33 | 8.10 | |
| Quorum sensing and biofilm formation | 0.35 | 0.55 | 0.16 | 0.40 | 0.61 | 0.54 | |
| Fermentation | 14.38 | 5.24 | 10.51 | 7.52 | 6.77 | 9.18 | |
| CO2 fixation | 0.74 | 2.17 | 3.60 | 4.46 | 7.78 | 2.33 | |
| Sugar alcohols | 4.17 | 4.98 | 3.24 | 6.96 | 5.68 | 7.35 | |
| Methanogenesis | 0.43 | 0.02 | 0.22 | 0.22 | 0.10 | ||
| Monosaccharides | 28.70 | 24.35 | 7.12 | 18.41 | 11.55 | 18.04 | |
| Carbohydrate | 0.13 | 0.07 | 0.01 | 0.09 | |||
| Subsystem | Butanol biosynthesis | 2.60 | 10.16 | 3.57 | 20.21 | 21.20 | 21.27 |
| Mixed acid fermentation | 2.72 | 8.54 | 5.79 | 3.54 | 7.92 | 3.90 | |
| Lactate fermentation | 65.93 | 33.33 | 29.29 | 25.66 | 27.19 | 16.08 | |
| Acetyl coenzyme A fermentation to butylate | 0.65 | 18.29 | 43.71 | 30.97 | 32.33 | 42.23 | |
| Acetoin and butanediol metabolism | 28.10 | 29.67 | 17.64 | 19.62 | 11.35 | 16.54 | |
Relative abundance means the relative proportion of sequencing reads performing specific metabolisms within each subsystem. Data in boldface type are statistically significant.
The number in parentheses is the project identifying number in the MG-RAST system.
Individual genome recruitment.
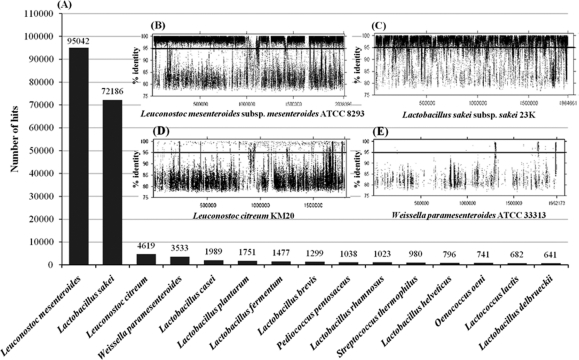
Genome recruitment analysis is an insightful way to examine the degree to which microorganisms in a given habitat are represented by completed genomic sequences (16). Therefore, the metagenomic sequence reads were compared to all complete and draft microbial genome sequences available from the NCBI (http://www.ncbi.nlm.nih.gov/sites/genome) using the criterion of 95% nucleotide identity. The two most highly recruited genomes were those of Leuconostoc mesenteroides and Lactobacillus sakei (Fig. 6 A). Prior reports have indicated that Le. mesenteroides is one of the prominent species in kimchi fermentation (23, 33). However, Lb. sakei has been better known as a psychrotrophic LAB associated with fresh meat and fish (4). The reads that aligned with the Le. mesenteroides subsp. mesenteroides ATCC 8293 and Lb. sakei subsp. sakei 23K genomes often featured extremely high sequence identities (Fig. 6B and C), suggesting that LAB closely related to these two reference strains were abundant in the fermented kimchi samples. It seems clear that close relatives of these specific strains contribute to a large fraction of the heterotrophic lactic acid fermentation in kimchi. Nevertheless, there were some significant differences between the genomes of the two reference strains and the LAB genomes of the kimchi samples, since certain reference sequences were not represented within the metagenome sequence, implying that the kimchi habitat may feature specific selective pressures and support diverse populations not yet isolated or sequenced.
FIG. 6.
Recruitment of whole clean reads against complete or draft genomes (A). Insets are the recruitment plots for the four top highest recruiting genomes, Leuconostoc mesenteroides subsp. mesenteroides ATCC 8293 (B), Lactobacillus sakei subsp. sakei 23K (C), Leuconostoc citreum KM20 (D), and Weissella paramesenteroides ATCC 33313 (E). Horizontal lines within the recruitment plots indicate the level of 95% nucleotide identity.
Although Leuconostoc citreum KM20 was originally isolated from Chinese cabbage kimchi (22), the degree to which it was a genomic match to the metagenome was relatively low (Fig. 6D). Phylogenetic analysis based on 16S rRNA gene sequences showed that the genus Weissella was one of the predominant bacterial groups during kimchi fermentation (Fig. 3). However, the draft genome of W. paramesenteroides ATCC 33313 was a poor match to the metagenome sequence (Fig. 6E). This result was surprising and suggests that the metagenome sequences from Weissella might be derived from other Weissella species, such as W. koreensis S-5623T, which has been isolated from Chinese cabbage kimchi (26).
Assembly of pyrosequencing-derived sequencing reads and assignment of assembled contigs.
A very important aspect of metagenomic sequencing is the possibility of assembling contigs derived from a single microorganism. Therefore, sequencing reads from the kimchi metagenome, which comprised a total of 560,907 clean reads, were assembled, resulting in a total of 27,835 contigs. However, many sequences on the contigs were found to be split due to frameshifts caused mostly by homopolymeric sequences, which are difficult to resolve by 454 pyrosequencing technology. The longest contig was 270,682 bp in size. One hundred twenty-seven contigs that were longer than 10 kb were classified according to their taxonomic affiliations by BLASTN comparisons to the GenBank nonredundant nucleotide (nr/nt) database and selection of the top BLASTN hits (see Table S1 in the supplemental material).
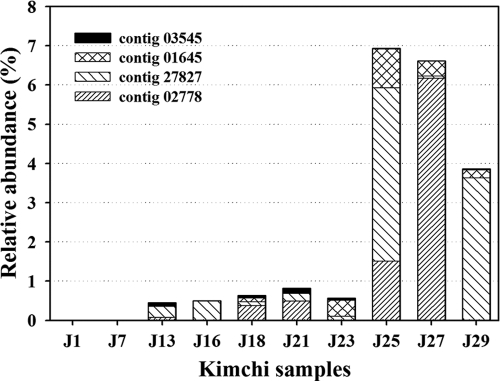
Most contigs were shown at the species level (>97% sequence identity) to be clearly derived from the predominant LAB, Leuconostoc mesenteroides subsp. mesenteroides ATCC8293 and Lactobacillus sakei subsp. sakei 23K. Surprisingly, four contigs, 02778, 27827, 01645, and 03545, of 139,667 bp, 137,689 bp, 79,287 bp, and 10,233 bp, respectively, were classified as putative bacteriophage fragments. Three of these contigs, 02778, 27827, and 01645, contained genes for a putative bacteriophage major capsid protein (MCP), which is used frequently for phage phylogenic analysis (3). Phylogenetic analysis based on the MCP sequences showed that contigs 02778, 27827, and 01645 were related to LAB phages (data not shown). Although contig 03545 did not contain a gene for the putative bacteriophage MCP, most of the genes in the contig sequence were matched with the Leuconostoc phage 1-A4 complete genome (32) with high identity (92%), indicating that contig 03545 was clearly able to be assigned as a Leuconostoc phage (see Table S1 in the supplemental material). CRISPR elements are sites containing multiple short palindromic repeats (23 to 47 bp) that flank short “spacers” composed of viral DNA (58); their presence in a bacterial genome represents key evidence of bacteriophage attack (58). Four CRISPR elements were detected from 127 contigs longer than 10 kb, suggesting that the bacterial cells might be suffering from bacteriophage infection.
Relative abundance of the phage-related contigs.
Bacteriophages are ubiquitous and abundant components of microbial communities and have significant influence upon bacterial diversity, horizontal gene transfer, and genetic diversity in environmental habitats (43). By counting the number of reads mapped to each phage-related contig and normalizing by the total read count, it was possible to estimate the relative abundance of phages. Figure 7 shows the frequency at which reads were observed for each phage-related contig in the kimchi microbial samples. Putative phage sequences were detected early in the J13 sample (day 13), and the numbers subsequently increased steadily throughout the fermentation. However, putative phage sequences increased suddenly in the J25 sample, and their abundance amounted to approximately 7% of the total reads. This large number of putative phage sequences in the metagenomic data sets was unexpected, because it was thought that most phages (if present) escaped from the cell fraction obtained by centrifugation. Possible explanations for phage detection are that the phages were replicating within the kimchi microbiome and free phage particles adhered to either particulate matter or bacterial cells at the time of harvesting (9, 16).
FIG. 7.
Relative abundance of putative phage contigs in whole unassembled clean reads during kimchi fermentation. Kimchi metagenomic data sets were matched against data for four putative phage contigs using the BLASTN algorithm with the criteria of a minimum percent identity of 95% and a minimum overlap of 90 bp.
In any event, the high abundance of phage sequences in the cellular fraction from kimchi indicates that a remarkably high proportion of bacterial cells were suffering from infection and perhaps from lytic phage attack. Although the frequency of putative phage genes was high at the end of the fermentation, such levels are consistent with previously reported ranges of phage-infected bacterial cells (9). It was inferred that the decrease in the bacterial population after day 25 (Fig. 1) and the major changes in the microbial community between days 23 and 25 (Fig. 2) were affected by phage-host dynamics that occur during kimchi curing. With the decrease of bacterial abundance after day 25, the putative phage sequences also slowly decreased. Previous studies have reported that the fermentation temperature and acidity are important determinants of the microbial community composition in kimchi fermentation (5, 27). However, our results showed that in addition to temperature and acidity, a new influence, that of bacteriophages, was another clearly important determinant of microbial community dynamics, as in sauerkraut fermentation (31, 32, 37), especially at the end stages of kimchi fermentation.
Conclusions.
Changes in the overall genetic features and metabolic potential of the kimchi-fermenting microbial community were investigated by examining a large pyrosequencing-derived data set, in conjunction with metabolite analysis. The metagenome and metabolites revealed that the kimchi microbiome had high metabolic potential with respect to heterotrophic lactic acid fermentations. Based on the abundances of 16S rRNA genes and the representation of whole-genome sequences in the metagenome, we conclude that active microbial populations in kimchi fermentation were dominated by members of three genera, Leuconostoc, Lactobacillus, and Weissella, which is consistent with many previous results (5, 23, 27). Besides microbial genome sequences, a surprisingly high abundance of phage DNA sequences were identified, indicating that bacteriophages influence the kimchi microbial community and may be a key determinant of kimchi microbial community dynamics. Metagenomic analysis of the kimchi samples over time made it possible to monitor not only microbial community composition but also overall features and the metabolic potential during fermentation. Future analyses of microbial abundances and interactions will likely provide insights into how the kimchi microbial community influences metabolite production and, ultimately, kimchi flavor.
Supplementary Material
Acknowledgments
We thank J. S. Chun, C. J. Cha, W. S. Park, B. H. Ryu, and Y. S. Ko for their valuable help and collaboration during this study.
This work was supported by the Technology Development Program for Agriculture and Forestry (TDPAF) of the Ministry for Agriculture, Forestry and Fisheries and the 21C Frontier Microbial Genomics and Application Center Program (grant MG05-0104-4-0) of the Ministry of Education, Science and Technology, Republic of Korea.
Footnotes
Published ahead of print on 11 February 2011.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Aziz, R. K., et al. 2008. The RAST server: rapid annotations using subsystems technology. BMC Genomics 9:75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bae, J. W., et al. 2005. Development and evaluation of genome-probing microarrays for monitoring lactic acid bacteria. Appl. Environ. Microbiol. 71:8825-8835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bamford, D. H., J. M. Grimes, and D. I. Stuart. 2005. What does structure tell us about virus evolution? Curr. Opin. Struct. Biol. 15:655-663. [DOI] [PubMed] [Google Scholar]
- 4.Chaillou, S., et al. 2005. The complete genome sequence of the meat-borne lactic acid bacterium Lactobacillus sakei 23K. Nat. Biotechnol. 23:1527-1533. [DOI] [PubMed] [Google Scholar]
- 5.Cho, J., et al. 2006. Microbial population dynamics of kimchi, a fermented cabbage product. FEMS Microbiol. Lett. 257:262-267. [DOI] [PubMed] [Google Scholar]
- 6.Chou, H. H., and M. H. Holmes. 2001. DNA sequence quality trimming and vector removal. Bioinformatics 17:1093-1104. [DOI] [PubMed] [Google Scholar]
- 7.Cole, J. R., et al. 2009. The ribosomal database project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 37:D141-D145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Costello, E. K., et al. 2009. Bacterial community variation in human body habitats across space and time. Science 326:1694-1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DeLong, E. F., et al. 2006. Community genomics among stratified microbial assemblages in the ocean's interior. Science 311:496-503. [DOI] [PubMed] [Google Scholar]
- 10.Dinsdale, E. A., et al. 2008. Functional metagenomic profiling of nine biomes. Nature 452:629-632. [DOI] [PubMed] [Google Scholar]
- 11.Edgar, R. C. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792-1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ercolini, D. 2004. PCR-DGGE fingerprinting: novel strategies for detection of microbes in food. J. Microbiol. Methods 56:297-314. [DOI] [PubMed] [Google Scholar]
- 13.Farrelly, V., F. A. Rainley, and E. Stackebrandt. 1995. Effect of genome size and rrn gene copy number on PCR amplification of 16S rRNA genes from a mixture of bacterial species. Appl. Environ. Microbiol. 61:2798-2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Figueiredo, I. M., et al. 2006. 1H NMR, a rapid method to monitor organic acids during cupuassu (Theobroma grandiflorum Spreng) processing. J. Agric. Food. Chem. 54:4102-4106. [DOI] [PubMed] [Google Scholar]
- 15.García Martín, H., et al. 2006. Metagenomic analysis of two enhanced biological phosphorus removal (EBPR) sludge communities. Nat. Biotechnol. 24:1263-1269. [DOI] [PubMed] [Google Scholar]
- 16.Ghai, R., et al. 2010. Metagenome of the Mediterranean deep chlorophyll maximum studied by direct and fosmid library 454 pyrosequencing. ISME J. 4:1154-1166. [DOI] [PubMed] [Google Scholar]
- 17.Gomez-Alvarez, V., T. K. Teal, and T. M. Schmidt. 2009. Systematic artifacts in metagenomes from complex microbial communities. ISME J. 3:1314-1317. [DOI] [PubMed] [Google Scholar]
- 18.Grissa, I., G. Vergnaud, and C. Pourcel. 2007. CRISPRFinder: a web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Res. 35:W52-W57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ha, J. H., W. S. Hawer, Y. J. Kim, and Y. J. Nam. 1989. Changes of free sugars in kimchi during fermentation. Korean J. Food Sci. Technol. 21:633-638. [Google Scholar]
- 20.Juck, D., T. Charles, L. G. Whyte, and C. W. Greer. 2000. Polyphasic microbial community analysis of petroleum hydrocarbon-contaminated soils from two northern Canadian communities. FEMS Microbiol. Ecol. 33:241-249. [DOI] [PubMed] [Google Scholar]
- 21.Kim, J., J. Chun, and H. U. Han. 2000. Leuconostoc kimchii sp. nov., a new species from kimchi. Int. J. Syst. Evol. Microbiol. 50:1915-1919. [DOI] [PubMed] [Google Scholar]
- 22.Kim, J. F., et al. 2008. Complete genome sequence of Leuconostoc citreum KM20. J. Bacteriol. 190:3093-3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim, M., and J. Chun. 2005. Bacterial community structure in kimchi, a Korean fermented vegetable food, as revealed by 16S rRNA gene analysis. Int. J. Food Microbiol. 103:91-96. [DOI] [PubMed] [Google Scholar]
- 24.Kim, N., K. R. Park, I. S. Park, Y. J. Cho, and Y. M. Bae. 2005. Application of a taste evaluation system to the monitoring of Kimchi fermentation. Biosens. Bioelectron. 20:2283-2291. [DOI] [PubMed] [Google Scholar]
- 25.Lee, J.-S., et al. 1997. Classification of isolates originating from kimchi using carbon-source utilization patterns. J. Microbiol. Biotechnol. 7:68-74. [Google Scholar]
- 26.Lee, J., et al. 2002. Weissella koreensis sp. nov., isolated from kimchi. Int. J. Syst. Evol. Microbiol. 52:1257-1261. [DOI] [PubMed] [Google Scholar]
- 27.Lee, J. S., et al. 2005. Analysis of kimchi microflora using denaturing gradient gel electrophoresis. Int. J. Food Microbiol. 102:143-150. [DOI] [PubMed] [Google Scholar]
- 28.Lee, D., S. Kim, J. Cho, and J. Kim. 2008. Microbial population dynamics and temperature changes during fermentation of kimjang kimchi. J. Microbiol. 46:590-593. [DOI] [PubMed] [Google Scholar]
- 29.Lee, E. J., et al. 2009. Quality assessment of ginseng by 1H NMR metabolite fingerprinting and profiling analysis. J. Agric. Food Chem. 57:7513-7522. [DOI] [PubMed] [Google Scholar]
- 30.Li, W. Z., and A. Godzik. 2006. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22:1658-1659. [DOI] [PubMed] [Google Scholar]
- 31.Lu, Z., F. Breidt, V. Plengvidhya, and H. P. Flaming. 2003. Bacteriophage ecology in commercial sauerkraut fermentations. Appl. Environ. Microbiol. 69:3192-3202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lu, Z., E. Altermann, F. Breidt, and S. Kozyavkin. 2010. Sequence analysis of Leuconostoc mesenteroides bacteriophage Φ1-A4 isolated from an industrial vegetable fermentation. Appl. Environ. Microbiol. 76:1955-1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Makarova, K., et al.2006. Comparative genomics of the lactic acid bacteria. Proc. Natl. Acad. Sci. U. S. A. 103:15611-15616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McFeeters, R. F., and K.-H. Chen. 1986. Utilization of electron acceptors for anaerobic mannitol metabolism by Lactobacillus plantarum: compounds which serve as electron acceptors. Food Sci. 3:73-81. [Google Scholar]
- 35.Meyer, F., et al. 2008. The metagenomics RAST server: a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics 9:386-394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mou, X., S. L. Sun, R. A. Edwards, R. E. Hodson, and M. A. Moran. 2008. Bacterial carbon processing by generalist species in the coastal ocean. Nature 451:708-711. [DOI] [PubMed] [Google Scholar]
- 37.Mudgal, P., F. Jr., Breidt, S. R. Lubkin, and K. P. Sandeep. 2006. Quantifying the significance of phage attack on starter cultures: a mechanistic model for population dynamics of phage and their hosts isolated from fermenting sauerkraut. Appl. Environ. Microbiol. 72:3908-3915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nam, Y. D., H. W. Chang, K. H. Kim, S. W. Roh, and J. W. Bae. 2009. Metatranscriptome analysis of lactic acid bacteria during kimchi fermentation with genome-probing microarrays. Int. J. Food Microbiol. 130:140-146. [DOI] [PubMed] [Google Scholar]
- 39.Niu, B., L. Fu, S. Sun, and W. Li. 2010. Artificial and natural duplicates in pyrosequencing reads of metagenomic data. BMC Bioinformatics 11:187-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Park, K. Y., et al. 2003. Kimchi and an active component, beta-sitosterol, reduce oncogenic H-Ras(v12)-induced DNA synthesis. J. Med. Food. 6:151-156. [DOI] [PubMed] [Google Scholar]
- 41.Park, E. J., et al. 2009. Application of quantitative real-time PCR for enumeration of total bacterial, archaeal, and yeast populations in kimchi. J. Microbiol. 47:682-685. [DOI] [PubMed] [Google Scholar]
- 42.Park, J. M., et al. 2010. Identification of the lactic acid bacteria in kimchi according to initial and over-ripened fermentation using PCR and 16S rRNA gene sequence analysis. Food Sci. Biotechnol. 19:541-546. [Google Scholar]
- 43.Parsley, L. C., et al. 2010. Census of the viral metagenome within an activated sludge microbial assemblage. Appl. Environ. Microbiol. 76:2673-2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Parsley, L. C., et al. 2010. Identification of diverse antimicrobial resistance determinants carried on bacterial, plasmid, or viral metagenomes from an activated sludge microbial assemblage. Appl. Environ. Microbiol. 76:3753-3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Qin, J., et al.2010. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464:59-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ritalahti, K. M., et al. 2006. Quantitative PCR targeting 16S rRNA and reductive dehalogenase genes simultaneously monitors multiple Dehalococcoides strains. Appl. Environ. Microbiol. 72:2765-2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rodriguez-Brito, B., et al. 2010. Viral and microbial community dynamics in four aquatic environments. ISME J. 4:739-751. [DOI] [PubMed] [Google Scholar]
- 48.Roh, S. W., et al. 2010. Investigation of archaeal and bacterial diversity in fermented seafood using barcoded pyrosequencing. ISME J. 4:1-16. [DOI] [PubMed] [Google Scholar]
- 49.Sanapareddy, N., et al. 2009. Molecular diversity of a North Carolina wastewater treatment plant as revealed by pyrosequencing. Appl. Environ. Microbiol. 75:1688-1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shendure, J., and H. Ji. 2008. Next-generation DNA sequencing. Nat. Biotechnol. 26:1135-1145. [DOI] [PubMed] [Google Scholar]
- 51.Shin, M. S., S. K. Han, J. S. Ryu, K. S. Kim, and W. K. Lee. 2008. Isolation and partial characterization of a bacteriocin produced by Pediococcus pentosaceus K23-2 isolated from kimchi. J. Appl. Microbiol. 105:331-339. [DOI] [PubMed] [Google Scholar]
- 52.Simon, C., A. Wiezer, A. W. Strittmatter, and R. Daniel. 2009. Phylogenetic diversity and metabolic potential revealed in a glacier ice metagenome. Appl. Environ. Microbiol. 75:7519-7526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Son, H. S., et al. 2008. 1H nuclear magnetic resonance-based metabolomic characterization of wines by grape varieties and production areas. J. Agric. Food Chem. 56:8007-8016. [DOI] [PubMed] [Google Scholar]
- 54.Song, Y.-O. 2004. The functional properties of kimchi for the health benefits. J. Food Sci. Nutr. 9:27-33. [Google Scholar]
- 55.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596-1599. [DOI] [PubMed] [Google Scholar]
- 56.Tringe, S. G., et al. 2005. Comparative metagenomics of microbial communities. Science 308:554-557. [DOI] [PubMed] [Google Scholar]
- 57.Tyson, G. W., et al. 2004. Community structure and metabolism through reconstruction of microbial genomes from the environment. Nature 428:37-43. [DOI] [PubMed] [Google Scholar]
- 58.Tyson, G. W., and J. F. Banfield. 2008. Rapidly evolving CRISPRs implicated in acquired resistance of microorganisms to viruses. Environ. Microbiol. 10:200-207. [DOI] [PubMed] [Google Scholar]
- 59.Venter, J. C., et al. 2004. Environmental genome shotgun sequencing of the Sargasso Sea. Science 304:66-74. [DOI] [PubMed] [Google Scholar]
- 60.Wilmes, P., S. L. Simmons, V. J. Denef, and J. F. Banfield. 2009. The dynamic genetic repertoire of microbial communities. FEMS Microbiol. Rev. 33:109-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wisselink, H. W., R. A. Weusthuis, G. Eggink, J. Hugenholtz, and G. J. Grobben. 2002. Mannitol production by lactic acid bacteria: a review. Int. Dairy J. 12:151-161. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.